Abstract
Mammals deploy a large array of odorant receptors (ORs) to detect and distinguish a vast number of odorant molecules. ORs vary widely in the type of odorant structures recognized and in the breadth of molecular receptive range (MRR), with some ORs recognizing a small group of closely related molecules and other ORs recognizing a wide range of structures. While closely related ORs have been shown to have similar MRRs, the functional relationships among less closely related ORs are unclear. We screened a small group of ORs with a diverse odorant panel to identify a new odorant-OR pairing (unsaturated aldehydes and MOR263-3). We then extensively screened MOR263-3 and a series of additional MORs related to MOR263-3 in various ways. MORs related by phylogenetic analysis (several other members of the MOR263 subfamily) had MRRs that overlapped with the MRR of MOR263-3, even with amino acid identity as low as 48% (MOR263-2). MOR171-17, predicted to be functionally related to MOR263-3 by an alternative bioinformatic analysis, but with only 39% amino acid identity, had a distinct odorant specificity. Our results support the use of phylogenetic analysis to predict functional relationships among ORs with relatively low amino acid identity.
Keywords: Olfactory receptors, molecular receptive range, ligand specificity, heterologous expression, electrophysiology, Xenopus oocytes
Introduction
Olfaction is an immense ligand recognition task involving detection of and discrimination among numerous airborne chemicals. Mammals use a large family of G-protein Coupled Receptors (GPCRs) as odorant receptors (ORs) (Buck & Axel 1991). Activation of ORs by their cognate odorants initiates a signal transduction pathway that ultimately results in action potential initiation (Bakalyar & Reed 1990, Jones & Reed 1989, Lowe & Gold 1993, Nakamura & Gold 1987). Each olfactory sensory neuron (OSN) is thought to express a single type of OR (Serizawa et al. 2003). The axons of all OSNs expressing the same OR coalesce at specific locations in the olfactory bulb to synapse with mitral and tufted cells, forming glomeruli, the first level of integration of olfactory information (Shepherd 2004). In mice, there are at least 1000 intact OR genes (Godfrey et al. 2004, Young et al. 2002, Zhang & Firestein 2002) and most appear to be expressed (Zhang et al. 2004), yielding an exceptionally large receptor array. However, the number of distinct odorant chemicals that may need to be distinguished is far larger, potentially numbering in the millions (Mombaerts 2004). To solve this problem, ORs are thought to be used in a combinatorial fashion, with each odorant being recognized by multiple ORs and each OR recognizing multiple odorants (Malnic et al. 1999).
The range of odorant structures that are recognized by an OR is termed the molecular receptive range (MRR) (Araneda et al. 2000, Mori et al. 1999). The MRRs of different ORs vary, not only in type of structures recognized, but also in breadth, with some ORs appearing to be very narrowly tuned while others are able to recognize a broad range of structures (Li et al. 2012, Reed 2004, Saito et al. 2009, Grosmaitre et al. 2009). Of course, deciphering the full extent of the MRR of a receptor is not feasible; due to practical limitations of various screening approaches (it is impossible to test all potential odorant compounds). However, the MRR can be estimated through careful design of odorant screening panels (Haddad et al. 2008, Li et al. 2012, Saito et al. 2009).
The MRRs of the OR family are presumably organized to adequately cover the portions of odor space that are relevant to a particular species. This raises the question of how we might organize the relationships among ORs with regard to their MRRs. The most straightforward approach is phylogenetic analysis, which groups ORs into subfamilies according to overall sequence comparisons (Godfrey et al. 2004, Zhang & Firestein 2002). This approach is clearly valid for closely related ORs (>75% amino acid identity), which have been shown to have closely related and overlapping MRRs (Abaffy et al. 2006, Bozza et al. 2002, Feinstein & Mombaerts 2004, Kajiya et al. 2001, Schmiedeberg et al. 2007). What about less closely related ORs? Even phylogenetically disparate ORs can have similar MRRs (Repicky & Luetje 2009). To what extent are various phylogenetic groupings (Godfrey et al. 2004, Zhang & Firestein 2002) predictive of functional relationships (similar MRRs)? An alternate organizational scheme, based on similarities and differences at 22 positions predicted to contribute to the structure of the odorant-binding site in these receptors, has been proposed (Man et al., 2007). However, this approach, as well as a comparison of residue properties across the entire OR sequence, accounted for little of the functional variation in a diverse receptor set (Saito et al., 2009). Here, we test the functional relationships among several mouse ORs (MORs) that are predicted to be related, to varying extents, by phylogenetic and alternate organizational schemes. We screened a small group of receptors with a broad panel of odorants to identify a new odorant-receptor pair (unsaturated aldehydes and MOR263-3). We then extensively screened MOR263-3, and several MORs related to MOR263-3 in various ways, in order to examine the functional relationships among these receptors.
Experimental Procedures
Materials
Xenopus laevis frogs were purchased from Nasco (Fort Atkinson, WI, USA). The care and use of X. laevis frogs in this study was approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. RNA transcription kits were from Ambion (Austin, TX, USA). Collagenase B was from Boehringer-Mannheim (Indianapolis, IN, USA). All other compounds and all odorants were from Sigma–Aldrich (St. Louis, MO, USA). CAS numbers for the odorants used in this study are listed in Supplementary Tables 1 and 2.
Expression constructs
We refer to MORs using the nomenclature of Zhang and Firestein (Zhang & Firestein 2002). The coding regions of MOR104-2 (AY073478.1, GI:18480253), MOR165-2 (AY073218.1, GI:18479733), MOR171-17 (AY073487.1, GI:18480271), MOR263-1 (AY073054.1, GI:18479405), MOR263-2 (AY073055.1, GI:18479407), MOR263-3 (AY073466.1, GI:18480229), MOR263-4 (AY073468.1, GI:18480233), MOR263-9 (AY073766.1, GI:18480829), MOR263-10 (AY073805.1, GI:18480907) were amplified by PCR from mouse genomic DNA (Clontech, Mountain View, CA, USA), subcloned into the pCI vector (Promega), and confirmed by sequencing. An N-terminal extension consisting of the N-terminal 20 amino acid residues of human rhodopsin (Abaffy et al. 2006, Saito et al. 2004) was also added to some receptor constructs. The coding regions of MOR104-2, MOR165-2, MOR263-1, MOR263-2, MOR263-4 and MOR263-10 were each combined with a T7 RNA polymerase promoter sequence and the N-terminal rhodopsin extension using PCR. The human Gαolf construct was purchased from the UMR cDNA Resource Center. The human cystic fibrosis transmembrane regulator (CFTR) construct was kindly provided by Dr. Ian Dickerson (University of Rochester). Complementary RNA (cRNA) encoding the various MORs, Gαolf and CFTR was synthesized using mMESSAGE mMACHINE Kits (Ambion).
Preparation of oocytes and cRNA injection
Oocytes were surgically removed from mature female Xenopus laevis frogs (Nasco, Fort Atkinson, WI, USA). Follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem) for 2 hrs at 22–25°C. Oocytes were injected with 46 nL of water containing cRNAs: 40 ng MOR, 10 ng Gaolf, 0.5 ng CFTR. Oocytes were incubated at 16°C in Barth's saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.5 and 100 μg / mL Ceftazidime) for 2 - 5 days prior to electrophysiological recording.
Electrophysiology and data analysis
Electrophysiology and data analysis were performed as described previously (Abaffy et al. 2006, Repicky & Luetje 2009). Briefly, odorant induced Cl− currents, resulting from cAMP-mediated activation of the co-expressed CFTR reporter channel (Uezono et al. 1993), were measured 2 - 5 days after cRNA injection using two-electrode voltage clamp in an automated parallel electrophysiology system (OpusExpress 6000A, Molecular Devices, Union City, CA, USA). Micropipettes were filled with 3M KCl and had resistances of 0.2-2.0 MΩ. The holding potential was −70 mV. Current responses were filtered (4-pole, Bessel, low pass) at 20 Hz (−3 db) and sampled at 100 Hz, and were then captured and stored using OpusXpress 1.1 software (Molecular Devices). Initial analysis was performed using Clampfit 9.1 software (Molecular Devices). Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). High concentration (0.5 – 1.0 M) stock solutions of each odorant were prepared in dimethylsulfoxide (DMSO) or ethanol. Each odorant, diluted in ND96, was applied for 15 s, followed by a 10 min wash with ND96. During initial screening, odorants were applied in mixtures: in Figure 1, 7 mixtures contained 7-8 odorants each; in Figure 4, 8 mixtures contained 17-20 odorants each and 4 sub-mixtures (derived from mixture 4) contained 5 odorants each. Odorants were screened at 30 μM, a concentration near the EC50 for many of the MORs that have been previously characterized in heterologous expression systems (Abaffy et al. 2006, Li et al. 2012, Saito et al. 2009). The use of odorant mixtures during initial screening saves a considerable amount of time and effort (Nara et al. 2011, Li et al., 2012). However, it is possible (albeit, somewhat unlikely) that the presence of an active odorant may be “masked” by other mixture components. The receptor activity of an individual odorant might be obscured by the presence of an antagonist molecule in the same mixture, or by physical interaction with another component of the mixture. Thus, there some chance that we have overlooked an active odorant in our screening. IBMX (1 mM) was used to activate the CFTR in a receptor-independent manner. This occurs both through the inhibition of phosphodiesterase and consequent increase in cAMP concentration, and through a direct action on the CFTR (Schultz et al. 1999). For concentration-response analysis of the activation of MOR263-3 by trans,trans-2,4-octadienal (ODL), each odorant response was normalized to the response of the same oocyte to application of 1 μM ODL. Normalized data were fit using Prism 5 (Graphpad, San Diego, CA, USA) according to the equation: I = Imax/(1+(EC50/X)n) where I represents the current response at a given concentration of odorant, X; Imax is the maximal response; EC50 is the concentration of odorant yielding a half-maximal response; n is the apparent Hill coefficient.
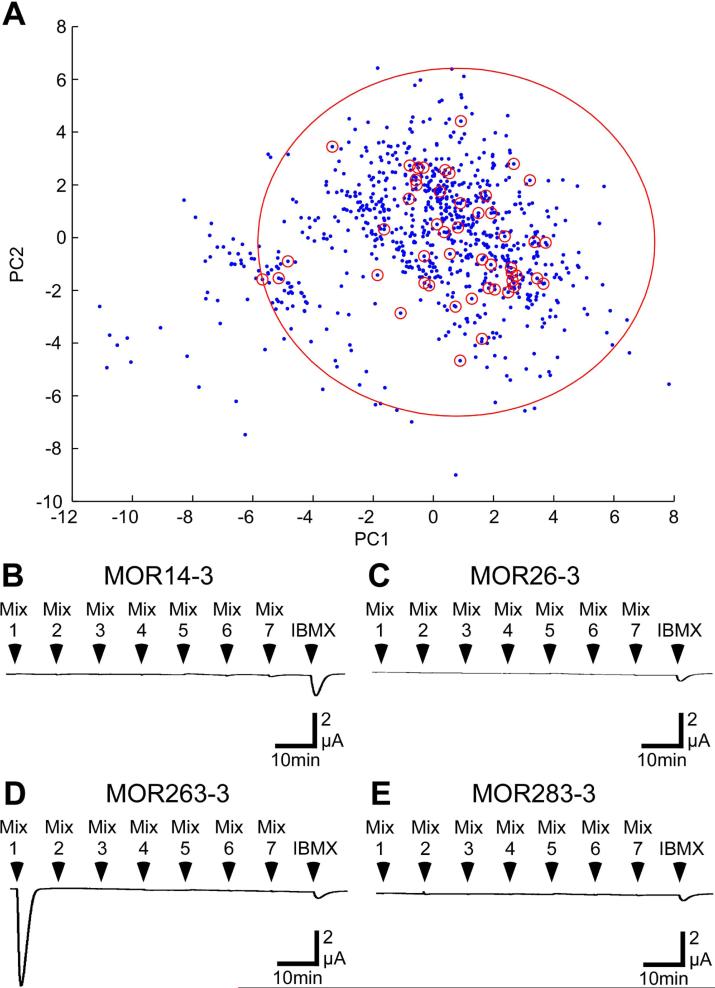
Figure 1. Screening MORs with odorant mixtures.
(A) Distribution of our odorant panel in odor space. Odor space was estimated using 961 odorants (blue dots) in a multi-dimensional odor space was based on 32 optimized physicochemical descriptors (Haddad et al., 2008). The first and second principal components were used to plot this odor space in 2 dimensions. The 54 odorants in our panel are indicated by small red circles. The large red circle is a 2D representation of the 32D hypersphere that was centered on the center of mass of the odorants in the screening set and enclosed all 54 odorants with a radius of 10.3. A hypersphere enclosing all 961 odorants in the odor space had a radius of 18.5. B-E) Oocytes injected with cRNA encoding MOR14-3 (B), MOR26-3 (C), MOR263-3 (D), or MOR283-3 (E), as well as Gαolf and CFTR, were challenged with 15 sec applications of seven odorant mixtures containing the 54 odorants in our panel, followed by 1mM IBMX. Each mixture contained 7-8 odorants, each at a concentration of 30 μM (the odorant composition of each mixture is listed in Supplementary Table 1).
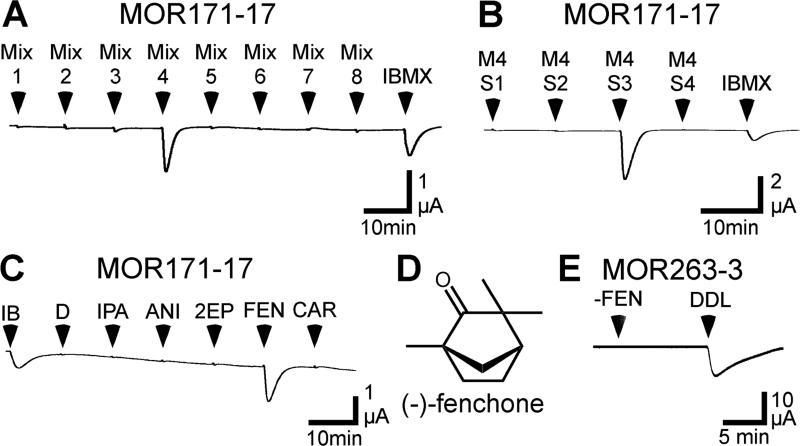
Figure 4. MOR171-17 responds to (−)-fenchone.
A) An oocyte expressing MOR171-17, Gαolf and CFTR was screened with 15 sec applications of 1mM IBMX and 8 mixtures containing a total of 155 odorants. Each mixture contained 17 - 20 odorants, each at a concentration of 30 μM (the odorant composition of each mixture is listed in the Supplemental Table 3). B) An oocyte expressing a MOR171-17, Gαolf and CFTR was challenged with 15 sec applications of 4 sub-mixtures derived from the mixture 4 and 1mM IBMX. M4S1 contained 3-heptanol, (+)-isopulegol, 3-phenyl-1-propanol, trans,trans-2,4-octadienal and cuminaldehyde. M4S2 contained trans,trans-2,4-heptadienal, phenylacetic acid, allyl tiglate, γ-octalactone and heptyl butyrate. M4S3 contained isobutyl phenylacetate, anisole, 2-ethylpyrazine, (−)-fenchone and (+)-carvone. M4S4 contained 3-nonanone, 2,3-pentanedione, 2-isobutylthiazole, 2-thiophenethiol and (−)-β-pinene. Each odorant was present at a concentration of 30 μM. C) An oocyte expressing a MOR171-17, Gαolf and CFTR was challenged with 15 sec applications of 1mM IBMX, 0.006% DMSO and 30 μM of each of the individual odorants from the M4S3: isobutyl phenylacetate (IPA), anisole (ANI), 2-ethylpyrazine (2EP), (−)-fenchone (FEN) and (+)-carvone (CAR). D) Structure of (−)-fenchone. E) An oocyte expressing a MOR263-3, Gαolf and CFTR was challenged with 15 sec applications of 30 μM (−)-fenchone and 30 μM trans,trans-2,4-decadienal.
The CFTR can be directly activated by a wide variety of structures (Ma et al. 2002). For this reason, all odorants shown to be active in our screens were tested, at our screening concentration of 30 μM, against control oocytes expressing the human M1 muscarinic receptor, Gαolf and CFTR, but no odorant receptors, to guard against false positives (Repicky & Luetje 2009). Most of the odorants did not yield a response under these conditions. For the few odorants that did yield small responses in control oocytes (< 30 nA), the responses seen in odorant receptor expressing oocytes were judged to be authentic receptor responses only if those responses were significantly greater than responses seen in the control oocytes (one-way ANOVA followed by Dunnett's post-test).
We estimated the distribution in odor space of the odorant set used for screening in Figure 1 by calculating the radius of a hypersphere centered on the center of mass of the odorants in the screening set (Saito et al. 2009). We estimated odor space using 961 odorants that are commonly used in olfaction studies (Haddad et al. 2008, Haddad et al. 2010). We obtained the molecular structure files from PubChem (http://pubchem.ncbi.nlm.nih.gov/) and used Dragon software (Talete) to compute physicochemical descriptors. This multi-dimensional odor space was based on 32 optimized physicochemical descriptors (Haddad et al. 2008). Principal component analysis was used to depict the odorants in two dimensions.
Results
Identification of odorant ligands for MOR263-3
The addition of an N-terminal sequence extension (usually 20 residues of N-terminal sequence of rhodopsin, known as the “rhodopsin-tag”) has generally been thought to be essential for successful functional expression of mammalian ORs in heterologous cells (Kajiya et al. 2001, Krautwurst et al. 1998, Reed 2004, Touhara et al. 1999, Wetzel et al. 1999). Indeed, we previously found that inclusion of the rhodopsin-tag was important for functional expression of several MORs (MOR23-1, MOR42-1 and MOR42-3) in Xenopus oocytes (Abaffy et al. 2006). However, concerns have been raised as to whether such fusion proteins might yield non-native functional data (Von Dannecker et al. 2006). This issue has been addressed in a mammalian cell (HEK293T) based assay (Zhuang & Matsunami 2007), in which several MORs were shown to be functionally expressed without an N-terminal sequence extension. It was also shown that the absence or presence of an N-terminal sequence extension (several were tested) had little effect on the odorant specificity of the MORs (Zhuang & Matsunami 2007). Thus, before attempting to de-orphanize additional MORs we decided to re-examine the need for the rhodopsin-tag when expressing MORs in Xenopus oocytes, as well as whether the presence of the rhodopsin-tag would alter odorant specificity in this expression system. In Supplementary Figure 1A, we show functional expression of four different MORs in Xenopus oocytes. Each MOR lacked an N-terminal sequence extension and was co-expressed with Gαolf and CFTR. Untagged MOR31-4 responded to undecanedioic acid, similar to what we previously observed for the rhodopsin-tagged version of the receptor, rhoMOR31-4 (Repicky & Luetje 2009); untagged MOR42-2 responded to heptanoic acid, similar to rhoMOR42-2 (Abaffy et al. 2006); untagged MOR174-9 responded to eugenol, similar to rhoMOR174-9 (Abaffy et al. 2006, Repicky & Luetje 2009, Saito et al. 2004); untagged MOR267-13 responded to lyral, similar to rhoMOR267-13 (Repicky & Luetje 2009). We examined the odorant specificities of two of these untagged MORs in more detail. In Supplementary Figure 1B-D, we show that the activation of untagged MOR174-9 by agonists (vanillin ≥ eugenol > ethyl vanillin) and antagonism by methylisoeugenol are identical to what we previously observed with rhoMOR174-9 (Abaffy et al. 2006, Repicky & Luetje 2009). Furthermore, this pattern of odorant specificity recapitulates what has been reported for MOR174-9 natively expressed in dissociated OSNs (Kajiya et al. 2001, Oka et al. 2006, Oka et al. 2004), indicating that an MOR expressed in Xenopus oocytes can display accurate ligand specificities whether the rhodopsin tag is included or not. In Supplementary Figure 1E-G, we show that untagged MOR42-2 responds well to monocarboxylic acids, but not to dicarboxylic acids. This is the same ligand specificity that we previously showed for rhoMOR42-2 (Abaffy et al. 2006). While the odorant specificity of these MORs appears unaffected by inclusion of the rhodopsin tag, the ability of these MORs to be functionally expressed without the tag suggested to us that we could proceed with our attempt to de-orphanize MORs in the absence of the rhodopsin tag (but see below).
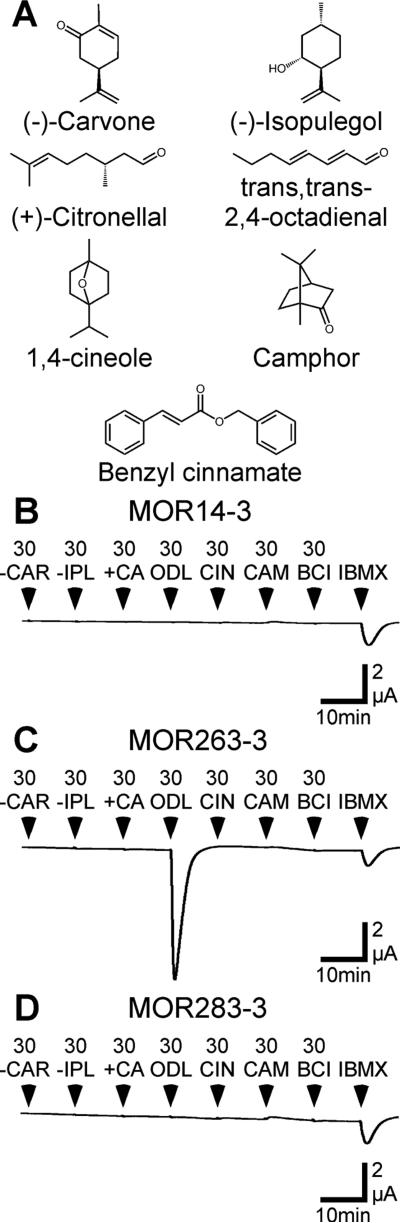
In Figure 1, we screened four previously uncharacterized and randomly chosen MORs (two from Class I, 14-3 and 26-3; two from Class II, 263-3 and 283-3) without inclusion of an N-terminal extension. We used a panel of 54 odorant (Supplementary Table 1) that represent a wide range of structures and are distributed across a large portion of an estimated odor space (Figure 1A). The radius of the 32D hypersphere enclosing these 54 odorants and centered on the center of mass of the odorants in our panel was 10.3, while the radius of the hypersphere enclosing all 961 odorants in our estimated odor space was 18.5. The odorants were applied in 7 mixtures, with each odorant present at a concentration of 30 μM, a concentration near the EC50 for many previously characterized ORs (Abaffy et al. 2006, Repicky & Luetje 2009, Li et al. 2012). Oocytes expressing MOR263-3 responded robustly to mixture 1 (Figure 1D). When we tested the individual components of mixture 1, we identified trans,trans-2,4-octadienal (ODL) as an agonist of MOR263-3 (Figure 2B). Concentration-response analysis yielded an EC50 of 21 ± 6 μM for activation of MOR263-3 by ODL (Supplementary Figure 2). MOR263-3 did not respond to any of the other compounds in mixture 1 and we did not observe any responses to these odorants with oocytes injected with cRNA encoding MOR14-3 or MOR283-3 (Figure 2C,D).
Figure 2. Identification of an odorant ligand for MOR263-3.
A) Structures of the components of odorant mixture 1. B-D) Oocytes injected with cRNA encoding MOR14-3 (B), MOR263-3 (C), or MOR283-3 (D), as well as Gαolf and CFTR, were challenged with 30 μM each of (−)-carvone (-CAR), (−)-isopulegol (-IPL), (+)-citronellal (+CA), trans, trans-2,4-octadienal (ODL), 1,4-cineole (CIN), camphor (CAM), benzyl cinnamate (BCI) and 1mM IBMX.
We further explored the MRR for MOR263-3 by screening with a panel of 28 compounds that are structurally related to ODL. This panel included aldehydes (unsaturated and saturated), ketones, alcohols, carboxylic acids and a hydrocarbon, all 6 to 11 carbons in length. In addition to ODL, MOR263-3 responded well to trans,trans-2,4-nonadienal, trans,trans-2,4-decadienal, trans-2-octenal and trans-3-nonen-2-one (Figure 3). Thus, MOR263-3 appears to be focused on a small group of unsaturated aldehydes and ketones.
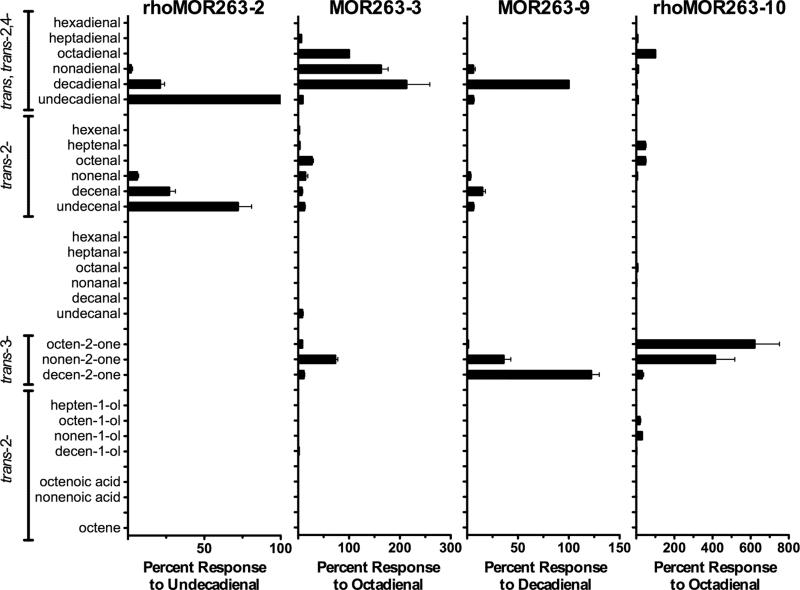
Figure 3. Responsiveness of MOR263 subfamily receptors to aliphatic odorants.
Oocytes expressing rhodopsin-tagged MOR263-2, untagged MOR263-3, untagged MOR263-9, or rhodopsin-tagged MOR263-10, as well as Gαolf and CFTR, were screened with a panel of trans,trans-2,4-aldehydes, trans-2-aldehydes, saturated aldehydes, trans-2-ketones, trans-2-alcohols, trans-2-carboxylic acids and a trans-2-alkene, ranging in length from 6 to 11 carbons. Each compound was applied for 15 s at a concentration of 30 μM. Responses were normalized to the response of each oocyte to 30 μM trans,trans-2,4-undecadienal (MOR263-2), trans,trans-2,4-octadienal (MOR263-3, MOR263-10), or trans,trans-2,4-decadienal (MOR263-9). Data are presented as the mean ± SEM (n = 3-18).
Receptive range analysis within the MOR263 subfamily
To determine whether other members of the MOR263 subfamily would respond to similar odorants, we cloned several additional members of this receptor subfamily: MOR263-1, MOR263-2, MOR263-4, MOR263-9 and MOR263-10. These receptors were also screened untagged (without the “rhodopsin-tag”) with the same focused panel of 28 compounds with which we characterized MOR263-3. One of these receptors, MOR263-9, responded well to trans,trans-2,4-decadienal, trans-3-nonen-2-one and trans-3-decen-2-one, displaying a pattern of odorant responsiveness similar to, but slightly shifted from, that of MOR263-3 (Figure 3). We did not observe any responses to these 28 odorants with the untagged versions of the other four receptors: MOR263-1, MOR263-2, MOR263-4 and MOR263-10 (data not shown). We also tested these four untagged receptors with the broad panel of 54 odorants that we used in Figure 1, but did not observe any responses (data not shown).
Our failure to observe functional responses for MOR263-1, MOR263-2, MOR263-4 and MOR263-10, when screening without an N-terminal sequence extension, prompted us to ask whether reintroduction of the rhodopsin tag would aid in functional expression. Indeed, functional responses were obtained with rhodopsin-tagged versions of two of these receptors: rhoMOR263-2 and rhoMOR263-10 (Figure 3). Again, the patterns of odorant responsiveness for these receptors were similar to, but slightly shifted from that of MOR263-3 and MOR263-9. MOR263-2 responded well to trans,trans-2,4-decadienal, trans,trans-2,4-undecadienal, trans-2-decenal and trans-2-undecenal, while MOR263-10 responded well to ODL, trans-2-heptenal, trans-2-octenal, trans-3-octen-2-one, and trans- 3-nonen-2-one. No responses were observed with rhodopsin-tagged versions MOR263-1 or MOR263-4 when screened with the 28 compounds from Figure 3 (data not shown). We also did not observe any responses when rhodopsin-tagged MOR263-4 was screened with the broad panel of 54 compounds from Figure 1 or when rhodopsin-tagged MOR263-1 was screened with a larger panel of 155 odorant compounds (Supplementary Table 2, Li et al., 2012), which contains the 54 compound panel, plus 101 additional compounds (data not shown). The four members of the MOR263 subfamily that we were able to functionally characterize (rhoMOR263-2, MOR263-3, MOR263-9 and rhoMOR263-10) have distinct but overlapping MRRs, responding primarily to unsaturated aldehydes and ketones.
The odorant specificity of MOR171-17 does not overlap with the odorant specificities of members of the MOR263 subfamily
An alternative approach to discerning the functional relationships of mammalian ORs has been proposed (Man et al. 2007), in which ORs are organized based on similarities and differences at 22 positions predicted to contribute to the structure of the odorant binding site in these receptors (Man et al. 2004). The members of the MOR263 subfamily are clustered within a multidimensional receptor space derived from this analysis (Man et al. 2007). Interspersed among them are several MORs (MOR104-2, MOR165-2 and MOR171-17), that while not closely related by overall sequence comparison (Supplementary Table 3), are predicted by this alternative analysis to be functionally related to the MOR263 subfamily. We cloned each of these MORs and screened them (untagged) against the panel of 28 compounds in Figure 3. No responses were observed (data not shown). We expanded our search for odorants that could activate these MORs by screening with our large panel of 155 odorants (Supplementary Table 2). The radius of a 32D hypersphere enclosing the odorants in this larger panel was 11.6 (Li et al. 2012), while the radius of the hypersphere enclosing all 961 odorants in our estimated odor space was 18.5. MOR171-17 responded robustly to mixture 4, containing 20 odorants (Figure 4A). When we screened the individual components of mixture 4, (−)-fenchone was found to activate MOR171-17 (Figure 4C), but not MOR263-3 (Figure 4E). Oocytes injected with cRNA encoding untagged MOR104-2 and MOR165-2 did not respond to any of the odorant mixtures (data not shown). Furthermore, oocytes injected with cRNA encoding rhodopsin-tagged versions of MOR104-2 or MOR165-2 also failed to respond to any of the 28 compounds in Figure 3 or any of the 8 mixtures containing 155 odorants (data not shown). This may be due to a failure of these ORs to be functionally expressed in the oocytes. Alternatively, MOR104-2 and MOR165-2 may have been functionally expressed but may only recognize odorants that are not within our screening panels.
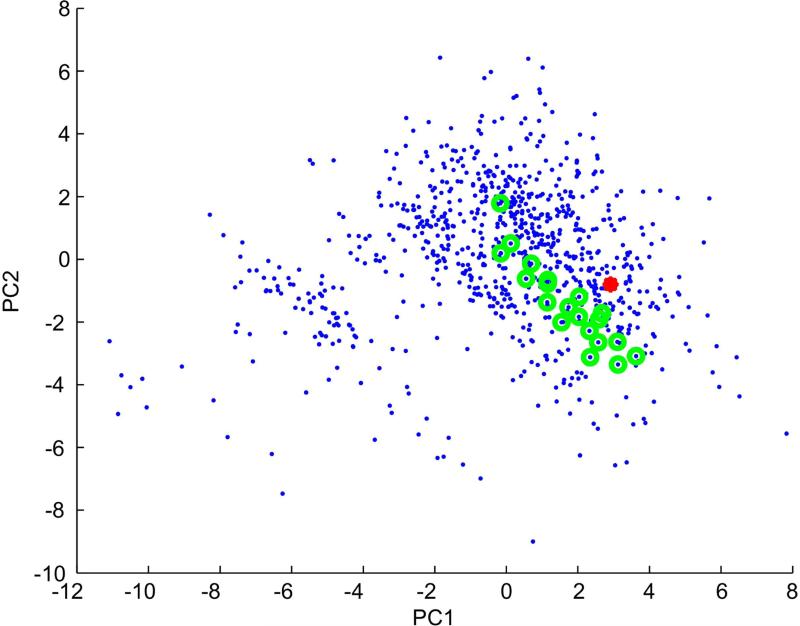
The four members of the MOR263 subfamily that we have functionally characterized display overlapping odorant specificities, with each receptor sharing odorants with one or more of the other subfamily members. In contrast, MOR171-17 failed to respond to any of the compounds that we have shown to activate members of the MOR263 subfamily. This result suggests that the alternate organizational scheme might not be predictive of OR function. To better assess the relationship, if any, between the MRRs of MOR263 subfamily members and the one odorant ((−)-fenchone) shown to activate MOR171-17, we plotted the locations of these odorants within our estimate of odor space (Figure 5). All of the odorants that we have found to activate members of the MOR263 subfamily are grouped in a diagonal band (green circles), while the MOR171-17 ligand (red dot) is located near, but outside the group of MOR263 subfamily odorants.
Figure 5. Distribution in odor space of odorants activating MOR263 subfamily members and MOR171-17.
Odor space was estimated using 961 odorants (blue dots) in a multi-dimensional odor space, based on 32 optimized physicochemical descriptors (Haddad et al., 2008). The first and second principal components were used to plot this odor space. Odorants shown (in Figure 3) to activate MOR263 subfamily members are indicated by green circles. (−)-fenchone, which activates MOR171-17, is indicated by a red dot.
Discussion
Our finding that several members of the MOR263 subfamily have distinct, but overlapping MRRs, forming a contiguous MRR subfamily is similar to what we previously observed for the MOR42 subfamily (Abaffy et al. 2006). The functional similarity among the MRRs of MOR263-3, MOR263-9 and MOR263-10 is not surprising, given the ≥ 90% amino acid identity among these three ORs. However, MOR263-2 differs substantially, with only 48% amino acid identity when compared with the other three ORs, and yet displays an overlapping MRR. When phylogenetic analyses are performed, division of ORs into clades or subfamilies depends on various criteria including bootstrap support and amino acid identity. The analysis that yielded the MORxxx-x nomenclature, using >50% bootstrap support and ≥40% amino acid identity within the clade (Zhang & Firestein 2002), groups MOR263-2, MOR263-3, MOR263-9 and MOR263-10 into the same subfamily. A subsequent analysis, using the same bootstrap support requirement and a ≥60% amino acid identity requirement (Godfrey et al. 2004), groups MOR263-3, MOR263-9 and MOR263-10 into the same subfamily (subfamily 25), but MOR263-2 into a separate subfamily (subfamily 28). Our functional results suggest that the former analysis (Zhang & Firestein 2002) is a good predictor of OR function, while the more stringent cutoff of the latter analysis (Godfrey et al. 2004) may be unnecessary. However, the number of receptors we screened in this study is small and more extensive screening of a wider array of OR subfamilies is necessary to determine whether this finding can be generalized.
We found that some ORs (MOR263-3, MOR263-9 and MOR171-17) could be functionally expressed in an untagged form, while other ORs (MOR263-2 and MOR263-10) required the rhodopsin tag for functional expression. There has been some concern that use of the tag might yield non-native functional data (Von Dannecker et al. 2006), but our findings (Supplementary Figure 1) and the work of others (Zhuang & Matsunami 2007) indicates that the rhodopsin tag does not alter the odorant specificity of ORs. Thus, our comparison of tagged and untagged ORs in Figure 3 is appropriate. However, because a subset of ORs require an N-terminal sequence extension for functional expression in heterologous systems and there is currently no obvious way to predict which ORs are in this group, inclusion of an N-terminal sequence extension such as the rhodopsin tag should remain a part of OR deorphanization efforts, as this will increase the likelihood of success. It should also be noted that the absence of N-terminal extensions in our initial screening (Figure 1) means that we may have overlooked functional responses for MOR14-3, MOR26-3 and MOR283-3 (which might have been apparent had these receptors been screened with the rhodopsin tag).
While there are some differences in carbon length preference, MOR263-2, MOR263-3 and MOR263-9 each respond particularly well to the most unsaturated (trans,trans-2,4-) aldehydes, less well to less unsaturated (trans-2-) aldehydes, but poorly or not at all to fully saturated aldehydes (Figure 3). This is interesting because the highly flexible saturated aldehydes can adopt numerous conformations, including all of the more limited set of conformations that can be adopted by unsaturated aldehydes. Our results suggest that the most active conformers reside within the set of conformations to which the most unsaturated aldehydes have access. The active set of conformers is a larger fraction of the accessible conformations for the most unsaturated aldehydes than it is for the less unsaturated aldehydes or for the saturated aldehydes. Thus, the conformational entropy penalty in the free energy of binding is greatest for the saturated aldehydes, accounting for the low or lack of sensitivity of the ORs to these compounds (Peterlin et al. 2008). If substantially higher concentrations of the fully saturated aldehydes were tested, low potency receptor responses could perhaps be observed.
An alternate organizational scheme has been proposed (Man et al. 2007) in which ORs are organized into a multi-dimensional receptor space based on the residue attributes at 22 positions predicted to contribute to the structure of the odorant binding site in these receptors (Man et al. 2004). Members of the phylogenetically defined MOR263 subfamily cluster together in this analysis. In addition, several non-phylogenetically related MORs (MOR104-2, MOR165-2 and MOR171-17) are located within the MOR263 subfamily cluster, suggesting a functional relationship. We were able to obtain functional data for one of these receptors. MOR171-17 failed to respond to any of the compounds that were active at members of the MOR263 subfamily. Instead, MOR171-17 was activated by (−)-fenchone. In our estimate of odor space (Figure 5), (−)-fenchone occupies a location that is somewhat distinct from the odorants that activate MOR263 subfamily members, but the proximity of (−)-fenchone to MOR263 subfamily odorants suggests that the alternative bioinformatic analysis (Man et al., 2007) may have some predictive power. However, the number of receptors we screened in this study is small. More extensive screening of a much larger group of ORs is clearly needed to determine whether this finding can be generalized. Combining a subset of physicochemical odorant descriptors and amino acid properties was successful in predicting odorant-receptor interactions in simulated screening of previously characterized receptors (Saito et al. 2009), but it is unclear whether this approach will allow ligand identification for previously uncharacterized receptors. It is also important to note that while our current results are supportive of phylogenetic analysis for predicting functional relationships, we have previously shown that MORs that are less related by phylogenetic (Zhang & Firestein 2002) and alternative (Man et al. 2007, Saito et al. 2009) analyses can, nonetheless, have similar MRRs (Repicky & Luetje 2009). Clearly, there are aspects of the functional organization of ORs that we have yet to grasp.
Supplementary Material
Acknowledgements
We thank Ana Castro and Benjamin Sherman for excellent technical assistance.
Funding
This work was supported by a grant from the National Institutes of Health [RO1 DC008119 to CWL] and a grant from the I-CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation [51/11 to RH].
Abbreviations
- CFTR
cystic fibrosis transmembrane regulator
- DMSO
dimethylsulfoxide
- GPCR
G-protein coupled receptor
- MOR
mouse odorant receptor
- MRR
molecular receptive range
- ODL
trans,trans-2,4-octadienal
- OR
odorant receptor
References
- Abaffy T, Matsunami H, Luetje CW. Functional analysis of a mammalian odorant receptor subfamily. J Neurochem. 2006;97:1506–1518. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29:14545–14552. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Khan R, Takahashi YK, Mori K, Harel D, Sobel N. A metric for odorant comparison. Nat Methods. 2008;5:425–429. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- Haddad R, Weiss T, Khan R, Nadler B, Mandairon N, Bensafi M, Schneidman E, Sobel N. Global features of neural activity in the olfactory system form a parallel code that predicts olfactory behavior and perception. J Neurosci. 2010;30:9017–9026. doi: 10.1523/JNEUROSCI.0398-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- Li J, Haddad R, Chen S, Santos V, Luetje CW. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J Neurochem. 2012;121:881–890. doi: 10.1111/j.1471-4159.2012.07740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- Ma T, Vetrivel L, Yang H, Pedemonte N, Zegarra-Moran O, Galietta LJ, Verkman AS. High-affinity activators of cystic fibrosis transmembrane conductance regulator (CFTR) chloride conductance identified by high-throughput screening. J Biol Chem. 2002;277:37235–37241. doi: 10.1074/jbc.M205932200. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man O, Willhite DC, Crasto CJ, Shepherd GM, Gilad Y. A framework for exploring functional variability in olfactory receptor genes. PloS one. 2007;2:e682. doi: 10.1371/journal.pone.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Gold GH. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature. 1987;325:442–444. doi: 10.1038/325442a0. [DOI] [PubMed] [Google Scholar]
- Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31:9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. Odorant receptor map in the mouse olfactory bulb: In vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006;52:857–869. doi: 10.1016/j.neuron.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. Embo J. 2004;23:120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin Z, Li Y, Sun G, Shah R, Firestein S, Ryan K. The importance of odorant conformation to the binding and activation of a representative olfactory receptor. Chem Biol. 2008;15:1317–1327. doi: 10.1016/j.chembiol.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RR. After the holy grail: establishing a molecular basis for Mammalian olfaction. Cell. 2004;116:329–336. doi: 10.1016/s0092-8674(04)00047-9. [DOI] [PubMed] [Google Scholar]
- Repicky SE, Luetje CW. Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids. J Neurochem. 2009;109:193–202. doi: 10.1111/j.1471-4159.2009.05925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Schultz BD, Singh AK, Devor DC, Bridges RJ. Pharmacology of CFTR chloride channel activity. Physiol Rev. 1999;79:S109–144. doi: 10.1152/physrev.1999.79.1.S109. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. The synaptic organization of the brain. Oxford University Press; New York: 2004. [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci U S A. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezono Y, Bradley J, Min C, et al. Receptors that couple to 2 classes of G proteins increase cAMP and activate CFTR expressed in Xenopus oocytes. Receptors Channels. 1993;1:233–241. [PubMed] [Google Scholar]
- Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CH, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J Neurosci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.