Abstract
Candida albicans is a major human fungal pathogen. One of the important features of C. albicans pathogenicity is the ability to form biofilms on mucosal surfaces and indwelling medical devices. Biofilm formation involves complex processes in C. albicans, including cell adhesion, filamentous growth, extracellular matrix secretion and cell dispersion. In this work, we characterized the role of the transcription factor Sfp1, particularly with respect to its function in the regulation of biofilm formation. The deletion of the SFP1 gene enhanced cell adhesion and biofilm formation in comparison to the wild-type strain. Interestingly, the sfp1-deleted mutant also exhibited an increase in the expression of the ALS1, ALS3 and HWP1 genes, which encode adhesin proteins. In addition, Sfp1 was demonstrated to function downstream of the Rhb1-TOR signaling pathway. Bcr1 and Efg1 are transcription factors that are critical for controlling biofilm formation, and Efg1 is also required for hyphal growth. Deleting either the BCR1 or EFG1 gene in the sfp1-null background led to reduced adhesin gene expression. As a result, the bcr1/sfp1 or efg1/sfp1 double deletion mutants exhibited dramatically reduced biofilm formation. The results indicated that Sfp1 negatively regulates the ALS1, ALS3 and HWP1 adhesin genes and that the repression of these genes is mediated by the inhibition of Bcr1 and Efg1.
Introduction
Candida albicans is a part of the normal microbial flora and typically inhabits the skin, the mucosal surfaces of the oral cavity, and the gastrointestinal and genitourinary tracts. However, this organism is also an opportunistic pathogen that can cause invasive and life-threatening infections, particularly in immunocompromised patients [1]. A critical feature that is closely related to the ability of C. albicans to cause infections is its ability to form surface-associated microbial communities called biofilms [2–4]. C. albicans can form biofilms on biotic or inert surfaces, such as the mucosal epithelia and a variety of indwelling medical devices. The increased use of implanted medical devices has enhanced the risk of C. albicans biofilm-related infections; thus, C. albicans has emerged as a major causative agent of nosocomial infections [2,4–6]. For example, biofilm formation on intravascular catheters is a key step in the development of hematogenously disseminated candidiasis [7]. Moreover, C. albicans is ranked third among the leading causes of intravascular catheter-related infections with high levels of mortality [5,8–11].
Complex processes for biofilm formation in C. albicans have been proposed. Biofilm formation begins with the adhesion of yeast cells to a supporting surface, which is followed by cell proliferation that forms a basal layer along the surface [12]. Filamentous growth is then induced, and the biofilm is enclosed by secreted extracellular matrix materials, forming a complex three-dimensional architecture [11–13]. Finally, cells disperse from the mature biofilm into the surroundings. Biofilms protect C. albicans from antifungals and may also help cells escape from the host immune system [14–17]. The antifungal resistance of C. albicans remains a great threat in clinical settings. Therefore, to understand C. albicans pathogenesis and develop novel antifungal strategies, it is necessary to elucidate different aspects of biofilm formation.
In addition, C. albicans biofilm formation involves complex signaling and gene regulation networks [11,18]. The identification and study of transcription factors related to biofilm formation should provide important insights into the molecular mechanisms that control biofilm formation. For example, the deletion of the BCR1 gene or the MSS11 gene, which both encode transcription factors, affects biofilm formation in C. albicans [19,20]. Moreover, using a computational approach, we constructed the gene regulatory networks of biofilm and planktonic cells [21]. By comparing the network structure and performing statistical analysis, we revealed the differences between the networks and identified several potential transcription factors related to biofilm formation [21]. Among those candidates, orf19.5953 exhibited the most statistical significance and was considered to be a transcription factor closely related to biofilm formation. In this study, we characterized the function of C. albicans orf19.5953. We demonstrated that C. albicans orf19.5953 is a functional homolog of Saccharomyces cerevisiae Sfp1. Moreover, the deletion of C. albicans SFP1 promotes biofilm formation in comparison to the wild-type (WT) strain. The SFP1-deleted mutant also exhibited higher expression of the ALS1, ALS3 and HWP1 genes, which encode cell wall adhesion proteins [22–24]. Finally, Sfp1 functions downstream of the Rhb1-Tor1 signaling pathway and may coordinate with the transcriptional factors Bcr1 and Efg1. Our findings highlight a previously unknown mechanism for signaling and transcription regulation during C. albicans biofilm formation.
Materials and Methods
Yeast strains and growth conditions
All of the C. albicans strains used in this study are listed in Table A in S1 File. The cells were routinely grown in YPD medium (2% glucose, 1% yeast extract and 2% peptone). Plates were prepared with 1.5% agar. For the assay of cell adhesion and biofilm formation, synthetic complete (SC) medium (0.67% yeast nitrogen base [YNB] with ammonium sulfate, 2.0% glucose, and 0.079% complete supplement mixture) was used.
DNA manipulation and strain construction
(1) Deletion and reintegration of the C. albicans SFP1, EFG1 and BCR1 genes
All deletion strains were generated from SC5314 using the SAT1-flipper method [25]. The primers used are listed in Table B in S1 File. The 5’ flanking region of SFP1 was amplified from the SC5314 genome using the primer pair CaSFP1uF-KpnI and CaSFP1uR-XhoI. The 3’ flanking region of SFP1 was amplified from the SC5314 genome using the primer pair CaSFP1dF-SacII and CaSFP1dR-SacI. The resulting 5’ and 3’ flanking regions of SFP1 were independently cloned into the pSFS2A vector [25] to generate pSFS2AdSFP1. The DNA fragment carrying the 5’ and 3’ flanking regions of SFP1 and the SAT1-flipper cassette was excised from pSFS2AdSFP1 via KpnI/SacI digestion. The linear DNA was purified and transformed into C. albicans cells for integration into the chromosome between the 5’ and 3’ flanking sequences of SFP1 via homologous recombination. The transformants were selected for nourseothricin resistance and verified by PCR. To remove the integrated SAT1-flipper cassette from the SFP1 locus, the cells were grown in YPM medium (2% maltose, 1% yeast extract and 2% peptone) to induce the MAL2 promoter-regulated recombinase for SAT1-FLIP excision. The heterozygous sfp1-deleted mutants (sfp1Δ/SFP1) were used for a second round of deletion cassette integration and excision to knock out the second allele of SFP1. Two independently generated heterozygous and homozygous sfp1-deleted mutants (sfp1Δ/sfp1Δ) were used for further study.
To construct the SFP1-reintegrated strain, the DNA fragment composed of the SFP1 promoter along with the full-length SFP1 coding sequence was amplified from the SC5314 genome using the primer pair CaSFP1uF-KpnI and CaSFP1-2R-XhoI. This fragment was cloned into pSFS2AdSFP1 upstream of the SAT1-flipper cassette to replace the original KpnI-XhoI fragment, generating pSFP1R. The DNA fragment carrying the full-length SFP1 gene, the SAT1-flipper cassette and the 5’ and 3’ flanking regions of SFP1 was excised from pSFP1R via KpnI/SacI digestion, purified and transformed into the homozygous SFP1-deleted strains. Nourseothricin selection and removal of the SAT1-cassette were performed as described above. The strains carrying the integration in the first allele of SFP1 were used to integrate SFP1 into the second allele. Two independent SFP1-reintegrated strains were used in different experiments.
Similar procedures were used to delete and reintegrate EFG1 and BCR1 in the homozygous sfp1-deleted background, generating the efg1Δ/efg1Δ/sfp1Δ/sfp1Δ and bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ double mutant strains. The primer pair EFG1-UR-F-KpnI and EFG1-UR-R-XhoI was used for the 5’ flanking region of the EFG1 gene, and the primer pair EFG1-DR-F-SacII and EFG1-DR-R-SacI was used for the 3’ flanking region of the EFG1 gene. In addition, the primer pair BCR1 UR F1-KpnI and BCR1 UR R2-XhoI was used for the 5’ flanking region of BCR1, and the primer pair BCR1 DR F1-SacII and BCR1 DR R2-SacI was used for the 3’ flanking region of BCR1.
(2) Overexpression of the C. albicans SFP1 gene
To construct a strain capable of tetracycline-induced SFP1 overexpression, the GFP gene was excised from the pNIM1 plasmid via BglII/SalI digestion [26] and replaced with the full-length SFP1 sequence, generating pNIM1-SFP1. A DNA fragment containing the tetracycline-inducible promoter (Ptet)-SFP1-SAT1 cassette was obtained from pNIM1-SFP1 via SacII/KpnI digestion. This linear DNA fragment was purified and transformed into the SC5314 genome for integration into one allele of the ADH1 locus.
One-hybrid assay
To generate the strains used in the one-hybrid assay, the SFP1 gene was PCR-amplified from the SC5314 genome using the primer pair SFP1-one-hybrid-1 and SFP1-one-hybrid-2 (Table B in S1 File). The PCR product was cloned into the pCIp-lexA-F1 plasmid [27,28] between the MluI and SphI restriction enzyme sites to generate pCIp-LexA-F-SFP1. The DNA fragment containing the lexA-SFP1 fusion was obtained from pCIp-LexA-F-SFP1 via StuI digestion, purified and introduced into the RPS1 locus of the COP1 and CCR1 strains [28] to yield COP-LexASFP1 and CCR-LexASFP1, respectively (Table A in S1 File). The COP1-derived strains use a LexA operator and an ADH1 basal promoter to drive the lacZ reporter, whereas the CCR1-derived strains lack the LexA operator.
The expression level of β-galactosidase was determined using an X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) overlay assay and a liquid β-galactosidase assay, as described previously [28], with some modifications. For the overlay assays, colonies were formed on agar plates during an overnight incubation and lysed with chloroform for 5 min. We decanted the chloroform solution and air-dried the plates for 10 min. The plates were overlaid with X-Gal—agarose (0.25 or 0.5 mg/ml of X-Gal, 0.1 M sodium phosphate buffer [pH 7.0] and 1% agarose). After the gel solidified, the plates were incubated overnight at 37°C until the blue color developed. For the liquid β-galactosidase assays, the cells were grown overnight in YPD at 30°C, subcultured into fresh YPD and grown to mid-log phase (~5 h). The cell pellets were collected, washed with sterile double-distilled water (ddH2O), and stored at -80°C until use. The cells were resuspended in 300 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4·7H2O, and 4 μl/ml of 98% mercaptoethanol, pH 7.0). The cell suspension was divided equally into three tubes, and 900 μl of Z buffer were added to each tube. The cells were lysed by adding 15 μl of 0.1% SDS and 30 μl of chloroform and vortexing the samples for 15 sec. Then, 0.2 ml of 4 mg/ml o-nitrophenyl-β-D-galactopyranoside (ONPG) in potassium phosphate buffer (pH 7.0) was added and the mixture was incubated at 37°C for 30 min. When the mixture turned yellow, the reaction was stopped via the addition of 400 μl of 1 M Na2CO3. The cell debris was removed by centrifugation, and the absorbance of the supernatants was measured at 420 and 550 nm. The β-galactosidase activity was calculated as follows: 1 Miller Unit = 1000 × [(OD420)–(1.75 × OD550)] / [(t) × (v) × (OD600)], where OD420 is the absorbance derived from ONPG, t is the duration of the reaction (in minutes), v is the volume of the supernatant used in the assay (in milliliters), OD600 is the cell density at the beginning of the reaction, and OD550 is the light scattering from cell debris. The assays were performed in triplicate, with at least three independent experiments for each tested strain.
RNA preparation and reverse transcription (RT) real-time quantitative PCR (qPCR)
The biofilm cells were scraped from the microplates and washed with ddH2O. The cell pellets were collected via centrifugation and stored at -80°C until use. Total RNA extraction and cDNA synthesis were performed as described previously [27]. The PMA1 transcripts were used as an internal control for the RNA input [29].
Real-time qPCR was performed using the StepOne Plus real-time PCR system (Applied Biosystems, Framingham, MA, USA). The primers used are listed in Table B in S1 File. Briefly, each 20 μl reaction mixture contained 80 ng of cDNA, 300 nM of each forward and reverse primer, and 10 μl of the Power SYBR green PCR master mixture (Applied Biosystems). The reactions were performed with 1 cycle at 95°C for 10 min, followed by 40 repeated cycles at 95°C for 15 sec and 60°C for 1 min. The PMA1 transcripts were used as an endogenous control for qPCR [29]. All experiments were repeated independently at least two times, and the average CT values were determined. The relative fold change in the expression of each gene was calculated using the 2–ΔΔCT method [30].
Cell susceptibility to rapamycin and hygromycin B
Cells from overnight cultures were harvested via centrifugation, washed and diluted to 3 × 107 cells per ml in sterile ddH2O. Five microliters of 10-fold serial dilutions were spotted onto YPD agar plates containing 25 ng/ml of rapamycin (catalog no. 553210; Merck KGaK, Darmstadt, Germany). The rapamycin stock solution (100 μg/ml) was prepared in methanol. In addition, cells were grown on YPD plates that contained the same volume of methanol (without rapamycin) and used as controls. For the assay of cell susceptibility to hygromycin B, YPD agar plates with or without 400 μg/ml of hygromycin B (catalog no. H7772-250MG; SIGMA) were used. Cell viability was recorded after incubation at 30°C for 5 days. The cell susceptibility assay was performed independently 3 times.
Measurement of cell adhesion and biofilm formation
The WT, SFP1-deleted and SFP1-reintegrated strains of C. albicans were grown overnight in YPD broth at 30°C. The overnight culture was subcultured into fresh SC medium and grown for 5 h. For the cell adhesion assay, 107 cells were suspended in 0.25 ml of SC medium/per well of a 24-well flat polystyrene microplate (Cat number 4430300, Orange Scientific, Braine-l’Alleud, Belgium) and incubated for 1 h at 37°C with 5% CO2. The plate was washed twice with sterile phosphate-buffered saline (PBS). Adhered cells were evaluated by assessing the reduction of 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) and by measuring the optical density at 600 nm (OD600). For the XTT reduction assay, XTT (1 mg/ml) and menadione (14.28 μM) were added to each well and the samples were incubated at 37°C for 20 minutes. The reaction was measured at 490 nm using a microplate reader [31]. For OD600 measurement, adherent cells were scraped from each well and resuspended in PBS and the OD600 was measured [32].
For the assay of biofilm formation, 3×105 cells were suspended in 1 ml of SC medium and incubated in each well of a 24-well polystyrene microplate at 37°C with 5% CO2. After 24 h, the wells were washed twice with sterile PBS. The degree of biofilm formation was determined using the XTT assay, as described above, except 0.125 mg/ml of XTT was used, and no menadione was added.
Biofilm evaluation using scanning electron microscopy (SEM) and confocal scanning laser microscopy (CSLM)
The examination of C. albicans biofilm structure using SEM was performed as described previously [20]. Briefly, 3×105 cells were grown on polystyrene coverslips (Thermanox plastic coverslip 174950, Thermo Scientific), which were placed in each well of a 24-well microplate containing 1 ml of SC medium. After biofilm formation, the coverslip was washed twice with PBS and fixed with 3.75% formaldehyde (in PBS) for 40 minutes. The post-fixed coverslip was treated with 1% osmium tetroxide for 5 minutes. After fixation, the samples were dehydrated with serial ethanol solutions (35% for 5 min; 50% for 5 min; 70% for 5 min; 80% for 5 min; 95% for 5 min; and 100% for 10 min twice), followed by overnight treatment with hexamethyldisilazane. The samples were then dried in a 60°C oven. Finally, the biofilms were examined and micrographs were collected using a SEM (Hitachi, S-4700, Type II).
To further examine the morphology and architecture of the biofilms, CSLM was used. The cells were grown on a polystyrene coverslip (Thermanox plastic coverslip 174950) in a 24-well microplate. After biofilm formation, the coverslip was washed with PBS and the cells were stained with 0.2 mg/ml of calcofluor (Sigma F-3543) for 1 h. The coverslip with the biofilm cells was placed on a regular microscope slide and covered with a regular microscope cover glass. A reinforcing ring was glued between the coverslip and the regular microscope cover glass to prevent the biofilm from becoming flattened. The morphology and architecture of the biofilms were examined using a Carl Zeiss LSM 510 confocal microscope with a 405 nm diode excitation laser.
Results
C. albicans orf19.5953 is a homolog of S. cerevisiae Sfp1
In our previous study, C. albicans orf19.5953 was identified as a promising transcription factor that may function in biofilm formation [21]. In the Candida Genome Database (http://www.candidagenome.org), C. albicans orf19.5953 is annotated as the homolog of S. cerevisiae Sfp1 and the functions of orf19.5953 are mostly uncharacterized.
In this work, to study the functions of Orf19.5953, we first aligned the amino acid sequences of C. albicans orf19.5953 and S. cerevisiae Sfp1 using Clustal W2 (Fig A in S1 File). The sequences of these two proteins share ~40% similarity and ~31% identity. In addition, both of these proteins contain two C2H2-type zinc finger domains in their C-terminal regions, which may be responsible for DNA binding (Fig A in S1 File).
S. cerevisiae Sfp1 acts as an activator to regulate ribosome protein gene expression [33]. To investigate the role of C. albicans Sfp1 in transcriptional regulation, the one-hybrid assay [28] was performed. In the one-hybrid assay, both the X-gal overlay and the liquid β-galactosidase assay were performed. Strains that express LexA-Gcn4 and LexA-Nrg1 were used as controls for the activator and repressor, respectively. As shown in Fig 1, LexA-Sfp1 can strongly activate the expression of the lacZ reporter to the same extent as LexA-Gcn4. Moreover, in the liquid β-gal assay, LexA-Sfp1 activated the lacZ reporter gene even more strongly than LexA-Gcn4 (Fig 1). These results indicate that C. albicans Sfp1 is a transcriptional activator. Therefore, based on the sequence comparison and one-hybrid analysis, we refer to orf19.5953 as C. albicans Sfp1.
Fig 1. C. albicans Sfp1 functions as a transcriptional activator, as demonstrated by a one-hybrid analysis.
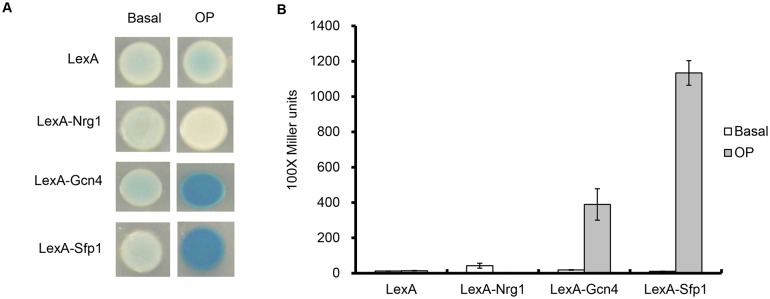
Sfp1 was fused to LexA and regulated by the ACT1 promoter. LexA-Sfp1 binds to the LexA operator (“OP”) upstream of the lacZ reporter gene. LacZ activity was measured using the X-Gal overlay assay (A) and the liquid β-galactosidase assay (B). A strain expressing only the LexA protein and a strain without the LexA operator (“basal”) upstream of the lacZ reporter gene were used as controls. The known activator Gcn4 and the repressor Nrg1 fused with LexA were used as positive and negative controls, respectively.
C. albicans Sfp1 is involved in the regulation of ribosomal gene expression and is related to the TOR signaling pathway
To further reveal the functions of C. albicans Sfp1, we generated the sfp1-deleted and SFP1-reintegrated strains (Fig A in S1 File). The successful construction of these strains was verified by Southern blot analysis and real-time PCR (Fig A in S1 File). The growth rates of the WT, sfp1Δ/sfp1Δ and SFP1-reintegrated strains were then determined. As shown in Fig B in S1 File, C. albicans sfp1Δ/sfp1Δ mutants exhibited slower growth than the WT and SFP1-reintegrated strains, in both YPD and SC broth. A similar delay in cell growth was also observed after SFP1 deletion in S. cerevisiae [34].
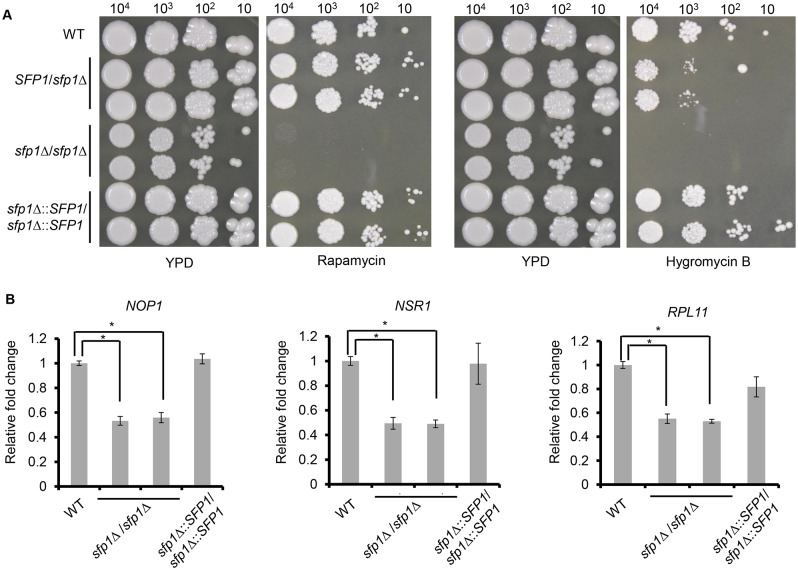
In addition, S. cerevisiae Sfp1p functions downstream of the target of rapamycin (TOR) kinase to regulate ribosomal protein (RP) and ribosome biogenesis (Ribi) gene transcription [33–36]. Moreover, the S. cerevisiae sfp1-deleted mutant is sensitive to hygromycin B, which is a drug that inhibits translation [37]. We hypothesize that C. albicans Sfp1 functions in a similar manner. To test this hypothesis, the WT, sfp1Δ/SFP1, sfp1Δ/sfp1Δ and SFP1-reintegrated strains were spotted onto agar plates with or without rapamycin. This drug is an inhibitor of the Tor1 kinase, which is the key enzyme of the C. albicans TOR signaling pathway. In Fig 2A, the sfp1Δ/sfp1Δ mutants were more sensitive to rapamycin than the WT, sfp1Δ/SFP1 and SFP1-reintegrated strains, suggesting that C. albicans Sfp1 is related to the TOR signaling pathway. Moreover, the WT, sfp1Δ/SFP1, sfp1Δ/sfp1Δ and SFP1-reintegrated strains were spotted onto agar plates with or without hygromycin B. Similar to the findings obtained for S. cerevisiae, our results demonstrated that the sfp1Δ/sfp1Δ strains were much more sensitive to hygromycin B than the WT, sfp1Δ/SFP1 and SFP1-reintegrated strains (Fig 2A).
Fig 2. C. albicans Sfp1 regulates ribosomal gene expression and is related to the TOR signaling pathway.
(A) Deletion of SFP1 increases susceptibility to rapamycin and hygromycin B. The cells were ten-fold serially diluted and spotted onto YPD agar plates with or without rapamycin (25 ng/ml) and hygromycin B (400 μg/ml). The plates were incubated at 30°C for 5 days. (B) Deletion of SFP1 affects ribosomal gene expression. The cells were grown in YPD medium overnight at 30°C, subcultured into fresh YPD medium and incubated until log phase (OD600 = 2). RNAs were isolated, cDNAs were generated and quantitative real-time PCR was performed. The expression levels of each gene are calculated as the mean ± standard deviation (SD) for three independent experiments. For each gene, the relative fold changes were displayed as the expression levels of an individual strain normalized to the WT strain (as 1). *, p < 0.05.
To determine the role of C. albicans Sfp1 in the regulation of the RP and Ribi genes, RNAs were isolated from exponential phase cells and used for RT real-time qPCR. The expression of the RP (RPL11) and Ribi (NOP1 and NSR1) genes was detected. As shown in Fig 2B, the expression of NOP1, NSR1 and RPL11 decreased in the sfp1Δ/sfp1Δ mutants compared to the WT and SFP1-reintegrated strains.
Sfp1 affects cell adhesion and biofilm formation
Using a computational approach, we hypothesized that Sfp1 is involved in biofilm formation [21]. Cell adhesion is the first step of biofilm formation. Therefore, the ability of WT, sfp1Δ/sfp1Δ and SFP1-reintegrated cells to adhere to a 24-well polystyrene microplate was compared using the XTT reduction assay. In metabolically active cells, the yellow tetrazolium salt XTT was reduced to a water-soluble orange-colored product whose absorbance can be measured at 490 nm [20]. In Fig 3A, the adhesion ability of the sfp1Δ/sfp1Δ strains was much higher than that of the WT and SFP1-reintegrated strains after 1 h incubation in SC medium. To further verify this finding, the adherent cells were scraped from the polystyrene surface and the optical density was measured at 600 nm. The results indicated that density of the adherent cells of the sfp1Δ/sfp1Δ strain was higher than that of the WT and SFP1-reintegrated strains (Fig 3B). Similar results were also obtained when cell adhesion was tested in YPD medium (Fig C in S1 File).
Fig 3. Deletion of SFP1 enhances C. albicans adhesion.
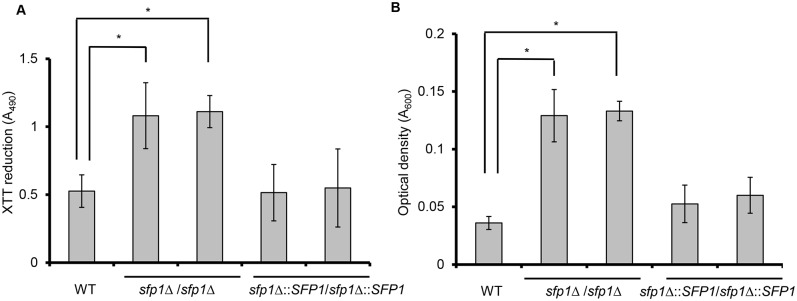
Cell adhesion was assessed in a 24-well polystyrene microplate in SC medium at 37°C with 5% CO2 for 1 h. The adherent cells were washed twice with PBS buffer. (A) Cell adhesion was determined using the XTT reduction assay. The results are presented as the mean ± SD from three independent experiments. *p < 0.05 for sfp1Δ/sfp1Δ vs. WT cells. (B) Cell adhesion was evaluated by measuring the density of the adherent cells. After washing, the adherent cells were scraped from the well, collected, and quantified by measuring the cell density (OD600). The results are presented as the mean ± SD from three independent experiments. *p < 0.05.
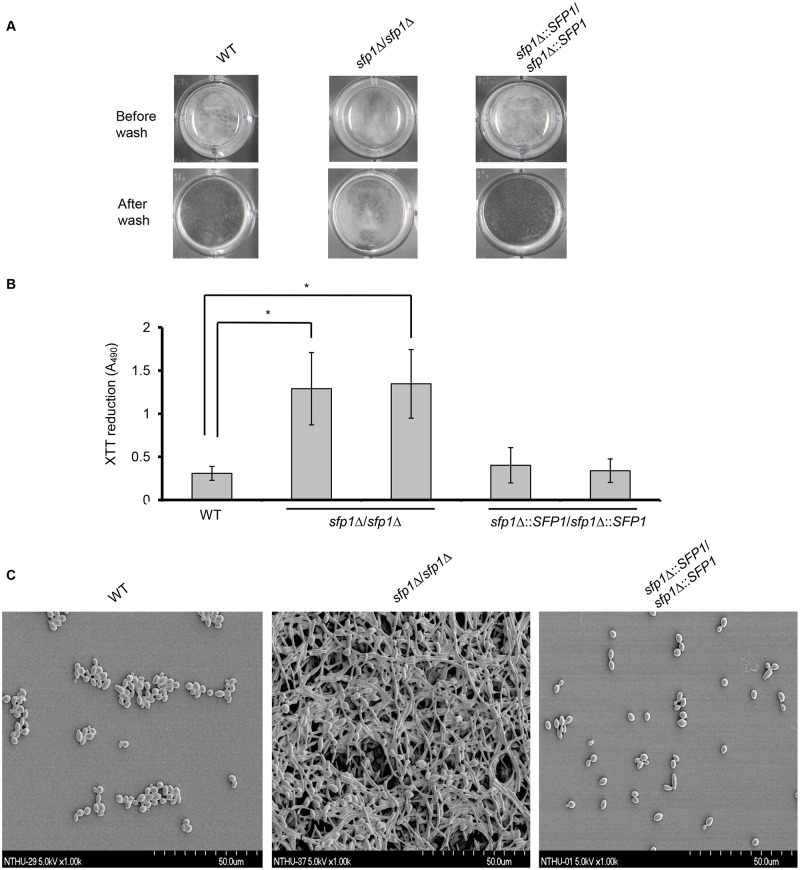
To further determine whether sfp1Δ/sfp1Δ strains can affect biofilm formation, cells were grown in SC medium in a 24-well polystyrene microplate for 24 h, and the mature biofilms in the plate were photographed and assayed using the XTT reduction method. The sfp1Δ/sfp1Δ strain exhibited much stronger biofilm formation than the WT and SFP1-reintegrated strains (Fig 4A and 4B). To further examine the structure of the formed biofilms, SEM was also used. The WT and SFP1-reintegrated strains exhibited a similar biofilm structure; the cells formed only a single layer of predominantly blastopore cells, which lacked a complex three-dimensional structure (Fig 4C). However, the structure of the sfp1Δ/sfp1Δ mutants differed considerably; the cells formed complex multilayered filamentous networks and internally embedded blastopore cells (Fig 4C). C. albicans biofilm formation was related to the growth medium used in a previous study [38]. Our finding that the deletion of SFP1 can lead to increased biofilm formation was also observed in YPD medium (Fig C in S1 File). Collectively, our results indicate that Sfp1 negatively affects cell adhesion and biofilm in C. albicans.
Fig 4. Deletion of SFP1 enhances C. albicans biofilm formation.
(A) Biofilms were formed on the surface of a 24-well polystyrene microplate in SC medium at 37°C with 5% CO2 for 24 h. (B) Biofilm formation was assessed using the XTT reduction assay. The results are presented as the mean ± SD from three independent experiments. *p < 0.05 for sfp1Δ/sfp1Δ vs. WT cells. (C) Biofilm structure was examined using scanning electron microscopy with a 1,000× magnification. Biofilm formation was carried out as described above, except the cells were grown on polystyrene coverslips for 24 h.
Sfp1 controls adhesin gene expression downstream of the Rhb1-TOR signaling pathway
The cell wall adhesin proteins Als1, Als3 and Hwp1 play critical roles in promoting C. albicans biofilm development [11,39]. The Tor1 kinase is involved in the regulation of adhesin gene expression [40]. Because the sfp1Δ/sfp1Δ mutants enhanced cell adhesion (Fig 3), we suspected that Sfp1 is involved in the regulation of the ALS1, ALS3 and HWP1 adhesin genes. RT real-time qPCR was performed, and dynamic patterns of gene expression were observed. For the ALS1 gene, very low levels of expression were detected in both the WT and sfp1Δ/sfp1Δ strains at time 0, when the cells are planktonic and nonadherent (Fig 5A). During the cell adherence stage (approximately 0.5 to 1 h), the ALS1 gene began to be induced in the sfp1Δ/sfp1Δ mutant compared to the WT strain (Fig 5A). Moreover, the expression of the ALS1 gene was enhanced in the sfp1Δ/sfp1Δ mutant during the stages of biofilm development and maturation (approximately 2 to 24 h). A similar enhancement of ALS3 and HWP1 gene expression was also detected in the sfp1Δ/sfp1Δ mutants compared to the WT strain during the development and maturation of C. albicans biofilms (Fig 5B and 5C).
Fig 5. Deletion of SFP1 enhances adhesin gene expression.
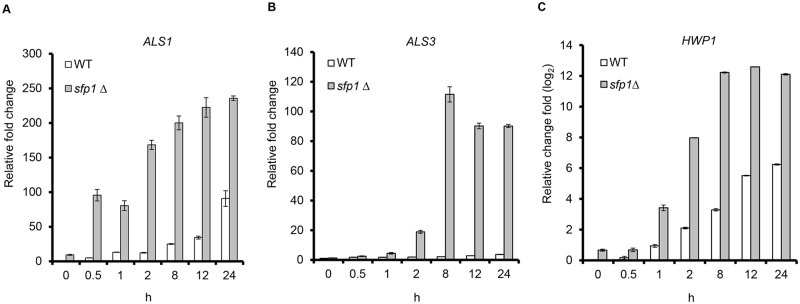
The expression of adhesin genes was compared between the WT and sfp1Δ/sfp1Δ strains during biofilm formation in SC medium at 37°C with 5% CO2. Real-time qPCR was used to detect the expression of adhesin genes. C. albicans PMA1 transcripts were used as an endogenous control. The results are presented as the mean ± SD from at least three independent experiments. For HWP1 gene expression, the fold change was represented on a log2 scale.
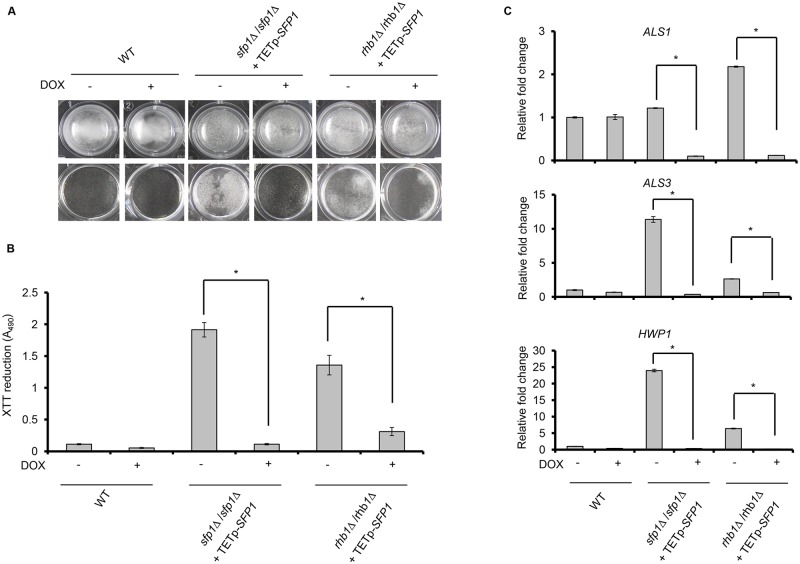
The association of Sfp1 with the TOR signaling pathway was proposed based on Fig 2. Interestingly, our previous studies linked the small GTPase Rhb1 to the Tor1 kinase in the control of filamentation and the secreted aspartyl protease 2 (Sap2), which are both important virulence factors of C. albicans [41,42]. Therefore, this finding suggests a possible connection between Sfp1 and Rhb1. To test this hypothesis, biofilm formation was compared among the sfp1Δ/sfp1Δ, rhb1Δ/rhb1Δ and sfp1Δ/sfp1/Δrhb1Δ/rhb1Δ strains. Fig D in S1 File showed that all the sfp1Δ/sfp1Δ, rhb1Δ/rhb1Δ and sfp1Δ/sfp1/Δrhb1Δ/rhb1Δ formed a robust biofilm in SC medium compared to the controls (WT, SFP1-reintegrated and RHB1-reintegrated strains). Therefore, these results were not helpful to detect epistatic relationship between SFP1 and RHB1 genes. Alternatively, we compared the strains with SFP1 overexpression in the sfp1Δ/sfp1Δ or rhb1Δ/rhb1Δ background. In the presence of the tetracycline derivative doxycycline a gene can be overexpressed under the control of a tetracycline-inducible promoter [26]. As shown in Fig 6A and 6B, the biofilm formation ability of the WT strain was not affected in the presence or absence of doxycycline. The sfp1Δ/sfp1Δ mutant exhibited enhanced biofilm formation, whereas the overexpression of SFP1 in the sfp1Δ/sfp1Δ background significantly decreased biofilm formation. Similar to the sfp1Δ/sfp1Δ mutant, enhanced biofilm formation was observed for the rhb1Δ/rhb1Δ strain without doxycycline, but doxycycline-induced SFP1 overexpression in the rhb1Δ/rhb1Δ background significantly suppressed biofilm formation. Finally, reduced expression of adhesin genes was also detected after SFP1 overexpression in the sfp1Δ/sfp1Δ and rhb1Δ/rhb1Δ strains (Fig 6C). Based on the results of this study and other studies [40], Sfp1 appears to function downstream of the Rhb1-Tor1 signaling pathway.
Fig 6. Rhb1 is involved in Sfp1-mediated regulation of biofilm formation.
Biofilm formation was compared between the sfp1Δ/sfp1Δ strain containing TETp-SFP1 and the rhb1Δ/rhb1Δ strain containing TETp-SFP1. Biofilms were formed for 24 h in SC medium at 37°C with 5% CO2, with or without 50 μg/ml of doxycycline (DOX). Doxycycline induces the overexpression of the SFP1 gene, which is under the control of a tetracycline-inducible promoter (TETp). (A) Biofilms were formed on the surface of a 24-well polystyrene microplate in SC medium at 37°C with 5% CO2 for 24 h. Biofilm cells are shown before washing (top) and after washing (bottom). Representative images from three independent experiments with similar results are shown. (B) Biofilm formation was measured using the XTT reduction assay. The results are presented as the mean ± SD from three independent experiments. *p < 0.05 for cells without DOX treatment vs. DOX-treated cells. (C) The expression of adhesin genes was compared in the presence and absence of SFP1 overexpression in the SFP1- or RHB1-deleted background. Real-time qPCR was performed, and C. albicans PMA1 transcripts were used as an endogenous control. The results are presented as the mean ± SD from at least three independent experiments. *p < 0.05 for cells without DOX treatment vs. cells with DOX treatment.
Sfp1 may regulate adhesin genes and biofilm formation through Bcr1 and Efg1
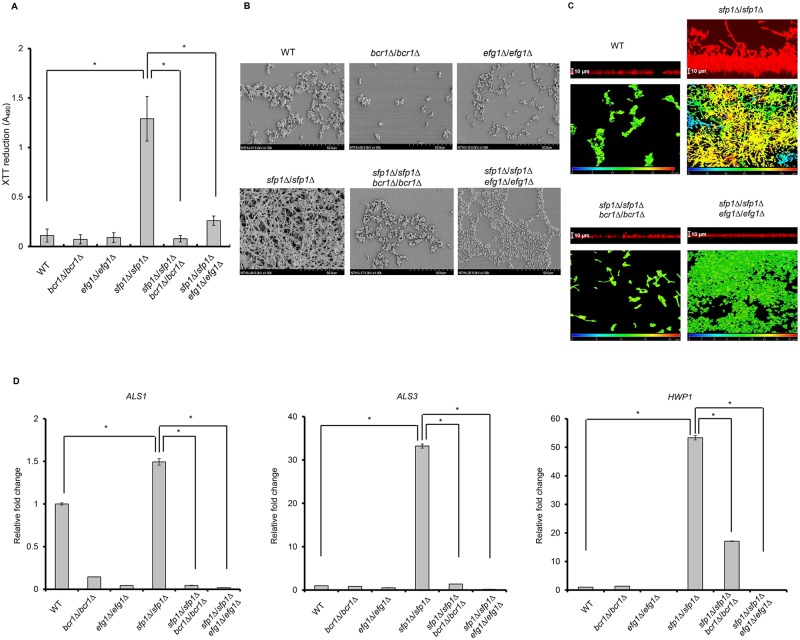
In addition to Tor1, the transcription factors Efg1 and Bcr1 are also known to control the ALS1, ALS3 and HWP1 genes and play a role in the activation of C. albicans biofilm formation [19,43–45]. Because Sfp1 can regulate the expression of the ALS1, ALS3 and HWP1 genes (Fig 5), the relationship between Sfp1, Efg1 and Bcr1 in adhesin gene regulation and biofilm formation is of interest. To address this question, we constructed bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ and efg1Δ/efg1Δ/sfp1Δ/sfp1Δ double mutants. Biofilm formation determined by the XTT reduction method showed that the sfp1Δ/sfp1Δ mutant significantly enhanced biofilm formation compared to the controls (WT, bcr1Δ/bcr1Δ and efg1Δ/efg1Δ strains). Interestingly, both the bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ and efg1Δ/efg1Δ/sfp1Δ/sfp1Δ double deletion mutants exhibited poor biofilm-forming abilities similar to that of the controls (Fig 7A). Moreover, SEM examination demonstrated that the sfp1Δ/sfp1Δ mutant formed a complex multilayered biofilm, while both the bcr1Δ/bcr1Δ and efg1Δ/efg1Δ mutants formed only a single layer of cells similar to that of WT (Fig 7B). However, the bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ and efg1Δ/efg1Δ/sfp1Δ/sfp1Δ double deletion mutants also formed a single layer of structure containing predominantly blastopore cells (Fig 7B). The biofilm structures were also examined using CSLM, and the biofilm of the sfp1Δ/sfp1Δ mutant exhibited a complex multilayered structure with a thickness of ~80 μm. However, the bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ and efg1Δ/efg1Δ/sfp1Δ/sfp1Δ double mutants exhibited reduced biofilm formation and only formed a very thin layer (~10 μm) of cell structure (Fig 7C). Because both Bcr1 and Efg1 are known to promote biofilm formation [44,46], our results suggest that Sfp1 suppresses biofilm formation via the negative regulation of Bcr1 and Efg1.
Fig 7. Sfp1 regulates biofilm formation through Bcr1 and Efg1.
(A) Biofilm formation was assessed using the XTT reduction assay. The results are presented as the mean ± SD from three independent experiments. *p < 0.05 for sfp1Δ/sfp1Δ vs. WT, bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ or efg1Δ/efg1Δ/sfp1Δ/sfp1Δ mutant cells. (B) The biofilm structure was examined using scanning electron microscopy. Pictures were taken at a 1,000× magnification. The cells were grown on polystyrene coverslips for 24 h to form biofilms. (C) The biofilm structure was examined using confocal scanning laser microscopy (CSLM). Biofilms were formed on the polystyrene coverslips in SC medium at 37°C with 5% CO2 for 24 h. After washing, the cells were stained with 0.2 mg/ml of calcofluor (Sigma F-3543, “Fluorescent brightener 28”) for CSLM visualization. Pictures were taken at a 400× magnification. (D) The expression of adhesin genes in biofilm cells was detected using RT real-time qPCR. PMA1 transcripts were used as an endogenous control. The results are presented as the mean ± SD from at least two independent experiments. *p < 0.05 for sfp1Δ/sfp1Δ vs. WT, bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ or efg1Δ/efg1Δ/sfp1Δ/sfp1Δ mutant cells.
To further reveal the relationships among Bcr1, Efg1 and Sfp1, the expression of adhesin genes was also examined using RT real-time qPCR. The ALS1, ALS3 and HWP1 genes were highly expressed in the sfp1Δ/sfp1Δ mutant, in agreement with the real-time qPCR analysis in Fig 5. However, the deletion of BCR1 or EFG1 in the sfp1Δ/sfp1Δ background significantly reduced the expression levels of all three adhesin genes (Fig 7D). Based on the biofilm structure and adhesin gene expression, Bcr1 and Efg1 appear to function as downstream effectors of Sfp1.
Discussion
The use of indwelling medical devices has become routine in the clinical setting. C. albicans easily adheres to and forms a biofilm on indwelling medical devices. Such biofilms require subsequent surgical removal and replacement of the infective devices. C. albicans cells can also adhere to and form a biofilm on mucosal surfaces, leading to resistance to antifungal agents and the initiation of infections. Therefore, C. albicans biofilms are a serious public health problem. Many recent studies focused on the regulation of biofilm formation. One study that was performed by Nobile et al. identified a transcriptional network that controls biofilm development [18]. This study combined ‘‘classical” genetics, genome-wide approaches, and RNA deep sequencing technology to comprehensively map the transcriptional circuitry that controls biofilm formation in C. albicans. In addition, this study also described a master circuit of six transcription regulators, including Bcr1, Tec1, Efg1, Ndt80, Rob1, and Brg1, that controls approximately 1,000 target genes and biofilm formation in vitro and in two animal models [18]. This master circuit led to many new predictions about the genes involved in biofilm formation, and some of those predicted genes have been confirmed to play roles in biofilm development. Another study that was performed by Wang et al. computationally screened for potential transcription factors that could regulate C. albicans biofilm [21]. In that study, gene expression profiles and ChIP-chip were used to establish biofilm and planktonic gene regulatory networks. The two networks were subsequently compared, and the relevance value was calculated to identify potential transcription factors related to biofilm formation. Among the identified candidates, the relevance value of Sfp1 (Orf19.5943) was the highest and that gene was considered the most relevant for biofilm formation. The aim of this study was to examine the functions of Sfp1, particularly the role of Sfp1 in biofilm formation.
Previous studies demonstrated that the growth medium may affect C. albicans biofilm formation under laboratory conditions [32,38,47,48]. In this study, we tested several growth media for biofilm formation, including Lee’s, Spider, SC and YPD media (Fig 4, Fig C in S1 File and Fig F in S1 File). Among the tested media, although the cells showed a growth delay (Fig B in S1 File), both the WT and sfp1Δ/sfp1Δ strains formed biofilms in the Lee’s and Spider media (Fig F in S1 File). However, the WT and sfp1Δ/sfp1Δ strains exhibited significant differences in biofilm formation in SC and YPD media (Fig 4 and Fig C in S1 File). Particularly, the WT and SFP1-reintegrated strains produced only a rudimentary biofilm in the SC medium, but the sfp1Δ/sfp1Δ mutant could form a robust biofilm with abundant filamentous cells (Fig 4). Mutant cells that are fixed in either the hyphal or yeast form can only develop into a rudimentary biofilm that is not as stable as a normal mature biofilm that contains both cell morphologies [49]. The sfp1Δ/sfp1Δ mutants exhibited highly enhanced biofilm formation compared to the WT strain, suggesting that Sfp1 negatively regulates biofilm formation under these experimental conditions. To reveal the mechanisms via which Sfp1 affects biofilm formation, we thus used the SC medium for most of our experiments.
Adhesion is the first key step for biofilm formation. Cell adhesion may be mediated by non-specific factors, including hydrophobicity and electrostatic forces of the cell surface, or by specific adhesins on the surface of C. albicans. Fig 3 demonstrated that the sfp1Δ/sfp1Δ mutants exhibit significantly enhanced cell adhesion in polystyrene microplates, indicating that Sfp1 suppresses cell adhesion. The SFP1 deletion may somehow alter the C. albicans cell wall architecture and composition, leading to changes in the non-specific properties of the cell surface. Coincidentally, the sfp1Δ/sfp1Δ mutants were resistant to the cell wall disrupting agents Congo red and calcofluor white and hyper-resistant to zymolyase, which has strong lytic activity against cell wall β-1,3-glucan (data not shown). These results implied that the SFP1 gene deletion alters cell wall structure and composition, leading to changes in adhesion-related cell surface properties.
However, other evidence suggests that Sfp1 can also regulate cell adhesion by regulating the expression of cell wall adhesin genes. The deletion of SFP1 increased ALS1 and HWP1 adhesin gene expression 0.5 h and 1 h after the cells adhered to the substrate (Fig 5). The easy detection of the ALS1 gene during the early adhesion stage of C. albicans yeast cells was reported previously [50]. Hwp1 is also required for covalent attachment to host epithelial cells and virulence [23]. Strains that lack either ALS1 or HWP1 can lose their abilities to attach to an abiotic surface and form a biofilm [18,51]. Moreover, ALS3 and HWP1 are expressed primarily in hyphal cells [50,52]. In Fig 5, the expression of ALS3 and HWP1 in the sfp1Δ/sfp1Δ strain began to be induced after cell attachment for 2 h, which is when the cells began to form germ tubes (Fig E in S1 File). Moreover, the Als1, Als3 and Hwp1 adhesins also play a complementary role in biofilm formation. A previous study demonstrated that both the hwp1Δ/hwp1Δ mutant and the als1Δ/als1Δ/als3Δ/als3Δ double mutant strain are defective in biofilm formation; however, a mixture of these two strains can form robust biofilms both in vitro and in vivo [39]. In addition, the hwp1Δ/hwp1Δ mutant produced a biofilm with significantly less biomass than the WT strain [39]. C. albicans mutants that lack Als3 produce scarce, defective biofilms on catheter material in vitro [46]. Although the functions of Als1 and Als3 in biofilm formation are somewhat overlapping, Als1 may not play as pivotal a role as Als3 [39]. An als1Δ/als1Δ mutant had only a partial defect in biofilm formation [18], whereas the als3 mutant displayed a severe defect in biofilm formation [46]. Following the early adhesion stage, the expression of all three adhesin genes increased gradually in the sfp1Δ/sfp1Δ strain (Fig 5). Together, these results suggest that SFP1 deletion can enhance biofilm development, possibly through the derepression of adhesin gene expression.
In addition to Sfp1, the transcription factors Bcr1 and Efg1 can also regulate adhesin gene expression [19,43,46,53,54] in a manner dependent on the Tor1 kinase [40]. Moreover, the bcr1Δ/bcr1Δ mutant failed to produce a mature biofilm and instead formed only a rudimentary biofilm composed of yeast and hyphal cells [19]. The efg1Δ/efg1Δ mutant also fails to form biofilms and instead generates a sparse monolayer of yeast cells without hyphae [44,55]. Efg1 was also demonstrated to be involved in both normoxic and hypoxic biofilm formation [45]. Interestingly, we found that the sfp1Δ/sfp1Δ strain was hypersensitive to rapamycin (Fig 2A) and rapamycin enhanced biofilm formation in the WT strain, leading to the formation of biofilms that were similar to those of the sfp1Δ/sfp1Δ mutant without rapamycin treatment (Fig G in S1 File). Taken together, these results and the fact that Sfp1 also regulates adhesin genes and affects cell adhesion and biofilm formation (Figs 3–5) suggest the existence of relationships among Sfp1, Efg1 and Bcr1. The bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ double mutant exhibited dramatically reduced adhesin gene expression in comparison to the sfp1Δ/sfp1Δ mutant (Fig 7D). More importantly, the biofilm of the bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ double mutant contained fewer adherent cells after a 24 h incubation than the biofilm of the sfp1Δ/sfp1Δ mutant, which contained a large number of attached cells (Fig 7A–7C). In addition, the bcr1Δ/bcr1Δ/sfp1Δ/sfp1Δ mutant had nearly no filamentous cells in the biofilm, in contrast to the sfp1Δ/sfp1Δ mutant (Fig 7B). Although Bcr1 is not required for hyphal growth, this transcription factor is required for the activation of several hyphal-specific genes, including ALS3 and HWP1. Our result is consistent with a previous biofilm assay, in which the WT strain formed a biofilm that consisted of abundant hyphal cells but the bcr1Δ/bcr1Δ strain generated a thin rudimentary biofilm that was comprised largely of yeast form cells [46]. Thus, our results suggested that Bcr1 appeared to induce adhesin expression in the absence of the SFP1 gene during biofilm formation in SC medium. In addition, the sfp1Δ/sfp1Δ/efg1Δ/efg1Δ exhibited reduced biofilm formation and was defective in filamentous growth compared with the sfp1Δ/sfp1Δ strain (Fig 7B). Moreover, the efg1Δ/efg1Δ/sfp1Δ/sfp1Δ double mutant also exhibited dramatically reduced adhesin gene expression in comparison to the sfp1Δ/sfp1Δ mutant (Fig 7D).
Taken together, these findings suggest a simple model for the functions of C. albicans Sfp1, as presented in Fig 8. Similar to S. cerevisiae Sfp1, C. albicans Sfp1 functions as an activator to regulate ribosomal gene expression. However, we further expand the function of Sfp1 to include a role in C. albicans biofilm formation, downstream of the Rhb1-Tor1 signaling pathway. Moreover, the functions of Sfp1 appear to be mediated by the negative regulation of the transcription factors Bcr1 and Efg1, which activate adhesin genes and promote biofilm formation (Fig 8). Although this study provides some new insights into the regulation of C. albicans biofilm formation, many questions must still be addressed. For example, the relationships among Sfp1, Ndt80 and Rob1 during biofilm formation remain unclear. Moreover, most of the transcription factors that have been identified to date are related to the activation of biofilm formation. In this study, we demonstrate that Sfp1 plays a negative role in biofilm formation. The identification of additional transcriptional repressors of biofilm formation is important. These studies will allow us to understand the complex interplay of negative and positive regulation in the context of biofilm formation.
Fig 8. A simple model of the role of Sfp1 in the regulation of ribosomal gene expression and biofilm formation.
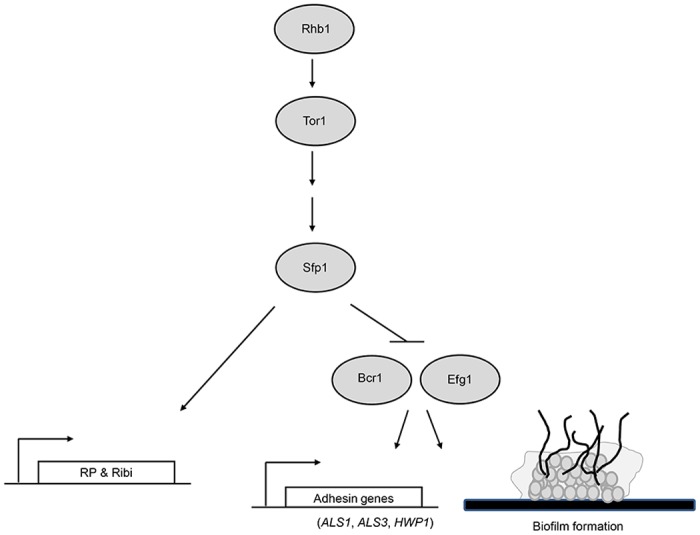
Sfp1 is related to the Tor1 signaling pathway and plays a role in the induction of RP and Ribi gene expression. Moreover, during the regulation of adhesin gene expression and biofilm formation, the small GTPase Rhb1 coordinates with Tor1 to function upstream of Sfp1, while the transcription factors Bcr1 and Efg1 function downstream of Sfp1.
Supporting Information
(DOCX)
Acknowledgments
We are grateful to A. J. P. Brown (University of Aberdeen, UK) and Joachim Morschhäuser (Universität Würzburg, Germany) for generously providing strains and plasmids.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was supported by grants NSC101-2311-B-007-010-MY3 and NSC100-2627-B-007-002 (to CYL) from the National Science Council (Taiwan). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007; 20: 133–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003; 11: 30–36. [DOI] [PubMed] [Google Scholar]
- 3. Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006; 8: 1382–1391. [DOI] [PubMed] [Google Scholar]
- 4. Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL. Candida biofilms: an update. Eukaryot Cell 2005; 4: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004; 17: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Douglas LJ. Medical importance of biofilms in Candida infections. Rev Iberoam Micol. 2002; 19: 139–143. [PubMed] [Google Scholar]
- 7. Hajjeh RA, Sofair AN, Harrison LH, Lyon GM, Arthington-Skaggs BA, Mirza SA, et al. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J Clin Microbiol. 2004; 42: 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crump JA, Collignon PJ. Intravascular catheter-associated infections. Eur J Clin Microbiol Infect Dis. 2000; 19: 1–8. [DOI] [PubMed] [Google Scholar]
- 9. Viudes A, Peman J, Canton E, Ubeda P, Lopez-Ribot JL, Gobernado M. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur J Clin Microbiol Infect Dis. 2002; 21: 767–774. [DOI] [PubMed] [Google Scholar]
- 10. Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009; 35: 340–355. 10.3109/10408410903241436 [DOI] [PubMed] [Google Scholar]
- 11. Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011; 9: 109–118. 10.1038/nrmicro2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006; 9: 588–594. [DOI] [PubMed] [Google Scholar]
- 13. Nett J, Andes D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006; 9: 340–345. [DOI] [PubMed] [Google Scholar]
- 14. Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007; 51: 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006; 55: 999–1008. [DOI] [PubMed] [Google Scholar]
- 16. Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001; 183: 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004; 72: 6023–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans . Cell 2012; 148: 126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005; 15: 1150–1155. [DOI] [PubMed] [Google Scholar]
- 20. Tsai PW, Chen YT, Yang CY, Chen HF, Tan TS, Lin TW, et al. The role of Mss11 in Candida albicans biofilm formation. Mol Genet Genomics 2014; 289: 807–819. 10.1007/s00438-014-0846-0 [DOI] [PubMed] [Google Scholar]
- 21. Wang YC, Lan CY, Hsieh WP, Murillo LA, Agabian N, Chen BS. Global screening of potential Candida albicans biofilm-related transcription factors via network comparison. BMC Bioinformatics 2010; 11: 53–63 10.1186/1471-2105-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Loza L, Fu Y, Ibrahim AS, Sheppard DC, Filler SG, Edwards JE Jr. Functional analysis of the Candida albicans ALS1 gene product. Yeast 2004; 21: 473–482. [DOI] [PubMed] [Google Scholar]
- 23. Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 1999; 283: 1535–1538. [DOI] [PubMed] [Google Scholar]
- 24. Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, et al. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 2006; 152: 2287–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans . Gene 2004; 341: 119–127. [DOI] [PubMed] [Google Scholar]
- 26. Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans . Eukaryot Cell 2005; 4: 1328–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell 2011; 10: 207–225. 10.1128/EC.00158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Russell CL, Brown AJ. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet Biol. 2005; 42: 676–683. [DOI] [PubMed] [Google Scholar]
- 29. Nailis H, Coenye T, Van Nieuwerburgh F, Deforce D, Nelis HJ. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol Biol. 2006; 7: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu PC, Chao CC, Yang CY, Ye YL, Liu FC, Chuang YJ, et al. Diverse Hap43-independent functions of the Candida albicans CCAAT-binding complex. Eukaryot Cell 2013; 12: 804–815. 10.1128/EC.00014-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. Characteristics of biofilm formation by Candida albicans . Rev Iberoam Micol. 2001; 18: 163–170. [PubMed] [Google Scholar]
- 32. Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett R J. Genetic Control of Conventional and Pheromone-Stimulated Biofilm Formation in Candida albicans . PLoS Pathog. 2013; 9: e1003305 10.1371/journal.ppat.1003305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A 2004; 101: 14315–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science 2002; 297: 395–400. [DOI] [PubMed] [Google Scholar]
- 35. Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004; 18: 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell 2009; 33: 704–716. 10.1016/j.molcel.2009.01.034 [DOI] [PubMed] [Google Scholar]
- 37. Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell 2003; 2: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kucharikova S, Tournu H, Lagrou K, Van Dijck P, Bujdakova H. Detailed comparison of Candida albicans and Candida glabrata biofilms under different conditions and their susceptibility to caspofungin and anidulafungin. J Med Microbiol. 2011; 60: 1261–1269. 10.1099/jmm.0.032037-0 [DOI] [PubMed] [Google Scholar]
- 39. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008; 18: 1017–1024. 10.1016/j.cub.2008.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans . PLoS Pathog. 2009; 5: e1000294 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsao CC, Chen YT, Lan CY. A small G protein Rhb1 and a GTPase-activating protein Tsc2 involved in nitrogen starvation-induced morphogenesis and cell wall integrity of Candida albicans . Fungal Genet Biol. 2009; 46: 126–136. 10.1016/j.fgb.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 42. Chen YT, Lin CY, Tsai PW, Yang CY, Hsieh WP, Lan CY. Rhb1 regulates the expression of secreted aspartic protease 2 through the TOR signaling pathway in Candida albicans . Eukaryot Cell 2012; 11: 168–182. 10.1128/EC.05200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Argimon S, Wishart JA, Leng R, Macaskill S, Mavor A, Alexandris T, et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans . Eukaryot Cell 2007; 6: 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans . FEMS Microbiol Lett. 2002; 214: 95–100. [DOI] [PubMed] [Google Scholar]
- 45. Stichternoth C, Ernst JF. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans . Appl Environ Microbiol. 2009; 75: 3663–3672. 10.1128/AEM.00098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006; 2: e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tumbarello M, Posteraro B, Trecarichi EM, Fiori B, Rossi M, Porta R, et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol. 2007; 45: 1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR. Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell 2013; 12: 1389–1402. 10.1128/EC.00127-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999; 48: 671–679. [DOI] [PubMed] [Google Scholar]
- 50. Green CB, Zhao X, Hoyer LL. Use of green fluorescent protein and reverse transcription-PCR to monitor Candida albicans agglutinin-like sequence gene expression in a murine model of disseminated candidiasis. Infect Immun. 2005; 73: 1852–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 2006; 5: 1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Staab JF, Ferrer CA, Sundstrom P. Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans . J Biol Chem. 1996; 271: 6298–6305. [DOI] [PubMed] [Google Scholar]
- 53. Fan Y, He H, Dong Y, Pan H. Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans . Mycopathologia 2013; 176: 329–335. 10.1007/s11046-013-9684-6 [DOI] [PubMed] [Google Scholar]
- 54. Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, et al. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol. 2002; 44: 61–72. [DOI] [PubMed] [Google Scholar]
- 55. Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997; 16: 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.