Abstract
At present, approximately 187 genera and over 1300 species of Microsporidia have been described, among which almost half infect aquatic species and approximately 50 genera potentially infect aquatic arthropods. Lake Baikal is the deepest and one of the oldest lakes in the world, and it has a rich endemic fauna with a predominance of arthropods. Among the arthropods living in this lake, amphipods (Crustacea) are the most dominant group and are represented by more than 350 endemic species. Baikalian amphipods inhabit almost all depths and all types of substrates. The age and geographical isolation of this group creates excellent opportunities for studying the diversity, evolution and genetics of host-parasite relationships. However, despite more than 150 years of study, data investigating the microsporidia of Lake Baikal remain incomplete. In this study, we used molecular genetic analyses to detect microsporidia in the hemolymph of several endemic species of amphipods from Lake Baikal. We provide the first evidence that microsporidian species belonging to three genera (Microsporidium, Dictyocoela and Nosema) are present in the hemolymph of Baikalian endemic amphipods. In the hemolymph of Eulimnogammarus verrucosus, we detected SSU rDNA of microsporidia belonging to the genus Nozema. In the hemolymph of Pallasea cancellous, we found the DNA of Microsporidium sp. similar to that in other Baikalian endemic amphipods; Dictyocoela sp. was found in the hemolymph of Eulimnogammarus marituji and Acanthogammarus lappaceus longispinus.
Introduction
Microsporidia are ancient eukaryotic intracellular parasites. Their life cycles include multiple stages, they can change hosts and they affect all eukaryotic organisms [1–3]. Microsporidia are also found in the form of spores with various diameters outside of the cell [4]. At present, approximately 187 genera and over 1300 species of Microsporidia have been described, among which almost half infect aquatic species and approximately 50 genera potentially infect aquatic arthropods [3, 5].
The greatest number of microsporidian species found in arthropods mostly infect insects, and only a few of these parasites have been detected in well-studied crustacean species [5]. Their widespread detection throughout the animal kingdom indicates the great evolutionary age of this group of parasites [1, 6].
Lake Baikal is the deepest and one of the oldest lakes in the world, and it has a rich endemic fauna with a predominance of arthropods [7]. Among the arthropods living in the lake, amphipods (Crustacea) are the most dominant group and are represented by more than 350 endemic species. Baikalian amphipods inhabit almost all depths and all types of substrates [8]. The age and geographical isolation of this group creates excellent opportunities for studying the diversity, evolution and genetics of host-parasite relationships. However, despite more than 150 years of study, data investigating the microsporidia of Lake Baikal remain incomplete. To date, only three studies have described species of microsporidia in this ecosystem [9]. The first discovery of Microsporidia in amphipods occurred in 1967: Nosema kozhovi in the Baikalian amphipod Brandtia lata lata [10]. The first molecular genetic studies of microsporidia were performed in the 21st century, with the first results obtained from a 2008 study of the diversity of microsporidia parasitizing the Baikalian amphipod Gmelinoides fasciatus. Six endemic microsporidia and one cosmopolitan species, Dictyocoela duebenum, were detected in this amphipod by SSU rDNA sequencing [11]. J.E. Smith and colleagues reported preliminary data regarding the discovery of 100 new species of microsporidia in 31 species of amphipods from Lake Baikal [12]. However, whether all of the species of Baikal amphipods were infected by microsporidia and the ratio of endemic to non-endemic species of microsporidia remain open questions.
It should be noted that in previous studies the microsporidian DNA was isolated from the entire body of Baikalian crustaceans [11]. Thus, data should be presented for both ecto- and endoparasite microsporidian species as well as contamination by microsporidian spores, which were accumulated in the digestion system. Microsporidia are found in the hemolymph of aquatic and terrestrial invertebrates in the form of spores and in hemocytes [13–16]. To avoid contamination (from digestive remains) and exosymbiotic microsporidia in the analyzed samples, the identification of species can be performed using the hemolymph. In the present study, we used molecular genetic techniques to detect microsporidia in the hemolymph of several endemic species of amphipods from Lake Baikal.
Materials and Methods
Sampling and location
Animals were collected from Lake Baikal close to the Bolshie Koty settlement (Irkutsk region, Eastern Siberia, Russia). No specific permissions were required for the samplings, locations or activities. The investigated amphipod species did not involve endangered or protected species.
Collections of littoral species of amphipods (Pallasea cancellus (Pallas, 1772), Eulimnogammarus verrucosus (Gerstf., 1858), E. marituji Baz., 1945) were performed on the southern shore of Lake Baikal (the village of Listvyanka: 51°50'58.67"N; 104°52'0.14"E and Bolshie Koty: 51°54'9.51"N; 105° 4'10.64"E) at a depth of 1 m using hand-nets. Deep-water species (Acanthogammarus lappaceus longispinus Tacht., 2000) were collected during a hydrobiological expedition in June 2014 (Ushkany Islands: N 53° 51/ 12//; E 108° 35/ 49//). No specific permissions were required for these locations and activities. Information regarding the collection points and the number of analyzed species is presented in Table 1 and Fig 1.
Table 1. The SSU rDNA sequences of microsporidia used for phylogenetic analysis.
| Species | GenBank | Type of host | Location | References |
|---|---|---|---|---|
| Microsporidium sp. PCN4 | KM977842 | Pallasea cancellus | Listvyanka (Lake Baikal) | this paper |
| Microsporidium sp. PCN7a | KM977843 | Pallasea cancellus | Listvyanka (Lake Baikal) | this paper |
| Microsporidium sp. PCN11 | KM977844 | Pallasea cancellus | Listvyanka (Lake Baikal) | this paper |
| Microsporidium sp. PCN12 | KM977845 | Pallasea cancellus | Listvyanka (Lake Baikal) | this paper |
| Microsporidium sp. PCN16 | KM977846 | Pallasea cancellus | Listvyanka (Lake Baikal) | this paper |
| Dictyocoela sp. All. 5 | KM977839 | Acanthogammarus lappaceus longispinus | Ushkany Islands (Lake Baikal) | this paper |
| Dictyocoela sp. BK17 | KP027301 | Eulimnogammarus marituji | Bolshie Koty | this paper |
| Nosema sp. Vr.28 | KM977840 | Eulimnogammarus verrucosus | Listvyanka (Lake Baikal) | this paper |
| Nosema sp. VR31 | KM977841 | Eulimnogammarus verrucosus | Listvyanka (Lake Baikal) | this paper |
| Dictyocoela sp. BLAC VER | FJ756216 | Eulimnogammarus verrucosus* | Lake Baikal | only GenBank |
| Dictyocoela sp. BLAP LAP1 | FJ756199 | Acanthogammarus lappaceus* | Lake Baikal | only GenBank |
| Dictyocoela sp. BLAP PAR8 | FJ756209 | Dorogostaiskia parasitica* | Lake Baikal | only GenBank |
| Microsporidium sp. BALB1 LAT3 | FJ755962 | Brandtia latissima latior* | Lake Baikal | only GenBank |
| Microsporidium sp. BALB1 CAB | FJ755959 | Garjajewia cabanisii* | Lake Baikal | only GenBank |
| Microsporidium sp. BPAR12 PAR1 | FJ756112 | Dorogostaiskia parasitica* | Lake Baikal | only GenBank |
| Microsporidium sp. BKES3 | FJ756022 | Pallaseopsis kessleri* | Lake Baikal | only GenBank |
| Microsporidium sp. BALB1 PLA2 | FJ755965 | Micruropus platycercus* | Lake Baikal | only GenBank |
| Microsporidium sp. BVIC2 VIC | FJ756173 | Acanthogammarus victorii* | Lake Baikal | only GenBank |
| Dictyocoela duebenum isolate 775 | FN434091 | Gammarus duebeni duebeni | Iceland: Atlantic Ocean | [28] |
| Microsporidium sp. JES2002G isolate chevn1 | AJ438962 | Gammarus chevreuxi | UK, River Avon | [24] |
| Nosema furnacalis | U26532 | Ostrinia furnacalis | Asia (?) | [29] |
| Nosema antheraeae | DQ073396 | Antheraea pernyi | China | [30] |
| Nosema heliothidis | FJ772435 | Helicoverpa armigera | China | only GenBank |
| Nosema trichoplusiae | U09282 | Trichoplusia ni | USA | [29] |
‘*’—the species of microsporidia founds in endemic amphipod of Lake Baikal (GenBank data).
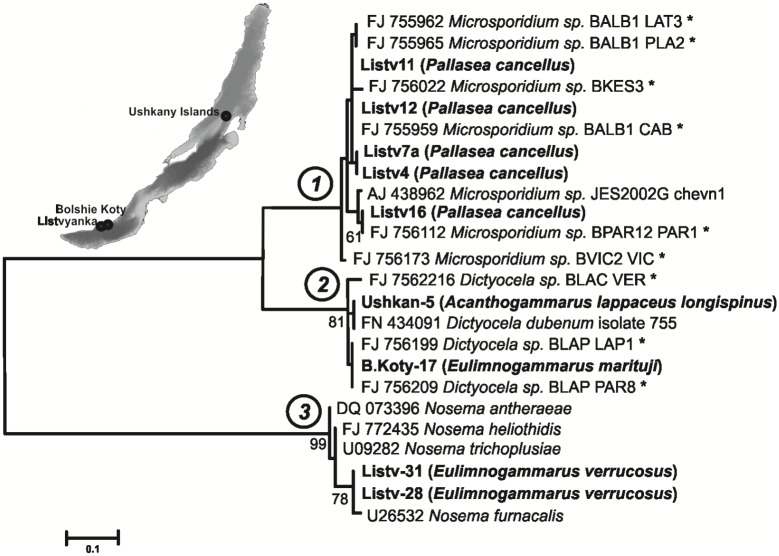
Fig 1. Maximum likelihood phylogenetic tree of Microsporidia.
Bootstrap values (> 50%) are presented for nodes and branch lengths are drawn to scale. The species of microsporidia noted with ‘*’ found in Baikalian endemic amphipods (GenBank data).
The identification of species and the hemolymph extraction were performed under laboratory conditions. Each sample (volumes ranging from 50–100 μl) contained a pool of hemolymph collected from several individuals of the same species. The number of individuals per sample depended on the size of the amphipods (2–5 individuals). Isolated samples were stored in liquid nitrogen.
DNA isolation and PCR
Total DNA isolation from hemolymph was performed using the "Riboprep” reagent kit (AmpliSens, Russia). The concentration and purity of the isolated DNA was determined using the UV spectrophotometer UNICO 2802 (UNICO, USA).
The small subunit of ribosomal DNA (SSU rDNA) from the microsporidia was selected as a molecular phylogenetic marker. Amplification was performed in two stages (nested PCR) with universal primers for microsporidia: V1f: 5'- CACCAGGTTGATTCTGCCTGAC-3' [17]; 1342r: 5'- ACGGGCGGTGTGTACAAAGAACAG-3' [18]; 18sf: 5'- GTTGATTCTGCCTGACGT-3' [19]; and 981r: 5'- TGGTAAGCTGTCCCGCGTTGAGTC-3' [20]. Each PCR was performed in a Gradient Thermocycler (Biometra, Germany) in a volume of 25 μl and contained 10X PCR buffer, 2.5 mM dNTPs, 5 U/ml SynTaq DNA polymerase, 25 mM MgCl2, 10 pmol primers, and deionized water. The conditions for the first round were 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 90 sec, and then 72°C for 7 min. Conditions for the second round of PCR were 95°C for 5 min, followed by 35 cycles of 95°C for 30 sec, 52°C for 30 sec, and 72°C for 1 min, followed by 72°C for 7 min.
The PCR products of the second round were visualized in 1% agarose gels, and products of the expected size were excised from the gel and purified with ethanol and sodium acetate [21].
Sequencing of the amplified DNA fragments was performed using a Genetic Analyzer 3500 xL (Applied Biosystems) with the BigDye Terminator Cycle Sequencing kit v.3.1.
Nucleotide sequence analysis
The sequences were aligned using the multisequence alignment program ClustalW within the BioEdit 7.0.5.3 environment [22]. The phylogenetic relationships between microsporidia representatives were determined using Mega 5.0 (Maximum Likelihood, HKY+G+I model). Bootstrap values were obtained for a consensus tree based on 1000 randomly generated trees using the same package [23].
Results
Nine nucleotide sequences of SSU rDNA (738 bp) from three species of microsporidia were obtained from the hemolymph of all of the endemic amphipod species: P. cancellus, E. verrucosus, E. marituji and A. lappaceus longispinus. The dendrogram (Fig 1) was reconstructed using these and additional sequences from GenBank (see Table 1).
Four microsporidian nucleotide sequences were found in the P. cancellus hemolymph. These sequences showed a maximum similarity to the following species in GenBank: FJ755959 Microsporidium sp. BALB1 CAB, FJ756022 Microsporidium sp. BKES3, FJ755962 Microsporidium sp. BALB1 LAT, FJ755965 Microsporidium sp. BALB1 PLA2, FJ756112 Microsporidium sp. BALB1 PAR1, and FJ756173 Microsporidium sp. BVIC2 VIC CAB; these hosts are Baikalian endemic amphipods. The exception is AJ438962 Microsporidium sp. JES2002G chevn1, which was found in Gammarus chevreuxi from the Avon River, UK. All of these species are located in a single cluster in the dendrogram (marked as cluster 1).
The microsporidian nucleotide sequences were detected in the Acanthogammarus lappaceus longispinus and Eulimnogammarus marituji hemolymph samples and clustered with those of microsporidia of the genus Dictyocoela (marked as cluster 2). One species was identified with a close similarity to the following sequences from the Baikalian amphipods: FJ756316 Dictyocoela sp. BLAC VER, FJ756209 Dictyocoela sp. BLAC PAR8 and FJ756199 Dictyocoela sp. BLAC LAP1. The Dictyocoela duebenum isolate 775 (FN434091) was found in Gammarus duebeni duebeni (Iceland).
The two SSU rDNA sequences from the genus Nosema were detected in the hemolymph of E. verrucosus (marked as cluster 3). This is the first description of this species in an endemic crustacean of Lake Baikal.
Thus, the microsporidian DNA sequences that we detected in amphipods from Lake Baikal can be divided into three clusters: Microsporidium, Dictyocoela and Nosema.
Discussion and Conclusion
This is the first study to demonstrate microsporidia in the hemolymph of Baikal endemic amphipods using molecular genetic methods. In all of the previous studies, microsporidian DNA was isolated from the entire body of the Baikalian crustaceans, thus providing data for both ecto- and endoparasite microsporidian species. The results of our study limited the number of microsporidia to endoparasite species only.
The microsporidian DNA detected in the P. cancellus hemolymph is similar to that of microsporidia found in some other endemic amphipods of Lake Baikal: Garjajewia cabanisii (FJ755959), Pallaseopsis kessleri (FJ756022), Brandtia latissima latior (FJ755962), Micruropus platycercus (FJ755965), Dorogostaiskia parasitica (FJ756112) and Acanthogammarus victorii (FJ756173). Therefore, one may assume that the majority of the detected species are likely to be endemic microsporidia. Therefore, one may assume that the majority of the detected species are likely to be endemic microsporidia excepting Gammarus chevreuxi (AJ438962) from the Avon River, UK. The nucleotide sequence of the SSU rDNA of this species is similar to those from several species of microsporidia found in fish [24].
It should be noted that the microsporidia of the genus Dictyocoela were found in two species of amphipods: A. lappaceus longispinus and E. verrucosus (GenBank data). It is known that some species of Dictyocoela stimulate feminization and change the sex ratio in their host populations [25]. There is hypothesis that this genus of microsporidia has a high degree of genetic diversity in populations of Eurasian amphipods and shows horizontal transmission [26].
Interestingly, we also detected some microsporidia that were similar to the genus Nosema. Many species in this genus primarily parasitize insects and belong to the class Terresporidia. According to Yu. S. Tokarev [27], “phylogenetic branching of the freshwater microsporidia among Terresporidia is indicative of multiple shifts of microsporidia between hosts of variable habitats, as well as of the secondary nature of adaptation of these species of parasites to the freshwater hosts.”
To summarize our data, we provide the first evidence that species of microsporidia belonging to three genera (Microsporidium, Dictyocoela and Nosema) reside in the hemolymph of Baikalian endemic amphipods. In the hemolymph of E. verrucosus, we detected the SSU rDNA of microsporidia belonging to the genus Nosema. In the hemolymph of P. cancellus, we found the DNA of Microsporidium sp. similar to that in other Baikalian endemic amphipods. Dictyocoela sp. was found in the hemolymph of E. marituji and A. lappaceus longispinus.
Acknowledgments
This research was supported by Ministry of education and science of RF as a part of "Goszadanie" (№ 6.382.2014/K, 1354(2014)), grant of Russian Science Foundation (№ 14-14-00400), grant of Russian Foundation for Basic Research (№ 14-04-00501, 14-34-50051, 15-04-06685) CRDF (FSCX-15-61168-0), program of strategic development at Irkutsk State University and grants of ISU for young researchers and DAAD. The support is gratefully acknowledged.
Data Availability
All relevant data are within the paper. Sequences ID's are available in Genbank.
Funding Statement
This research was supported by Ministry of education and science of RF as a part of "Goszadanie" (№ 6.382.2014/K, 1354(2014)), grant of Russian Science Foundation (№ 14-14-00400), grant of Russian Foundation for Basic Research (№ 14-04-00501, 14-34-50051, 15-04-06685), CRDF (FSCX-15-61168-0), program of strategic development at Irkutsk State University and grants of ISU for young researchers and DAAD. The support is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dunn AM, Smith JE. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes Infect. 2001;3: 381–388. [DOI] [PubMed] [Google Scholar]
- 2. Issi IV. Microsporidian parasitic systems: descriptions and terminology questions. Parazitologia. 2002;36: 478–492. Russian. [PubMed] [Google Scholar]
- 3. Didier ES, Weissb LM. Microsporidosis: current status. Curr Opin Infect Dis. 2006;19: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56: 93–116. [DOI] [PubMed] [Google Scholar]
- 5. Stentiford GD, Feist SW, Stone DM, Bateman KS, Dunn AM. Microsporidia: diverse, dynamic and emergent pathogens in aquatic systems. Trends Parasitol. 2013;29: 567–578. 10.1016/j.pt.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 6. Smith JE. The ecology and evolution of microsporidian parasites. Parasitology. 2009;136: 1901–1914. 10.1017/S0031182009991818 [DOI] [PubMed] [Google Scholar]
- 7. Kozhova OM, Izmesteva LR. Lake Baikal: Evolution and Biodiversity. The Netherlands: Backhuys Publishers; 1998. [Google Scholar]
- 8. Takhteev VV. Baicalogy. Novosibirsk: Nauka; 2012. Russian. [Google Scholar]
- 9. Timoshkin OA Index of animal species inhabiting Lake Baikal and its catchment area Book 1. Novosibirsk: Nauka; 2001. Russian. [Google Scholar]
- 10. Lipa JJ. Nosema kozhovi sp. n., a new microsporidian parasite of Brandtia lata lata (Crustacea, Gammaridae) of Baical Lake. Acta Protozool. 1967;5: 93–97. [Google Scholar]
- 11. Kuzmenkova ZV, Sherbakov DU, Smith JE. Diversity of microsporidia, that parasitize on Baikal amphipods Gmelinoides fasciatus from different populations. The bulletin of Irkutsk State University; Series”Biology, Ecology”. 2008;1: 56–61. Russian. [Google Scholar]
- 12.Smith JE, Yang Q, Kamaltynov RM, Sherbakov DY. Evolutionary interactions between microsporidia and their hosts: Lessons from an ancient lake. 41st Annual Meeting of the Society for Invertebrate Pathology and 9th International Conference on Bacillus thuringiensis Aug 3–7. University of Warwick, Coventry, United Kingdom. 2008;71. Available: http://www.sipweb.org/docs/2008abstracts.pdf
- 13. Vorontsova YL, Tokarev YS, Sokolova YY, Glupov VV. Microsporidiosis in the wax moth Galleria mellonella (Lepidoptera: Pyralidae) caused by Vairimorpha ephestiae (Microsporidia: Burenellidae). Parazitologia. 2004;38: 239–250. Russian. [PubMed] [Google Scholar]
- 14. Vilcinskas A, Stoecker K, Schmidtberg H, Rohrich CR, Vogel H. Invasive Harlequin ladybird carries biological weapons against native competitors. Science. 2013;340: 862–863. 10.1126/science.1234032 [DOI] [PubMed] [Google Scholar]
- 15. Issi IV, Dolgikh VV, Sokolova YY, Tokarev YS. Factors of the pathogenicity of microsporidia, intracellular parasites of insects. Plant Protection News. 2005;3: 16–25. Russian. [Google Scholar]
- 16. Meyer T, Burton T. Diseases of wild and cultured shellfish in Alaska. Alaska: Department of Fish and Game; 2009. pp. 94–98. [Google Scholar]
- 17. Weiss LM, Zhu X, Cali A, Tanowitz HB, Wittner M. Utility of microsporidian rRNA in diagnosis and phylogeny: a review. Folia Parasitol. 1994;41: 81–90. [PubMed] [Google Scholar]
- 18. McClymont HE, Dunn AM, Terry RS, Rollinson D, Littlewood DT, Smith JE. Molecular data suggest that microsporidian parasites in freshwater snails are diverse. Int J Parasitol. 2005;35: 1071–1078. [DOI] [PubMed] [Google Scholar]
- 19. Baker MD, Vossbrinck CR, Didier ES, Maddox JV, Shadduck JA. Small subunit ribosomal DNA phylogeny of various microsporidia with emphasis on AIDS-related forms. J Eukaryot Microbiol. 1995;42: 564–570. [DOI] [PubMed] [Google Scholar]
- 20. MacNeil C, Dick JTA, Hatcher MJ, Terry RS, Smith JE, Dunn AM. Parasite-mediated predation between native and invasive amphipods. Proc R Soc Lond B Biol Sci 2003;270: 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22. Hall A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41: 95–98. [Google Scholar]
- 23. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 24. Terry RS, Smith JE, Sharpe RG, Rigaud T, Littlewood DTJ, Ironside JE, et al. Widespread vertical transmission and associated host sex-ratio distortion within the eukaryotic phylum Microspora. Proc R Soc Lond B Biol Sci 2004;271: 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gismondia E, Rigaud T, Beisel J-N, Cossu-Leguille C. Microsporidia parasites disrupt the responses to cadmium exposure in a gammarid. Environ Pollut. 2012. January;160(1):17–23. 10.1016/j.envpol.2011.09.021 [DOI] [PubMed] [Google Scholar]
- 26. Wilkinson TJ, Rock J, Whiteley NM, Ovcharenko MO, Ironside JE. Genetic diversity of the feminising microsporidian parasite Dictyocoela: New insights into host-specificity, sex and phylogeography. Int J Parasitol. 2011;41: 959–966. 10.1016/j.ijpara.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 27. Tokarev YS. Molecular phylogeny of entomopathogenic microsporidia, a review of the last five years of study. Evraziat Entomol Z. 2010;9: 571–576. Russian. [Google Scholar]
- 28. Krebes L., Blank M., Frankowski J., Bastrop R. Molecular characterisation of the Microsporidia of the amphipod Gammarus duebeni across its natural range revealed hidden diversity, wide-ranging prevalence and potential for co-evolution. Infect Genet Evol. 2010;10: 1027–1038. 10.1016/j.meegid.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 29. Fries IM, Feng F, da Silva AJ, Slemenda SB, Pieniazek NJ. Nosema ceranae n. sp. (Microsporidia, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996;32: 356–365. [Google Scholar]
- 30. Wang LL, Chen KP, Zhang Z, Yao Q, Gao GT, Zhao Y. Phylogenetic analysis of Nosema antheraeae (Microsporidia) isolated from Chinese oak silkworm, Antheraea pernyi . J. Eukaryot. Microbiol. 2006;53: 310–313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper. Sequences ID's are available in Genbank.