Abstract
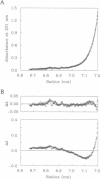
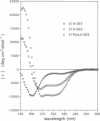
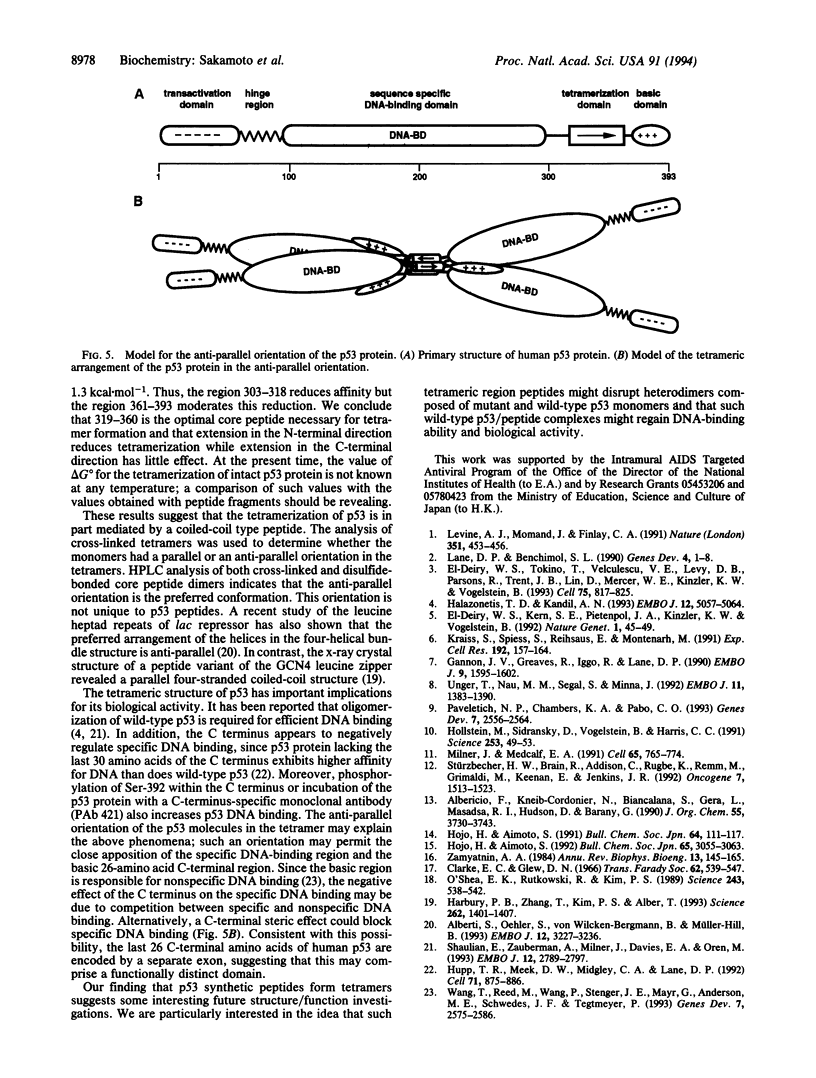
Human p53 is a tumor-suppressor gene product associated with control of the cell cycle and with growth suppression, and it is known to form homotetramers in solution. To investigate the relationship of structure to tetramerization, nine peptides corresponding to carboxyl-terminal sequences in human p53 were chemically synthesized, and their equilibrium associative properties were determined by analytical ultracentrifugation. Secondary structure, as determined by circular dichroism measurements, was correlated with oligomerization properties of each peptide. The sedimentation profiles of peptides 319-393 and 319-360 fit a two-state model of peptide monomers in equilibrium with peptide tetramers. Successive deletion of amino- and carboxyl-terminal residues from 319-360 reduced tetramer formation. Further, substitution of alanine for Leu-323, Tyr-327, and Leu-330 abolished tetramerization. Circular dichroism studies showed that peptide 319-351 had the highest alpha-helix content, while the other peptides that did not form tetramers had low helical structure. These studies define a minimal region and identify certain critical residues involved in tetramerization. Cross-linking studies between monomer units in the tetramer suggest that the helices adopt an anti-parallel arrangement. We propose that conformational shifts in the helical structure of the p53 tetramerization domain result in a repositioning of subunits relative to one another. This repositioning provides an explanation relating conformational changes at the carboxyl terminus with changes in sequence-specific DNA binding by the highly conserved central domain.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberti S., Oehler S., von Wilcken-Bergmann B., Müller-Hill B. Genetic analysis of the leucine heptad repeats of Lac repressor: evidence for a 4-helical bundle. EMBO J. 1993 Aug;12(8):3227–3236. doi: 10.1002/j.1460-2075.1993.tb05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon J. V., Greaves R., Iggo R., Lane D. P. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 1990 May;9(5):1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993 Dec 15;12(13):5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993 Nov 26;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. Regulation of the specific DNA binding function of p53. Cell. 1992 Nov 27;71(5):875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Kraiss S., Spiess S., Reihsaus E., Montenarh M. Correlation of metabolic stability and altered quaternary structure of oncoprotein p53 with cell transformation. Exp Cell Res. 1991 Jan;192(1):157–164. doi: 10.1016/0014-4827(91)90170-y. [DOI] [PubMed] [Google Scholar]
- Lane D. P., Benchimol S. p53: oncogene or anti-oncogene? Genes Dev. 1990 Jan;4(1):1–8. doi: 10.1101/gad.4.1.1. [DOI] [PubMed] [Google Scholar]
- Levine A. J., Momand J., Finlay C. A. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Milner J., Medcalf E. A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991 May 31;65(5):765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Chambers K. A., Pabo C. O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993 Dec;7(12B):2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- Shaulian E., Zauberman A., Milner J., Davies E. A., Oren M. Tight DNA binding and oligomerization are dispensable for the ability of p53 to transactivate target genes and suppress transformation. EMBO J. 1993 Jul;12(7):2789–2797. doi: 10.1002/j.1460-2075.1993.tb05940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stürzbecher H. W., Brain R., Addison C., Rudge K., Remm M., Grimaldi M., Keenan E., Jenkins J. R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992 Aug;7(8):1513–1523. [PubMed] [Google Scholar]
- Unger T., Nau M. M., Segal S., Minna J. D. p53: a transdominant regulator of transcription whose function is ablated by mutations occurring in human cancer. EMBO J. 1992 Apr;11(4):1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Reed M., Wang P., Stenger J. E., Mayr G., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993 Dec;7(12B):2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- Zamyatnin A. A. Amino acid, peptide, and protein volume in solution. Annu Rev Biophys Bioeng. 1984;13:145–165. doi: 10.1146/annurev.bb.13.060184.001045. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992 Apr;1(1):45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993 Nov 19;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]