Abstract
Capsaicin bestows spiciness by activating TRPV1 channel with exquisite potency and selectivity. Capsaicin-bound channel structure was previously resolved by cryo-EM at 4.2-to-4.5 Å resolution, however important details required for mechanistic understandings are unavailable: capsaicin was registered as a small electron density, reflecting neither its chemical structure nor specific ligand-channel interactions. We obtained the missing atomic-level details by iterative computation, which were confirmed by systematic site-specific functional tests. We observed that the bound capsaicin takes “tail-up, head-down” configurations. The vanillyl and amide groups form specific interactions to anchor its bound position, while the aliphatic tail may sample a range of conformations, making it invisible in cryo-EM images. Capsaicin stabilizes the open state by “pull-and-contact” interactions between the vanillyl group and the S4-S5 linker. Our study provided a structural mechanism for the agonistic function of capsaicin and its analogs, and demonstrated an effective approach to obtain atomic level information from cryo-EM structures.
Spicy foods are enjoyable for many people over the globe. In fact, we humans are the only species that deliberately seeks spiciness in foods1. Spiciness is generally a repulsive chemesthetic sensation elicited by capsaicinoids in plants that is thought to serve as deterrent to herbivores while allow avians, which are insensitive to them2, to ingest the seeds for wider dispersal. For humans, studies have shown that capsaicin, the leading member of capsaicinoids, not only acts as an analgesic for pain3, a promoter of energy expenditure to assist weight control4 and vasodilation to facilitate heat dissipation5, but also exhibits promising antitumor activity6. The noxious property of capsaicin is also exploited, as capsaicin injection has been serving as a standard animal model for pain study. The molecular basis for these actions has started to emerge since the cloning of its receptor, transient receptor potential vanilloid 1 (TRPV1) ion channel7.
Being a polymodal receptor for a wide spectrum of physical and chemical stimuli such as heat, proton and toxins8, TRPV1 exhibits exquisite affinity (sub-μM), sensitivity (near unity open probability) (Fig. 1a), and selectivity for capsaicin (which does not activate the homologous TRPV2-6 channels). Understanding this outstanding agonist recognition process at molecular level will shed new light on the general protein-ligand interaction mechanism while at the same time guide pharmaceutical efforts to regulate this important pain target in a modality-specific manner. Based on capsaicin-insensitive chicken TRPV1, it was found that Y512 and S513 on S3 (all residue numbering here is based on mouse TRPV1) are important for capsaicin activation2. With less sensitive rabbit TRPV1, M548 and T551 on S4 were identified as additional key residues9. Cryo-EM structures revealed that these residues are scattered around a small electron density close to S3 and S4 segments (Fig. 1b), which likely represents a bound capsaicin molecule10,11. These structural and functional studies established the location of capsaicin-binding pocket. The cryo-EM structures set the stage to unveil the detailed capsaicin-channel interaction mechanism but, at 4.2-to-4.5 Å resolution of capsaicin and its binding pocket11, they are insufficient to show atomic interactions. In particular, the electron density observed inside the pocket is too small to account for the mass of capsaicin, hence it remains largely elusive how capsaicin is positioned and coordinated. Regarding capsaicin-induced activation, cysteine accessibility measurements suggested that the lower part of S6 moves to open the activation gate12. The cryo-EM data support such movement of S6 and further suggest that it may be caused by an outward movement of the S4-S5 linker10,11. What dynamic molecular interactions stabilize capsaicin inside the pocket and provide activation energy to drive these downstream conformational rearrangements, however, are unknown.
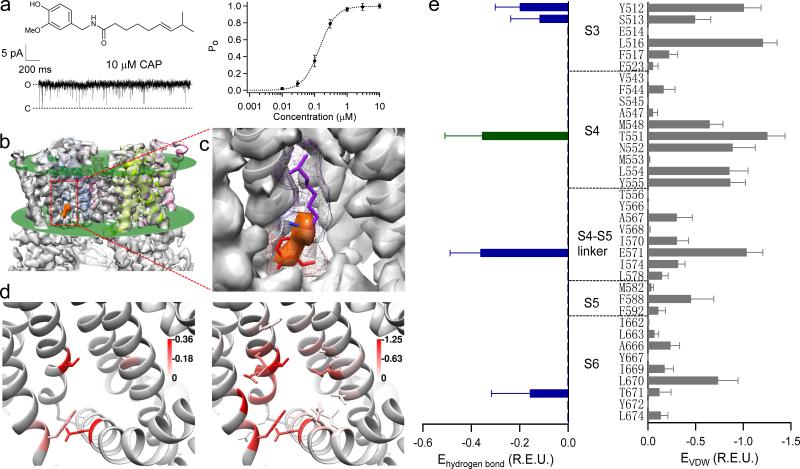
Figure 1. The formation of capsaicin-binding pocket.
(a) Chemical structure of capsaicin and its concentration-response curve on TRPV1. Saturating concentration of capsaicin (10 μM) almost fully activates TRPV1 as shown in the single-channel trace. (b) The small electron density for capsaicin11 (orange colored) locates inside the membrane, whose boundaries are indicated by green planes. (c) A zoomed in view of capsaicin-binding pocket. A representative configuration of docked capsaicin is shown with its vanillyl, amide and aliphatic chain groups colored in red, blue and purple, respectively. Its molecular surface is shown as a mesh. (d) and (e) Distributions of average hydrogen bonds (left panels) and VDW interactions (right panels) between capsaicin (top 10 docking models) and the channel (liganded open state, PDB ID: 3J5R). Green and blue bars indicate potential hydrogen bonds involving the amide group and the vanillyl group of capsaicin, respectively. Energy unit is Rosetta Energy Unit (R.E.U.).
To address these fundamental questions, here we employed an iterative approach that combined structural computation and functional analyses with cryo-EM information (Supplementary Results, Supplementary Fig. 1). We first used molecular docking to probe the conformation of ligand-channel complex and then quantitatively ranked all potential interactions between capsaicin and the channel by stabilization energy (see online Methods for details). We then tested each of these predictions by perturbing the structure of the ligand (with a series of synthesized capsaicin analogs) and/or the channel (with point mutations), and analyzing the coupling energy of each interaction with thermodynamic mutant cycle analysis. From these tests, we determined binding configurations of capsaicin, composition of binding energies, ligand-interacting channel residues and their contribution to ligand stabilization and activation transitions. Such information in turn guided our computational efforts to unveil the series of conformational changes starting from initial binding of capsaicin to final opening of the S6 activation gate, as well as the energetic driving forces behind these steps.
RESULTS
Potential capsaicin-TRPV1 interactions probed by docking
We docked capsaicin molecule into the binding pocket in liganded state (PDB ID: 3J5R) without using the small electron density observed in cryo-EM structure11 as constraints. Since the binding pocket locates inside cell membrane11 (Fig. 1b), to accurately dock capsaicin, implicit membrane environment was simulated using RosettaMembrane energy function in RosettaLigand application13-16. The top 10 docking models with lowest binding energy exhibited good structural convergence in the vanillyl and amide groups, which overlap nicely with the observed density (Fig. 1c and Supplementary Fig. 2). The vanillyl group (Fig. 1c, red) points downward to the S4-S5 linker at lipid-water interface, while the aliphatic tail (Fig. 1c, purple) points upward to the upper S4 segment. The capsaicin molecule occupies a considerably larger volume than the electron density: while vanillyl and amide groups could account for almost all of the density, the tail of capsaicin has no corresponding density, which we will address later. The docking results support existing findings of capsaicin-binding pocket and serve as a starting point for our quantitative analysis below.
To reveal the molecular interactions between capsaicin and the channel, we decomposed the calculated binding energy and examined the spatial distributions of each energy component on a per-residue basis. Ligand binding is usually determined by van der Waals (VDW) interactions, hydrogen bond networks, as well as electrostatic interactions17. Capsaicin molecule is not charged, hence electrostatic interactions likely contribute little. Indeed, while total VDW (approximated by Lennard-Jones potential18) and hydrogen bond energies ranged from -5 to -15 in Rosetta energy unit (R.E.U.), total electrostatic interaction scored less than 0.7. The calculated per-residue distribution profiles of VDW and hydrogen bonding interactions exhibit distinct patterns (Fig. 1d and e). Hydrogen bonds are sparse and specific: only one residue T551 can bond with the amide oxygen atom of capsaicin, whereas other four residues may bond with its vanillyl group. In contrast, VDW interactions are widely distributed over almost all the residues lining the capsaicin-binding pocket. Such distinct distributions of different interaction energies prompt us to employ a “divide-and-conquer” strategy to functionally test these predictions by altering the nature of atomic interactions one part at a time.
Tail of capsaicin: non-specific VDW interactions
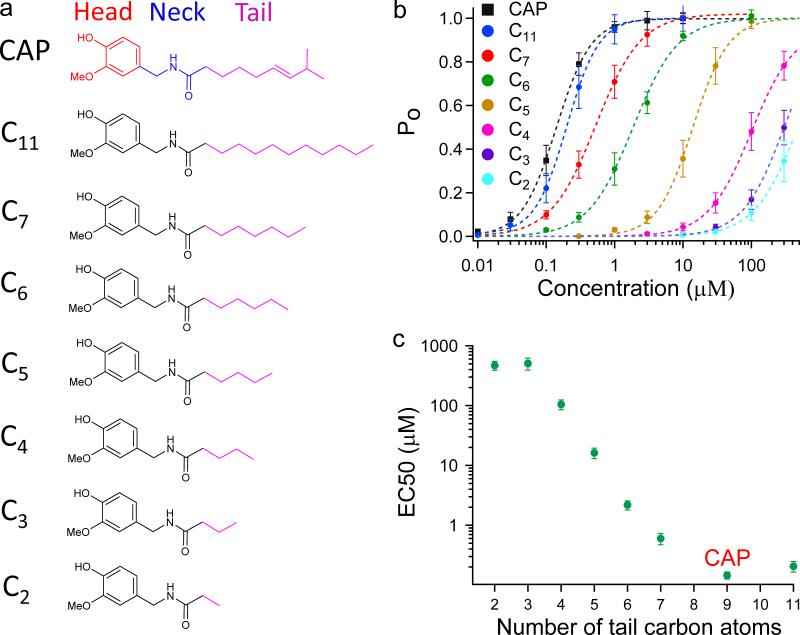
We named the vanillyl group, amide group and aliphatic chain (3E)-2-methyloct-3-ene (9 carbon atoms) of capsaicin the Head, Neck and Tail, respectively (Fig. 2a). Both the docking results and the chemical nature of the tail suggest that it cannot form hydrogen bond with the channel; instead, extensive VDW interactions are involved. To test this hypothesis, we aimed to systematically alter VDW interactions by sequentially shortening (or lengthening) the Tail. We synthesized a series of capsaicin analogs (Compound 1 to 7) with varied number of carbon atoms in Tail, ranging from 2 (C2) to 11 (C11) (Fig. 2a). With patch-clamp recording, we observed that the EC50 value of TRPV1 activation increased progressively with the shortening of Tail, whereas the maximum current responses remained similar to that of capsaicin (Fig. 2b). Channel open probability (Po) was determined by normalizing the capsaicin analog response to 2-aminoethoxydiphenyl borate (2-APB) response (for which Po was determined from single-channel recordings, Supplementary Fig. 3). The concentration-response relationships suggest the affinities of these analogs were gradually decreased. Indeed, the EC50 value was increased by ~ 3,000 fold by cutting 6 carbons (C3) from capsaicin (Fig. 2c and Supplementary Table 1). This change is more likely due to weakening of VDW interactions, not changes in the analog's ability to stay in membrane, because even after offsetting the ~ 12 fold change in lipid partition coefficient (LogP values being 2.35 for capsaicin and 1.26 for C3, Supplementary Table 3), the ~ 250 fold increase in EC50 translates to a large energetic effect of 5.5 kT. Therefore, we believe that the aliphatic Tail indeed largely contributes to binding though VDW interactions.
Figure 2. VDW interactions between the Tail and channel.
(a) Functional groups of capsaicin and chemical structures of its analogs (Compound 1 to 7) with varied length of the Tail. (b) Concentration-response curves of capsaicin analogs. (c) EC50 values of these analogs are plotted against length of the Tail. At least four independent trials were performed for each chemical at each concentration. All statistical data are given as mean ± SEM.
Unlike hydrogen bond, VDW interactions are usually isotropic in biological system without preferred bond angles19. So for the Tail, is there any structural preference for its VDW interactions with the channel? Molecular docking suggests that while Head and Neck of capsaicin converged well, the Tail showed considerable structural flexibility with much larger RMSD over top 10 models (Fig. 3a and Supplementary Fig. 2). To experimentally test such prediction, we applied thermodynamic mutant cycle analysis20-23. Briefly, if one part of capsaicin specifically interacts with one residue on the channel, the effect of changing this part on capsaicin and mutating the interacting residue should be non-additive. However, if the two parts are independent of each other, these changes should have additive effects (Fig. 3b and online Methods). Specific interaction is assumed only when the calculated coupling energy is larger than 1.5 kT22,24 (equivalent to 0.89 kcal/mol at 24°C). By switching between capsaicin and C6, we probed interactions between the tip of Tail and several residues inside capsaicin binding pocket. For residue E571 in the S4-S5 linker, the coupling energy is calculated as (equivalent to 0.40 kcal/mol at 24°C), which is much lower than 1.5 kT. This indicates that E571 does not interact specifically with the Tail. Similar analysis on several other residues around the binding pocket all yielded a coupling energy smaller than 1.5 kT (Fig. 3c). Interestingly, residue F544 on the upper half of S4 segment exhibited the largest coupling energy (1.35 ± 0.51 kT, equivalent to 0.80 ± 0.30 kcal/mol at 24°C). The distribution of coupling energy indicates that the Tail may not have specific interaction with residues inside the binding pocket, nonetheless it may prefer an up-pointing configuration allowing it to experience a strongly hydrophobic environment (Fig. 3d).
Figure 3. The Tail is flexible and forms non-specific interactions with the channel.
(a) Structural flexibility of capsaicin docked inside its binding pocket. Colors indicate RMSD of top 10 models with lowest binding energies. (b) Schematic diagram of thermodynamic mutant cycle analysis (top panel, EC50_1 indicates EC50 value measured from the condition where WT channels are activated by CAP.) and representative concentration-response curves for determining coupling energy between the Tail and E571 (lower panel). (c) Summary of coupling energy measurements. Residue with coupling energy at least 90% of the 1.5 kT threshold is colored in orange; those with less energy are colored in different shades of blue. At least four independent trials were performed for each chemical at each concentration. (d) Coupling energy is mapped onto the binding pocket. Color scheme is the same as in (c). The tip of the Tail, which is removed in C6, is presented as ball and-stick in magenta. All statistical data are given as mean ± SEM.
Both our computational and functional analyses suggest the Tail makes non-specific VDW interactions with TRPV1. Indeed, a flexible Tail can explain the much smaller electron density for capsaicin observed in cryo-EM studies11: the density of the Tail was averaged out because of its high mobility, while the Head and Neck adopted relatively fixed positions so only their electron densities were registered (Fig. 1c and Supplementary Fig. 2). The relatively fixed Head and Neck positions indicate they are stabilized by specific interactions, which have been suggested as hydrogen bonds by docking and will be tested in the following experiments.
Neck of capsaicin: specific interaction for binding
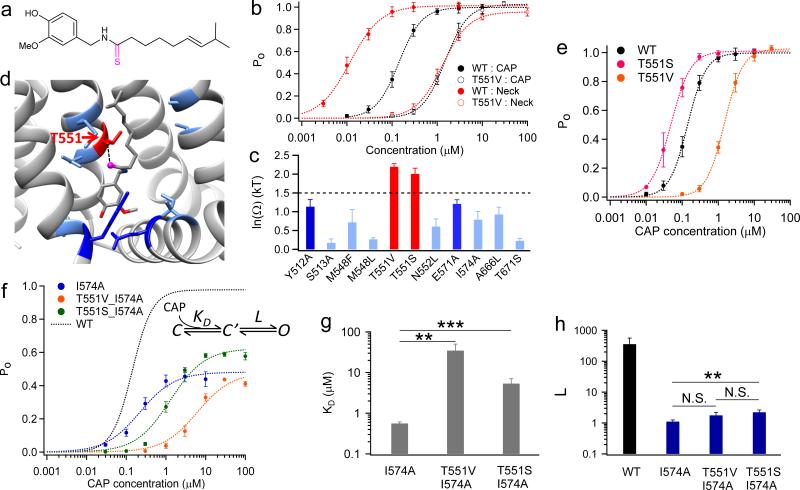
Unlike the aliphatic tail, the Neck of capsaicin, made of an amide group, is polar. We observed from docking that the oxygen atom makes specific interaction with T551. To test this prediction, we again employed thermodynamic mutant cycle analysis. Mutation T551V and a Neck analog (Compound 8) with an O→S replacement (Fig. 4a) both substantially affected EC50 (Fig. 4b). However, they exhibited no additive effect (Fig. 4b), indicating the existence of a specific interaction. Indeed, the coupling energy between T551 and the Neck oxygen is 2.19 ± 0.08 kT (equivalent to 1.29 ± 0.05 kcal/mol at 24°C), much larger than the 1.5 kT threshold for specific interaction. We further calculated the coupling energy for nine other residues (Fig. 4c). The distribution of coupling energy clearly shows that only at T551 there was a significant coupling with the Neck of capsaicin. Even the nearby residues M548 and N552 exhibited much weaker coupling (Fig. 4c and d), arguing for the sensitivity of our analysis for specific ligand-channel interactions. Moreover, when we altered the Neck by substituting the –NH– group with an oxygen atom, EC50 of this analog was ~ 1000 fold higher than that of capsaicin (Supplementary Fig. 4a and b). In fact this analog, named capsiate, is a capsaicinoid rich in non-spicy sweet peppers25. Therefore, we believe that T551 specifically interacts with the Neck of capsaicin, serving as an anchor for its binding.
Figure 4. The Neck interacts specifically with T551.
(a) Chemical structure of the capsaicin analog (Compound 8) with an altered Neck structure. (b) Representative concentration-response curves using analog in (a) and T551. (c) Summary of coupling energy at nine residues. Residue with coupling energy larger than the 1.5 kT threshold is colored in red. At least four independent trials were performed for each chemical at each concentration. (d) Coupling energy is mapped onto the binding pocket. Color scheme is the same as in (c). The oxygen atom of the Neck, which is changed in the analog in (a), is presented as ball-and-stick in magenta. (e) Concentration-response curves of TRPV1 WT, T551V and T551S. (f) Gating scheme of ligand binding and channel gating and concentration-response curves of double mutants T551V_I574A (colored in orange) and T551S_I574A (colored in green). (g and h) Comparison of KD (g) and L (h) values of mutants. L of WT was determined from Po_max by single- channel recordings as . N.S., **, and *** denote P > 0.05, P < 0.01, and P < 0.001, respectively. All statistical data are given as mean ± SEM.
Is this specific interaction mediated by hydrogen bond, as our docking results suggested? This is found likely because, while preventing hydrogen bond by T551V caused a major right-shift of the concentration-response curve, the conserved mutation T551S led to a slight left-shift of the concentration-response curve (Fig. 4e). Further evidence came from capsazepine, a competitive antagonist to capsaicin26. Capsazepine differs substantially from capsaicin in the Head and Tail, but shares a similar Neck group. Like capsaicin, we also observed opposite effects of T551V/S on channel inhibition by capsazepine (Supplementary Fig. 4c to e). All these observations agree with the existence of a hydrogen bond. Moreover, at single-channel level we found that increasing capsaicin concentration could compensate for the weakened binding by T551V mutation, yielding a near unity Po just like WT (Supplementary Fig. 5a and b). This observation suggests that the Neck-T551 interaction mainly contributes to ligand binding.
As seen in the general ligand gating scheme (Fig. 4f), a ligand-channel interaction can contribute to binding (reflected by KD) or promoting conformation transition for gating (reflected by L). Both effects contribute to the measured apparent affinity: . Distinguishing the two effects from EC50 measurements is thus in general difficult27 but becomes possible when L is small, because under such condition, EC50 is dominated by KD. This condition is met when the maximum open probability is substantially below unity, as . Since both WT and T551 mutants exhibited high Po_max (Fig. 1a and Supplementary Fig. 5a and b), to get a definitive answer for the role of Neck–T551 interaction, we used a mutant I574A in the S4-S5 linker that (for reasons that will become clear later) reduced Po_max at saturating capsaicin to 0.54 ± 0.03 (Supplementary Fig. 5c and d, see also Supplementary Fig. 3). To confirm that the recordings were from a single channel, the number of channels was statistically estimated (Supplementary Table 4). Introducing T551V or T551S mutation to the I574A background retained the low Po_max, allowing both KD and L to be reliably estimated. (None of these mutations altered channel conductance, Supplementary Fig. 6.) We found that in this background T551V largely shifted the concentration-response curve to higher concentrations without changing Po_max, clearly demonstrating a nearly pure KD effect (Fig. 4f, orange). The ~ 50 fold increase in KD (Fig. 4g) translates to a prominent energetic effect of 3.9 kT (equivalent to 2.30 kcal/mol at 24°C). At our recording temperature (~24°C) hydrogen bond strength could range from 2.5 to 16 kT28,29, therefore the energetic effect of T551V mutation is compatible with breaking a stable hydrogen bond. The conserved mutation T551S caused a right shift of the concentration-response curve, again reflecting weakened binding, accompanied by an increase in Po_max (Fig. 4f, green). This increase in Po_max may underline the left shift of concentration-response curve of T551S (Fig. 4e). Contrary to major changes in KD, while I574A greatly reduced L compared to WT, a second mutation of T551V or T551S did not largely affect L (Fig. 4h). Therefore, these observations strongly suggest that interaction between T551 and the Neck mostly contributes to ligand binding.
Head of capsaicin: for both binding and activation gating
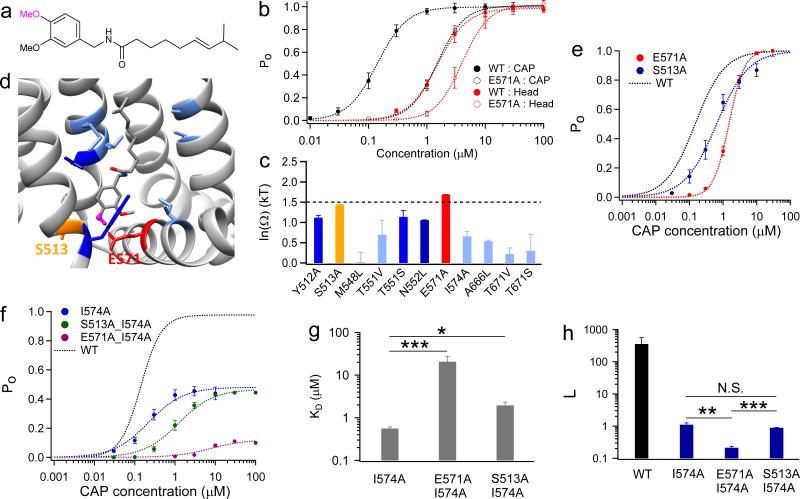
In contrast to a single specific interaction by the Neck, the Head of capsaicin may interact with four residues (Y512, S513, E571 and T671) through hydrogen bonding (Fig. 1e). By thermodynamic mutant cycle analysis with a Head analog (Compound 9) (Fig. 5a), we observed that two out of the four predicted sites exhibited coupling energies significantly larger (E571) or close to (S513) the 1.5 kT threshold (Fig. 5b and c). Interestingly, both residues are located at the intracellular lipid-water interface (Fig. 5d), confirming that the Head of capsaicin points downward to this interface.
Figure 5. The Head makes specific interactions with channel residues at lipid-water interface.
(a) Chemical structure of the capsaicin analog (Compound 9) with changes in the Head. (b) Example concentration-response curves for determining the coupling energy between the Neck and E571. (c) Summary of the coupling energy at seven residues. Residues are colored following the same scheme as previous figures. At least four independent trials were performed for each chemical at each concentration. (d) Coupling energy is mapped onto the binding pocket. Color scheme is the same as in (c). The hydroxyl group of the Head, which is replaced by a methyl group, is presented as ball-and-stick in magenta. (e) Concentration-response curves of TRPV1 WT, S513A and E571A. (f) Concentration-response curves of the double mutants S513A_I574A (colored in green) and E571A_I574A (colored in purple). (g and h) Comparison of KD (g) and L (h) values of mutants. N.S., *, **, and *** denote P > 0.05, P < 0.05, P < 0.01, and P < 0.001, respectively. All statistical data are given as mean ± SEM.
What is the nature of interactions between the Head and these two residues? We observed that E571A and S513A exhibited a right-shift in concentration-response curves (Fig. 5e). When introduced individually into the I574A background, S513A caused a small (< 10 fold) right shift of concentration-response curves without changing Po_max (Fig. 5f, green), therefore reflecting mostly a binding effect. Strikingly, E571A not only largely shifted its curve to higher concentration, but also Po_max was significantly reduced (Fig. 5f, purple). This strong gating effect was further confirmed by single-channel recordings (Supplementary Fig. 5c and d). By fitting these data to the gating scheme in Fig. 4f, we noticed that E571A led to ~ 50 fold (3.9 kT, equivalent to 2.30 kcal/mol at 24°C) increase in KD, which could be caused by losing a hydrogen bond (Fig. 5g). This mutation also significantly reduced L, underlining the large gating effect (Fig. 5h). In contrast, S513A increased KD by merely ~ 5 fold, suggesting that the Head does not form hydrogen bond with S513. It was demonstrated that the mutation S513Y can virtually eliminate capsaicin activation2. Based on our observations here, we believe that the bulky side chain of tyrosine in S513Y may collide with the Head to prevent capsaicin binding.
Unlike T551 and the Neck of capsaicin, interaction between E571 and capsaicin Head serves more than just stabilizing ligand binding. The much reduced Po_max by E571A argues that this interaction is strongly involved in gating of the channel by capsaicin27. In fact, both single-channel recording and noise analysis (Supplementary Fig. 7) suggest I574A and E571A are the only two mutations that substantially decreased Po_max (Supplementary Fig. 8). Interestingly, these residues are juxtaposed on the S4-S5 linker, a domain known to transduce activation conformational changes in both voltage-gated channels30,31 and TRP channels32,33. Therefore, they provide crucial clues to understand how binding of capsaicin induces channel activation.
Capsaicin-channel interactions in different gating states
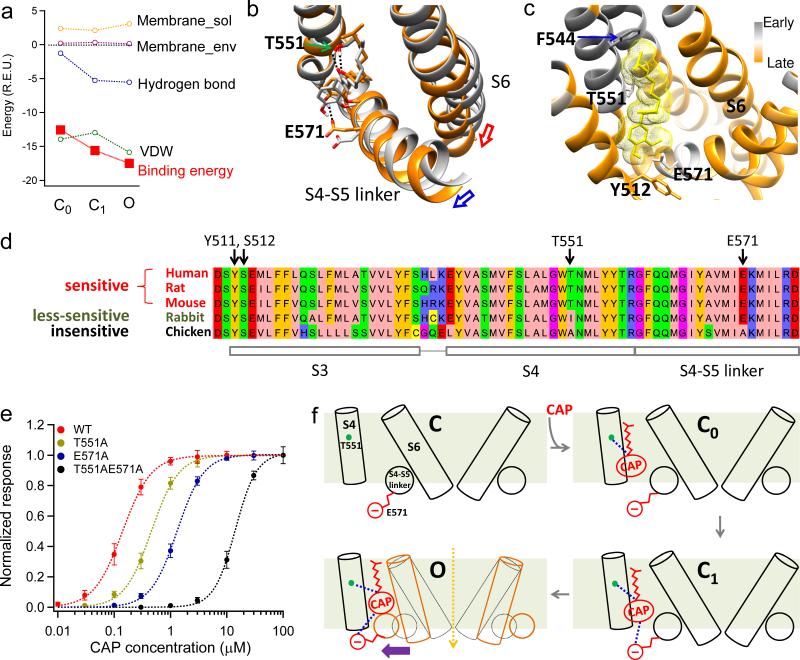
From the experiments discussed above, we identified residues that are involved in (or excluded from) specific interactions with capsaicin. With such knowledge as guidance and constraints, we aimed to simulate the process of capsaicin binding and gating. By docking with or without restrictions for structural adjustments of the channel, we captured two transitional closed states with capsaicin bound (C0, initial binding with channel structure fixed in closed state; C1, closed state of the channel but with induced fitting of the pocket upon capsaicin binding) and the open (O) state (Fig. 6a). The binding energies of capsaicin for each state were calculated and decomposed by Rosetta in the same way as that shown in Fig. 1d. We observed that in C0 the Tail has already contacted the upper half of S4 (Supplementary Fig. 9 and Supplementary Movie 1), suggesting that formation of VDW interactions is one of the early events in capsaicin binding. Additionally, in C0 a hydrogen bond was formed between the Neck and T551 (Fig. 6b and Supplementary Fig. 9). Therefore we believe that the binding of capsaicin has already been secured in C0 by such VDW and hydrogen bond interactions. As the system transits from C0 to O, a second hydrogen bond was formed in C1 between the Head and E571 (Fig 6a; Supplementary Fig. 9). The C0 to C1 transition was mainly driven by this additional hydrogen bond formation. From C1 to O, hydrogen bonding energy remained similar, whereas VDW interactions became more favorable for the open state. Together the increases in combined energy drove the channel from C0 through C1 to O (Fig. 6a).
Figure 6. Mechanisms for capsaicin induced channel activation.
(a) Components and total binding energies calculated from the initial closed state (C0), second closed state (C1) and open state (O), respectively. For each state, energy decomposition was done by applying Rosetta's residue_energy_breakdown utility to the docking model (see Online Methods for details). (b), Capsaicin-induced conformational changes between C0 (grey) and O (orange) states. Hydrogen bonds are indicated by black dotted lines. Capsaicin goes deeper into its binding pocket in O state. (c) fprogress values calculated by iNEM modeling are mapped onto the capsaicin binding pocket. Residues moving early are colored in grey, while those moving late are colored in orange. Residues that may interact with the Tail of capsaicin such as F544 move relatively early, indicating the Tail contacts the binding pocket first. Lower S6 segment moves late as it forms the activation gate, which opens after capsaicin binding. Y512 cradles capsaicin inside the binding pocket in the open state, so it also moves late. (d) T551 and E571 are key residues responsible for species difference in capsaicin sensitivity. Protein sequences of TRPV1 in five species with known capsaicin sensitivity are aligned by AlignX in Vector NTI Advance suite and visualized by Jalview47 with Zappo color scheme. (e) Concentration-response curves of TRPV1 WT, T551A, E571A and T551A_E571A. (f) A schematic diagram summarizing capsaicin binding and activation of the channel. All statistical data are given as mean ± SEM.
From these three states we observed that movement of S4-S5 linker towards the bound capsaicin is the key for activation: it drags S6 with it to open the activation gate (Supplementary Movie 1), as suggested in the cryo-EM studies11. Our analysis also revealed mechanisms for such sequential transitions: in C0 capsaicin binding is established while the S4-S5 linker stochastically switches between inward (closed) and outward (activated) conformations. In C1 and O the outward conformation is stabilized by hydrogen bond between the hydroxyl group of Head and E571, as well as VDW interactions between the methyl group of Head and the linker (Fig. 6b and Supplementary Fig. 10). Indeed, the two mutations (E571A and I574A) that can decrease Po_max are clustered on the linker (Supplementary Fig. 8), supporting the role of linker in mediating ligand induced activation. Moving to the O state, the S6 follows the outward movement of S4-S5 linker to open the pore. Interestingly, an alternative approach based on interpolated elastic network modeling (iNEM), which has successfully predicted temporal sequence of events in ligand-gated ion channels34-36, produced a compatible picture. We observed that residues that interact with the Tail move first, while opening of the S6 gate and movement of Y512 occur later (Fig. 6c).
Such a mechanism of TRPV1 activation well explains preciously observed species difference in capsaicin sensitivity. Y512 and S513 were reported as important sites for capsaicin activation2 but are well conserved among TRPV1 in species with different capsaicin sensitivity (Fig. 6d). Instead, the two hydrogen bond partners, T551 and E571, may offer an answer for such species difference. In the weakly capsaicin-sensitive rabbit TRPV19, T551 is replaced by an isoleucine, thus eliminating one hydrogen bond with Neck of capsaicin. For capsaicin-insensitive chicken TRPV12, both T551 and E571 are replaced by an alanine, eliminating two hydrogen bonds with Neck and Head of capsaicin (Fig. 6d). Indeed, we found that the EC50 value of a mutant containing both mutations was largely increased by more than 100 fold, making capsaicin a very poor agonist (Fig. 6e and Supplementary Table 1). As for Y512, both cryo-EM structures11 and the present study show that its bulky side chain flips up in the O state. This flip will close the mouth of binding pocket, cradling capsaicin molecule inside (Supplementary Fig. 11). Replacing Y512 with a smaller alanine substantially weakened binding whereas a similarly bulky phenylalanine did not2 (Supplementary Table 1). The fact that the aromatic phenylalanine does not partake in hydrogen bonding further confirms that the contribution of Y512 to binding affinity is likely steric.
DISCUSSION
Our results strongly suggest that inside its binding pocket, capsaicin molecule takes “Tail-up, Head-down” configurations. Its aliphatic tail interacts with the channel through non-specific VDW forces to contribute to binding affinity, while the specificity for ligand binding is bestowed by hydrogen bonds between its vanillyl Head, amide Neck and the channel (E571 and T551, respectively). Upon binding, capsaicin induces a series of structural rearrangements to stabilize the S4-S5 linker in its outward (activated) conformation mainly by “pulling” at E571 through hydrogen bond and VDW contact with the linker. As S4-S5 linker swings outward, S6 follows behind to open the activation gate. We believe that such a framework of “Tail-up, Head-down” binding and “pull-and-contact” gating should be generally applicable to TRPV1 activation by capsaicinoids37 (Fig. 6f).
Indeed, the extremely potent TRPV1 agonist resiniferatoxin (RTX) has an identical head group as capsaicin38 for efficacious gating, whereas its “tail” diterpene moiety is substantially larger to allow its registration in cryo-EM structure11. The “tail” clearly contributes to tight binding, probably as it allows more points of interaction with the binding pocket. The fact that the bulky tail of RTX can fit in the binding pocket is nicely compatible with the conclusion that the smaller capsaicin tail can sample multiple conformations in the spacious pocket. Many naturally occurring capsaicinoids found in spicy peppers exhibit the same chemical architecture: they contain identical Head and Neck with capsaicin to preserve activity but have a variable Tail that affects the perceived “hotness”37,39. It is further known that introducing a single halogen atom (Cl, Br or I) to the Head of either capsaicin or RTX40-42, or removing either the hydroxyl or methyl groups from the Head39 turns these TRPV1 agonists into competitive antagonists. Such dramatic switch occurs most likely because these changes compromise the Head's ability to engage in “pull-and-contact” interaction with S4-S5 linker (by either directly affecting inter-molecular contact or indirectly affecting hydrogen-bonding strength) while leaving the Tail and Neck intact for binding.
Our study demonstrated that computation is a useful gadget in structural biology toolbox, especially when combined with complementary functional methods. Recent advance in cryo-EM methodologies has greatly increased the power to determine protein structures, particularly for membrane proteins that are generally difficult to resolve by crystallography43. Despite the much improved spatial resolution, often atomic details important for understanding biological process are beyond the resolution limit of current cryo-EM technology. Computation methods based on atomic force fields, powered by rapid increase in computation power, is a promising approach to fill the gap by revealing atomic interactions44,45. Combination of cryo-EM with computational techniques makes it now practical to achieve mechanistic understanding of protein functions at the atomic level46.
Online Methods
Molecular biology and cell transfection
Murine TRPV1 was used in this study. To facilitate identification of transfected cells, enhanced yellow fluorescence protein (eYFP) was genetically linked to the C terminus of the channel, which does not alter the function of TRPV148. Point mutations were made by QuickChange II mutagenesis kit (Agilent Technologies). All mutations were confirmed by sequencing.
HEK293T cells were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 20 mM L-glutamine and 10% fetal bovine serum. Cells were transiently transfected with cDNA constructs by Lipofectamine 2000 (Life technologies) following manufacturer's protocol. Patch-clamp recordings were performed 1-2 days after transfection.
Chemicals
Capsaicin Tail analogs (Compound 1 to 7) were synthesized at Zhejiang University following the previously published method49. Characterization of these compounds by NMR matched with prior literature (Supplementary Notes). Capsaicin Neck and Head analogs (Compound 8 and 9, respectively) were synthesized as previously reported50,51 (Reaxys database: Rx-ID 1827424 for Compound 8; Rx-ID 311196 and 9064049 for Compound 9) at Chinese Academy of Sciences, Dalian Institute of Chemical Physics. Characterization of these compounds by NMR and MS matched with prior literature (Supplementary Notes).
To determine the partition coefficient (LogP) of those capsaicin analogs, shake-flask method, the classical and most reliable method of logP determination, was applied. Each compound was dissolved in a volume of octanol and water. The UV/VIS spectroscopy was used to measure the concentration of the dissolved compounds. Capsaicin, capsazepine, and other chemicals were purchased (Sigma-Aldrich). Purity was > 99%.
Molecular docking
RosettaLigand application15,52,53 from Rosetta program suite version 3.4 was used to dock capsaicin to TRPV1. Cryo-EM structure models of the transmembrane domains of rat TRPV1 at both the apo state and capsaicin bounded state10,11 (PDB ID: 3J5P and 3J5R, respectively) were first relaxed in membrane environment using the RosettaMembrane application13,31,54 and models with lowest energy scores were chosen for docking of capsaicin. Docking was comprised of three stages, which progressed from low-resolution conformational sampling and scoring to full atom optimization using all-atom energy function. In the first, low-resolution stage, capsaicin molecule was initially placed roughly in the center of the space defined by S3, S4, S4-S5 linker, and S6 segments. Its “center of mass” was constrained to move within a 10 Å diameter sphere, where it was allowed to move freely during the docking process. The electron density of capsaicin observed in the Cryo-EM structure model (3J5R) was not used as constraints for docking. A total of 200 capsaicin conformers were generated using Open Eye OMEGA55 software version 2.4.3 (OpenEye Scientific Software). They were then randomly rotated as a rigid body and scored for shape compatibility with the channel. The best-scoring models were filtered by RMSD to eliminate near-duplicates and one of the remaining models was selected at random to continue to the next stage. The second, high-resolution stage employed the Monte Carlo minimization protocol in which the ligand position and orientation were randomly perturbed by a small deviation (0.1 Å and 3°); channel residue side chains were repacked using a rotamer library; the ligand position, orientation, and torsions and protein side-chain torsions were simultaneously optimized using quasi-Newton minimization and the end result was accepted or rejected based on the Metropolis criterion. The side-chain rotamers were searched simultaneously during “full repack” cycles and one at a time in the “rotamer trials” cycles. The full repack made ~106 random rotamer substitutions at random positions and accepted or rejected each based on the Metropolis criterion. Rotamer trials chose the single best rotamer at a random position in the context of the current state of the rest of the system, with the positions visited once each in random order. The ligand was treated as a single residue and its input conformers served as rotamers during this stage. The third and final stage was a more stringent gradient-based minimization of the ligand position, orientation, and torsions and the channel torsion angles for both side chains and backbone.
As capsaicin binds to the transmembrane region of TRPV1, the molecular docking approach has to take into account the energetic effects of the lipid membrane. The membrane environment was setup using RosettaMembrane energy function13,31,54 in a XML style script in RosettaScripts14 (Supplementary Information). The script also allowed us to control the details of docking, such as whether the backbone of TRPV1 was fixed or flexible to mimic binding induced conformational changes. A total of 20,000 models were generated for each docking trial. To determine the best docking model, these models were first screened with total energy score (Rosetta energy term name: score). Top 1,000 models with lowest total energy score were selected. They were further scored with the binding energy between capsaicin and the channel. Binding energy was calculated as the difference in total energy between the capsaicin bounded state and the corresponding apo state models. Top 10 models with lowest binding energy (interface_delta_X) were identified as the candidates. These models exhibited good structural convergence (RMSD ~ 2 Å) in the head and neck regions of capsaicin molecule.
To quantitatively analyze the docking results, binding energy was decomposed as mainly van der Waals (VDW) energy and hydrogen bond energy (if_X_hbond_sc). VDW was calculated as the sum of attractive and repulsive components (if_X_fa_atr and if_X_fa_rep, respectively). As capsaicin is not charged, electrostatic interaction likely contributes little so it was not analyzed separately. Indeed, while VDW and hydrogen bond energies usually ranged from -5 to -15 Rosetta Energy Units (R.E.U.), electrostatic interaction (if_X_fa_pair) scored less than 0.7.
To reveal the spatial distribution of binding interaction between capsaicin and the channel, VDW and hydrogen bond energies were further mapped on a per residue basis to the channel by Rosetta's residue_energy_breakdown utility. Average values of VDW energy and hydrogen bond energy were calculated based the top 10 models with the lowest binding energy.
Experimentally derived information was used in iterative refinements of molecular docking. For this approach, once a specific interaction between atoms of the channel and the ligand was identified, a constraint was placed in the new round of energy minimization. For example, thermodynamic mutant cycle analysis guided by initial docking results identified that T551 on TRPV1 interacts with the oxygen atom on the neck region of capsaicin. Therefore, in subsequent docking experiments a filter was applied to require the side chain of this residue to be within 4 Å of the oxygen atom on capsaicin.
All the molecular graphics of capsaicin and TRPV1 were rendered by UCSF Chimera56 software version 1.9.
Elastic network modeling
Interpolated elastic network modeling was performed using iENM web server36(http://enm.lobos.nih.gov). 3D coordinates of same TRPV1 subunit in closed state structure (3J5P) and open state structure (3J5R) were submitted as the starting and ending conformation, respectively. The distance cutoff for elastic interaction between alpha carbon atoms was set as 13 Å. Based on this cutoff, two harmonic potentials were constructed for the starting and ending conformations, respectively. The server solved the saddle points of a general potential functions composed of these two harmonic potentials. The calculated fprogress values reflected the temporal sequence of movements. They were mapped on the capsaicin binding pocket in closed state.
Electrophysiology
Patch-clamp recordings were done with a HEKA EPC10 amplifier driven by PatchMaster software (HEKA). Single-channel recordings were performed under inside-out configuration, while macroscopic recordings were done in whole-cell mode. Serial resistance was compensated by at least 60% for whole-cell recordings. Membrane potential was hold at +80 mV. Patch pipettes were prepared from borosilicate glass and fire-polished to resistance of ~8 and ~2 MΩ for single-channel and whole-cell recordings, respectively. For both configurations, symmetrical solutions contained 130 mM NaCl, 0.2 mM EDTA and 3 mM Hepes (pH 7.2) were used for bath and pipette. All recordings were performed under room temperature (~24°C). Temperature variation was less than 1 °C as monitored by a thermometer. Current signal was sampled at 10 kHz and filtered at 2.9 kHz.
To apply capsaicin or other drugs during patch-clamp recording, a rapid solution changer with a gravity-driven perfusion system was used (RSC-200, Bio-Logic). Each solution was delivered through a separate tube so there was no mixing of solutions. Pipette tip with a membrane patch or a whole-cell patch was placed right in front of the perfusion outlet during recording to ensure the solution exchange was complete.
Data analysis
Data from whole-cell recordings were analyzed in Igor Pro (WaveMatrix). EC50 values were derived from fitting a Hill equation to the concentration-response relationship. Changes in EC50 by point mutation may be caused by either perturbation of ligand bind or channel gating or both. To distinguish these possibilities, dissociation constant (KD) for ligand binding was estimated assuming the follow gating scheme,
where L is the equilibrium constant for the final closed-to-open transition.
To perform double-mutant cycle analysis, EC50 values of four channel-ligand combinations (WT channel, CAP: EC50_1; Mutant channel, CAP: EC50_2; WT channel, CAP analog: EC50_3; Mutant channel, CAP analog: EC50_4) were determined separately. The strength of coupling was determined by the coupling energy (kT multiplied by LnΩ, where k is Boltzmann constant and T is temperature in Kelvin.). LnΩ was calculated as24
From single-channel recordings, the open probability (Po) was calculated after events detection with a hidden Markov approach in the software QuB57 (http://www.qub.buffalo.edu/). A dead time of 0.32 ms was imposed. To statistically estimate the number of channels in single-channel recordings, the probability of having two or three channels in a patch but exhibiting only one current level was calculated as previously described58. Briefly, for a patch of N independent channels, assuming that the channels transit between closed and open state with rates α and β, then given a channel open event, the probability (π) of next transition to closed state was from this particular channel was calculated as
The probability of have N channels was then approximated by P =π n0−1, where n0 was the number of single opening events.
For recordings with two channels, Po was calculated as . Recordings with more than two channels were discarded. As a complementary approach, Po was also estimated from noise analysis59 of whole-cell recordings as described previously60. To estimate channel conductance, current amplitude was estimated from all-point histogram of single-channel recordings.
Statistics
All statistical data are given as mean ± SEM. Student's t-test was applied to examine the statistical significance. *, **, and *** denote P < 0.05, P < 0.01, and P < 0.001, respectively.
Supplementary Material
Acknowledgements
We thank members of the Zheng lab for assistance and constructive discussion. This work was supported by funding from National Institutes of Health (R01NS072377) and National Natural Science Foundation of China (31328011) to J.Z., National Natural Science Foundation of China (31328011, 81301720) to W.C., National Basic Research Program of China (2013CB910204), National Natural Science Foundation of China (81371302), China Scholarship Council to W.Y., and American Heart Association (14POST19820027) to F.Y.
Footnotes
Author Contributions F.Y. and X.X. conducted most of experiments including molecular docking, patch-clamp recordings and data analysis; W.Y., P.Y, Z.S. and W.C. synthesized and analyzed capsaicin analogs; V.Y.Y. supervised molecular docking and revised the manuscript; J.Z. and F.Y. prepared the manuscript; J.Z. conceived and supervised the project, participated in data analysis.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Nilius B, Appendino G. Spices: the savory and beneficial science of pungency. Reviews of physiology, biochemistry and pharmacology. 2013;164:1–76. doi: 10.1007/112_2013_11. doi:10.1007/112_2013_11. [DOI] [PubMed] [Google Scholar]
- 2.Jordt SE, Julius D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 2002;108:421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- 3.Julius D. TRP channels and pain. Annual review of cell and developmental biology. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. doi:10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 4.Ludy MJ, Moore GE, Mattes RD. The effects of capsaicin and capsiate on energy balance: critical review and meta-analyses of studies in humans. Chemical senses. 2012;37:103–121. doi: 10.1093/chemse/bjr100. doi:10.1093/chemse/bjr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavva NR, et al. The vanilloid receptor TRPV1 is tonically activated in vivo and involved in body temperature regulation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:3366–3374. doi: 10.1523/JNEUROSCI.4833-06.2007. doi:10.1523/JNEUROSCI.4833-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Laviada I, Rodriguez-Henche N. The potential antitumor effects of capsaicin. Progress in drug research. Fortschritte der Arzneimittelforschung. Progres des recherches pharmaceutiques. 2014;68:181–208. doi: 10.1007/978-3-0348-0828-6_8. [DOI] [PubMed] [Google Scholar]
- 7.Caterina MJ, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. doi:10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 8.Zheng J. Molecular mechanism of TRP channels. Comprehensive Physiology. 2013;3:221–242. doi: 10.1002/cphy.c120001. doi:10.1002/cphy.c120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavva NR, et al. Molecular determinants of vanilloid sensitivity in TRPV1. The Journal of biological chemistry. 2004;279:20283–20295. doi: 10.1074/jbc.M312577200. doi:10.1074/jbc.M312577200. [DOI] [PubMed] [Google Scholar]
- 10.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. doi:10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. doi:10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salazar H, et al. Structural determinants of gating in the TRPV1 channel. Nature structural & molecular biology. 2009;16:704–710. doi: 10.1038/nsmb.1633. doi:10.1038/nsmb.1633. [DOI] [PubMed] [Google Scholar]
- 13.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. doi:10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleishman SJ, et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PloS one. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. doi:10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. Journal of molecular biology. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. doi:10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Lemmon G, Meiler J. Rosetta Ligand docking with flexible XML protocols. Methods in molecular biology. 2012;819:143–155. doi: 10.1007/978-1-61779-465-0_10. doi:10.1007/978-1-61779-465-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronowska AK. In: Thermodynamics - Interaction Studies - Solids, Liquids and Gases. Juan Carlos Moreno-Pirajan, editor. InTech; Rijeka: 2011. pp. 5–12. Ch. 1. [Google Scholar]
- 18.Jones JE. On the Determination of Molecular Fields. II. From the Equation of State of a Gas. Proc. R. Soc. Lond. A. 1924;106:15. [Google Scholar]
- 19.Lalatonne Y, Richardi J, Pileni MP. Van der Waals versus dipolar forces controlling mesoscopic organizations of magnetic nanocrystals. Nature materials. 2004;3:121–125. doi: 10.1038/nmat1054. doi:10.1038/nmat1054. [DOI] [PubMed] [Google Scholar]
- 20.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. Journal of molecular biology. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. doi:10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 21.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 22.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 23.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. Journal of molecular biology. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 25.Kobata KT,T, azawa S, Iwai K, Watanabe T. Novel Capsaicinoid-like Substances, Capsiate and Dihydrocapsiate, from the Fruits of a Nonpungent Cultivar, CH-19 Sweet, of Pepper (Capsicum annuum L.). Journal of agricultural and food chemistry. 1998;46:3. doi:10.1021/jf980135c. [Google Scholar]
- 26.Bevan S, et al. Capsazepine: a competitive antagonist of the sensory neurone excitant capsaicin. British journal of pharmacology. 1992;107:544–552. doi: 10.1111/j.1476-5381.1992.tb12781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colquhoun D. Binding, gating, affnity and efficacy. British journal of pharmacology. 1998;125:923–947. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franks F. Water: a matrix of life. 2nd edn The Royal Society of Chemistry; 225: Cambridge: 2000. [Google Scholar]
- 29.Sheu SY, Yang DY, Selzle HL, Schlag EW. Energetics of hydrogen bonds in peptides. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12683–12687. doi: 10.1073/pnas.2133366100. doi:10.1073/pnas.2133366100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. The Journal of general physiology. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E93–102. doi: 10.1073/pnas.1118434109. doi:10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boukalova S, Marsakova L, Teisinger J, Vlachova V. Conserved residues within the putative S4-S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. The Journal of biological chemistry. 2010;285:41455–41462. doi: 10.1074/jbc.M110.145466. doi:10.1074/jbc.M110.145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voets T, Owsianik G, Janssens A, Talavera K, Nilius B. TRPM8 voltage sensor mutants reveal a mechanism for integrating thermal and chemical stimuli. Nature chemical biology. 2007;3:174–182. doi: 10.1038/nchembio862. doi:10.1038/nchembio862. [DOI] [PubMed] [Google Scholar]
- 34.Zheng W, Auerbach A. Decrypting the sequence of structural events during the gating transition of pentameric ligand-gated ion channels based on an interpolated elastic network model. PLoS computational biology. 2011;7:e1001046. doi: 10.1371/journal.pcbi.1001046. doi:10.1371/journal.pcbi.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puljung MC, DeBerg HA, Zagotta WN, Stoll S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9816–9821. doi: 10.1073/pnas.1405371111. doi:10.1073/pnas.1405371111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tekpinar M, Zheng W. Predicting order of conformational changes during protein conformational transitions using an interpolated elastic network model. Proteins. 2010;78:2469–2481. doi: 10.1002/prot.22755. doi:10.1002/prot.22755. [DOI] [PubMed] [Google Scholar]
- 37.Steinberg X, Lespay-Rebolledo C, Brauchi S. A structural view of ligand-dependent activation in thermoTRP channels. Frontiers in physiology. 2014;5:171. doi: 10.3389/fphys.2014.00171. doi:10.3389/fphys.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szallasi A, Blumberg PM. Resiniferatoxin, a phorbol-related diterpene, acts as an ultrapotent analog of capsaicin, the irritant constituent in red pepper. Neuroscience. 1989;30:515–520. doi: 10.1016/0306-4522(89)90269-8. [DOI] [PubMed] [Google Scholar]
- 39.Thomas KC, et al. Structure-activity relationship of capsaicin analogs and transient receptor potential vanilloid 1-mediated human lung epithelial cell toxicity. The Journal of pharmacology and experimental therapeutics. 2011;337:400–410. doi: 10.1124/jpet.110.178491. doi:10.1124/jpet.110.178491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahl P, Foged C, Tullin S, Thomsen C. Iodo-resiniferatoxin, a new potent vanilloid receptor antagonist. Molecular pharmacology. 2001;59:9–15. doi: 10.1124/mol.59.1.9. [DOI] [PubMed] [Google Scholar]
- 41.Appendino G, et al. Halogenation of a capsaicin analogue leads to novel vanilloid TRPV1 receptor antagonists. British journal of pharmacology. 2003;139:1417–1424. doi: 10.1038/sj.bjp.0705387. doi:10.1038/sj.bjp.0705387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Appendino G, et al. The taming of capsaicin. Reversal of the vanilloid activity of N-acylvanillamines by aromatic iodination. Journal of medicinal chemistry. 2005;48:4663–4669. doi: 10.1021/jm050139q. doi:10.1021/jm050139q. [DOI] [PubMed] [Google Scholar]
- 43.Liao M, Cao E, Julius D, Cheng Y. Single particle electron cryo-microscopy of a mammalian ion channel. Current opinion in structural biology. 2014;27:1–7. doi: 10.1016/j.sbi.2014.02.005. doi:10.1016/j.sbi.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexander NS, et al. Energetic analysis of the rhodopsin-G-protein complex links the alpha5 helix to GDP release. Nature structural & molecular biology. 2014;21:56–63. doi: 10.1038/nsmb.2705. doi:10.1038/nsmb.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen MO, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. doi:10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 46.Lindert S, et al. EM-fold: de novo atomic-detail protein structure determination from medium-resolution density maps. Structure. 2012;20:464–478. doi: 10.1016/j.str.2012.01.023. doi:10.1016/j.str.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. doi:10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng W, Yang F, Takanishi CL, Zheng J. Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. The Journal of general physiology. 2007;129:191–207. doi: 10.1085/jgp.200709731. doi:10.1085/jgp.200709731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbero GF, et al. Application of Hansch's model to capsaicinoids and capsinoids: a study using the quantitative structure-activity relationship. A novel method for the synthesis of capsinoids. Journal of agricultural and food chemistry. 2010;58:3342–3349. doi: 10.1021/jf9035029. doi:10.1021/jf9035029. [DOI] [PubMed] [Google Scholar]
- 50.Gannett PM, et al. The Capsaicinoids - Their Separation, Synthesis, and Mutagenicity. J Org Chem. 1988;53:1064–1071. doi:Doi 10.1021/Jo00240a024. [Google Scholar]
- 51.Challis AAL, Clemo GR. Some Amines and Amides Derived from Vanillin. J Chem Soc. 1947:613–618. doi:Doi 10.1039/Jr9470000613. [Google Scholar]
- 52.Davis IW, Raha K, Head MS, Baker D. Blind docking of pharmaceutically relevant compounds using RosettaLigand. Protein science : a publication of the Protein Society. 2009;18:1998–2002. doi: 10.1002/pro.192. doi:10.1002/pro.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 54.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. doi:10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. Journal of chemical information and modeling. 2010;50:572–584. doi: 10.1021/ci100031x. doi:10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophysical journal. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. doi:10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakmann B, Neher E. Single-channel recording. 2nd edn Springer; 2009. [Google Scholar]
- 59.Sigworth FJ. The variance of sodium current fluctuations at the node of Ranvier. The Journal of physiology. 1980;307:97–129. doi: 10.1113/jphysiol.1980.sp013426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang F, Cui Y, Wang K, Zheng J. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7083–7088. doi: 10.1073/pnas.1000357107. doi:10.1073/pnas.1000357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.