Abstract
Background
Psoriatic arthritis (PsA) is an inflammatory arthritis whose pathogenesis is poorly understood; it is characterized by bone erosions and new bone formation. The diagnosis of PsA is mainly clinical and diagnostic biomarkers are not yet available. The aim of this work was to clarify some aspects of the disease pathogenesis and to identify specific gene signatures in paired peripheral blood cells (PBC) and synovial biopsies of patients with PsA. Moreover, we tried to identify biomarkers that can be used in clinical practice.
Methods
PBC and synovial biopsies of 10 patients with PsA were used to study gene expression using Affymetrix arrays. The expression values were validated by Q-PCR, FACS analysis and by the detection of soluble mediators.
Results
Synovial biopsies of patients showed a modulation of approximately 200 genes when compared to the biopsies of healthy donors. Among the differentially expressed genes we observed the upregulation of Th17 related genes and of type I interferon (IFN) inducible genes. FACS analysis confirmed the Th17 polarization. Moreover, the synovial trascriptome shows gene clusters (bone remodeling, angiogenesis and inflammation) involved in the pathogenesis of PsA. Interestingly 90 genes are modulated in both compartments (PBC and synovium) suggesting that signature pathways in PBC mirror those of the inflamed synovium. Finally the osteoactivin gene was upregulared in both PBC and synovial biopsies and this finding was confirmed by the detection of high levels of osteoactivin in PsA sera but not in other inflammatory arthritides.
Conclusions
We describe the first analysis of the trancriptome in paired synovial tissue and PBC of patients with PsA. This study strengthens the hypothesis that PsA is of autoimmune origin since the coactivity of IFN and Th17 pathways is typical of autoimmunity. Finally these findings have allowed the identification of a possible disease biomarker, osteoactivin, easily detectable in PsA serum.
Introduction
Psoriatic arthritis (PsA) is primarily characterised by enthesitis and by synovitis, leading to bone erosions and new bone formation [1]; 10% to 30% of patients with skin psoriasis are affected by the disease, with an estimated prevalence of 1%.
Genetic studies indicate that PsA has a heritable component [2] and many genes have been implicated in psoriasis and PsA [3]. However only a few genes have been associated to both psoriasis and PsA [4].
PsA is characterized by different clinical phenotypes: oligoarticular or polyarticular asymmetrical peripheral joint inflammation or axial involvement. In the last few years several criteria have been used for the classification of PsA. The most frequently classification criteria used are those proposed by Moll and Wright [5] and more recently, are the classification criteria for PsA (CASPAR) [6]. The diagnosis of PsA is mainly performed on a clinical basis and after the exclusion of other seronegative arthritides and up to now there are no diagnostic tests available. Diagnostic work up is based on medical history, physical examination, blood tests, and imaging of the joints. Plain radiographs are used to evaluate the joint damage. Magnetic resonance imaging (MRI) is able to detect joint damage earlier and to assess the extent of joint involvement more accurately than plain radiographs. Indeed MRI is able both to quantify the extent of the inflammatory process within the affected joints and to detect enthesitis even in apparently unaffected joints and in the absence of clinical symptoms. Enthesitis is considered the primary event in the pathogenesis of the disease [7]. Moreover MRI and scintigraphy can be used for an early detection of sacroiliitis and axial disease. In addition these imaging techniques are widely used to evaluate the efficacy of novel therapies for PsA [8,9].
In psoriatic skin lesions the typical cell infiltrate contains activated keratinocytes, T and B lymphocytes, macrophages and neutrophils. Both CD4 and CD8 T cells have been associated with skin and joint damage [10,11] typical of PsA.
The synovial tissue in PsA is characterized by an abundant T cell infiltrate, marked angiogenesis, and synovial hyperplasia with increased expression and/or secretion of cytokines and proteases that contribute to amplify the local inflammation and may explain the “erosive behavior” of the synovium leading to joint destruction.
The cytokine tumor necrosis factor-alpha (TNF-alpha) is a very important inflammatory mediator and has been implicated in PsA pathogenesis. TNF-alpha inhibitors are widely used in PsA therapy and are usually quite effective in reducing the extent of skin lesions and of musculoskeletal symptoms, however a high percentage of PsA patients does not respond to TNFalpha antagonists. Considering the expense and side effects of anti-TNF biological agents, the identification of biomarkers that could be used to predict which patients will respond to biological treatment is an important goal in medicine.
Therapies that target the TNFα induce a significant clinical improvement in approximately 70% of patients [12]. However, the extent of clinical improvement is often far from complete remission and the majority of PsA patients experience a flare of the disease within the first 2 years [12]. Therefore, the identification of new molecules that play a pivotal role in the pathogenesis of the disease is fundamental for the development of new therapies. Moreover prognostic and diagnostic biomarkers need to be identified. To this aim, gene expression profiling may be of great help since it has been used to classify lymphoid malignancies [13] and to dissect pathways involved in several autoimmune and inflammatory diseases. In this regard gene array studies have generated useful insight in several diseases including systemic lupus erythematosus (SLE) [14], rheumatoid arthritis (RA) [15], and multiple sclerosis (MS) [16]
Gene expression studies in PBMC of subjects with rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) have allowed the identification of specific gene 'signatures' that are differentially expressed in patients when compared to healthy subjects [14,15]. In a few cases differentially expressed genes (DEGs) have been shown to correlate with particular features of the disease (such as presence or absence of systemic organ involvement in RA), or with capacity to respond to immunosuppressive treatment.
In this study we used a gene array strategy to identify transcriptional profiles that distinguish PsA patients from healthy control subjects. For this purpose we analyzed for the first time gene expression in paired peripheral blood cells (PBC) and synovial biopsies of 10 PsA patients. We reasoned that the combined analysis of transcription data obtained in PBC and in the target tissue of PsA, the synovial membrane, could provide a deeper understanding of the genetic pathways that regulate the disease.
Using this approach we have been able to shed light on some aspects of disease pathogenesis by dissecting different aspects of this complex pathology. Interestingly DEGs recapitulate most of the typical features of PsA, such as bone remodeling with new bone formation, synovial hyperplastic growth with neoangiogenesis, local and systemic inflammatory response, activation of innate and adaptive immune reponses.
Moreover our results provide evidence for an autoimmune origin of PsA, since gene expression profiles are characterized by the combined overexpression of type I IFN inducible genes [17] and of Th17 related transcripts. Indeed coactivity of IFN and Th17 pathways is typical of autoimmunity and has been described both in human diseases and in animal models [18–20].
Moreover, using the same strategy, we could identify a novel biomarker, osteoactivin, that can be easily detected in PsA sera. Indeed increased serum levels of osteoactivin, seem to be typical of PsA and, in the absence of other disease markers, the detection of high levels of this molecule may represent an interesting tool in the diagnostic work up of PsA.
Patients and Methods
Patients
We studied a cohort of 60 patients (38 males and 22 females, mean age: 44 years) affected by PsA, attending the Unit of Autoimmune Diseases, at the University Hospital of Verona, Italy. The patients enrolled in the study were selected as already described elsewhere [21]. All patients fulfilled the CASPAR criteria for the diagnosis of PsA: inflammatory musculoskeletal involvement (inflammatory arthritis, enthesitis or lumbar pain) combined with at least 3 features: 1) evidence of current psoriasis, personal history of psoriasis, family history of psoriasis in unaffected patiens; 2) affected nails (onycholysis, pitting); 3) dactylitis; 4) negative rheumatoid factor; 5) radiographic evidence of new juxta-articular bone formation (excluding osteophytes) [22].
All the patients underwent clinical examination and laboratory evaluation comprehensive of inflammatory markers, such as CRP and erythrocytes sedimentation rate (ESR); rheumatoid factor (RF) and anti-CCP antibody detected by ELISA test; antinuclear antibody detected by indirect immunofluorescence on HEp-2 cells; and genetic screening for the association with the allele HLA-B27. All patients underwent the following instrumental investigations: ultrasonography with Power Doppler to investigate subclinical enthesopaty and synovitis in asymptomatic patients; conventional radiography, magnetic resonance imaging (MRI) and scintigraphy. The radiological features of peripheral PsA included asymmetric distribution, participation of distal interphalangeal joints, periostitis, bone density preservation, bone ankylosis and pencil-in-cup deformity.
A group of 10 subjects with recent onset PsA (within one year from the diagnosis) was selected within the entire cohort of PsA patients and utilized for the gene array study. These patients were not treated with anti-TNF agents or with disease-modifying antirheumatic drugs (DMARDs). The clinical features of the patients are reported in Table 1 that also includes a description of the PsA patients selected for the gene array study. Sixty control subjects matched for sex and age served as control group. Moreover 60 patients with rheumatoid arthritis (RA), 60 patients with Ankylosing spondylitis (AS) and 20 patients with osteoarthritis (OA) were used as controls. RA patients had the American College of Rheumatology classification criteria for RA [23]. In all RA patients the disease was at an active stage and blood was obtained before therapy with biological agents. AS patients met the SAS classification criteria for AS [24].
Table 1. Clinical features of the patients with PsA included in the study.
| Patients | 60 | |
| Sex | male | 38 |
| female | 22 | |
| Age at diagnosis (years) | 44±8 | |
| Involvement | axial | 25 |
| peripheral | 35 | |
| Enthesitis | 29 | |
| Dactylitis | 23 | |
| Psoriasis | 47 | |
| Association with HLA-B27 | 18 | |
| Patients utilized for the gene array study | 10 | |
| Sex | male | 6 |
| female | 4 | |
| Age at diagnosis (years) | 43±6 | |
| Involvement | axial | 4 |
| peripheral | 6 | |
| Enthesitis | 7 | |
| Dactylitis | 4 | |
| Psoriasis | 7 | |
| Association with HLA-B27 | 3 | |
All the participants to the study signed a written informed consent. The local Ethical Committee of the Azienda Ospedaliera Universitaria of Verona, Verona, Italy had approved the study protocol. All the investigations have been performed according to the principles contained in the Helsinki declaration.
Synovial biopsies
In the 10 PsA patients selected for the gene array study synovial membranes were obtained with needle biopsies.
In 10 control healthy donors synovial biopsies were collected during orthopedic knee arthroscopy following traumatic events.
Gene array
Tissue samples from every single patient were frozen in liquid nitrogen immediately after dissection and stored at -70°C until homogenization. Sample preparation for gene array analysis was carried out as previously described [25–27]. Briefly, frozen samples were homogenized in TRI REAGENT (1mL per 50–100mg of tissue) in a Potter-type mechanical homogenizer with Teflon pestle.
PAXgene Blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland) were used for blood collection and total RNA was extracted according to the protocol supplied by the manufacturer.
Preparation of cRNA hybridization and scanning of arrays for each samples were performed following the manufacturer instructions (Affymetrix, Santa Clara, CA, USA) by Cogentech Affymetrix microarray unit (Campus IFOM IEO, Milan, Italy) using the Human Genome U133A 2.0 Gene Chip (Affymetrix). The Human Genome U133A Gene Chip is a single array which contains 14,500 well-characterized human genes and more than 22,000 probe sets and 500,000 distinct oligonucleotide features. The gene expression profiles were analyzed using the Gene Spring software, version 12.1 (Agilent Technologies, SantaClara, CA, USA) that calculated a robust multi-array average of background-adjusted, normalized, and log-transformed intensity values applying the Robust Multi-Array Average algorithm (RMA).
With this software the mean optical background level for each array was subtracted from the signal intensity for each probe. The normalized data were transformed to the log2 scale. A signal log2 ratio of 1.0 corresponds to an increase of the transcript level by two-fold change (2 FC) and -1.0 indicates a decrease by two-fold change (-2 FC). A signal log2 ratio of zero would indicate no change. The unpaired t-test was performed to determine which genes were modulated at a significant level (p≤0.01) and p values were corrected for multiple testing by using Bonferroni correction. Finally, statistically significant genes were chosen for final consideration when their expression was at least 2.0 fold different in the test sample versus control sample. Genes that passed both the p-value and the FC restriction were submitted to functional classification analysis according to the Gene Ontology (GO) annotations.
The microarray results have been reported according to the MIAME guidelines and deposited in the public repository ArrayExpress http://www.ebi.ac.uk/arrayexpress; accession number E-MTAB-3201.
FACS analysis
Peripheral blood mononuclear cells (PBMCs) were obtained from 20 healthy donors and 30 patients affected by PsA following a density-gradient centrifugation on Lymphoprep (Nycomed Pharma, Oslo, NO) and two washes with PBS.
Cells collected from patients and normal controls were cultured in 2 mL tubes containing 1 mL of RPMI 1640 + FCS 10% (Lonza, Basel, CH) at a concentration of 1*106 cells/mL. Cells were stimulated for 4 hours with Dynabeads Human T-Activator CD3/CD28 (Life Technologies, Carlsbad, CA, USA) or with heat-inactivated Candida albicans. IL-17 production was assessed using the IL-17 Secretion Assay (Miltenyi Biotec, Bergish Gladbach, D) according to the manufacturer’s instruction. Briefly, cells were washed in 2 mL of cold buffer, centrifuged at 300xg for 5 minutes at 4°C and the pellet resuspended in 90 μL of cold medium. An incubation with IL-17 Catch Reagent was carried out for 5 minutes in ice and cells were washed as before. PBMCs were incubated in 1 mL of warm medium at 37°C for 45 minutes under slow continuous rotation. Cells were then washed and resuspended in in 75 μL of cold buffer and 10 μL of IL-17 Detection Antibody APC, 10 μL of anti-CD3 PerCP (Becton Dickinson, Franklin Lakes, NJ, USA) and 5 μL of anti-CD4 APC-H7 (Becton Dickinson) antibodies were added. Incubation was performed in ice for 10 minutes. Finally cells were washed and resuspended in an appropriate volume of PBS and acquired on a FACSCanto II cytometer (Becton Dickinson). Analysis was performed with FlowJo 9.3.3 software (Tree Star, Ashland, OR, USA).
Real Time RT-PCR
Total RNA was isolated from PBMC using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), following manufacturer’s instructions. First-strand cDNA was generated using the SuperScript III First-Strand Synthesis System for RT-PCR Kit (Invitrogen), with random hexamers, according to the manufacturer’s protocol. PCR was performed in a total volume of 25 μl containing 1× Taqman Universal PCR Master mix, no AmpErase UNG and 2.5 μl of cDNA; pre-designed, Gene-specific primers and probe sets for each gene (SPP1: Hs 00959010_m1; CXCL13: Hs 00757930_m1; CCL18 Hs 00268113_m1; LAMP3: Hs 00180880_m1; CADM1: Hs 00942509_m1) were obtained from Assay-on-Demande Gene Expression Products (Applied Biosystems).
As described in details previously [25–27], Real Time PCR reactions were carried out in a two-tube system and in singleplex. The Real Time amplifications included 10 minutes at 95°C (AmpliTaq Gold activation), followed by 40 cycles at 95°C for 15 seconds and at 60°C for one minute. Thermocycling and signal detection were performed with 7500 Sequence Detector (Applied Biosystems). Signals were detected according to the manufacturer’s instructions. This technique allows the identification of the cycling point where PCR product is detectable by means of fluorescence emission (Threshold cycle or Ct value). As previously reported, the Ct value correlates to the quantity of target mRNA [26]. Relative expression levels were calculated for each sample after normalization against the housekeeping genes GAPDH, beta-actin and 18s ribosomal RNA (rRNA), using the ΔΔCt method for comparing relative fold expression differences [25,28]. The data are expressed as fold change. Ct values for each reaction were determined using TaqMan SDS analysis software. For each amount of RNA tested triplicate Ct values were averaged. Because Ct values vary linearly with the logarithm of the amount of RNA, this average represents a geometric mean.
Detection of soluble mediators in sera and synovial fluids
Serum levels of osteoactivin/GPNMB [29] were detected using a commercially available ELISA kit (Biorbyt Ltd, Cambridge, UK). The ELISA kits for osteopontin, CCL20, CCL18, MMP-3 and IL-23 were all purchased from R & D Systems (Phoenix, AZ, USA) and used according to the manufacturer’s instructions. IL-17 and IL-23 were detected in synovial fluids (SFs) of 20 patients with PsA and in 20 patients with OA, used as controls. The ELISA kit for IL-17 was purchased from eBioscience (San Diego, CA, USA) and used according to the manufacturer’s instruction.
SF was distributed into heparinized tubes, and the cells removed by centrifugation (400 × g, 10 min). Before each test. synovial fluids were treated with hyaluronidase at a concentration of 10 U/ml for 80 min at 37°C followed by centrifugation (12,000 × g, 5 min). This treatment was used to reduce the viscosity of synovial fluid.
Statistical Analysis
Statistical testing was performed using SPSS Statistics 2 software (IBM,United States). Data obtained from the analysis of the soluble mediators were submitted to statistical testing using the non parametric Mann-Whitney test. For the analysis of IL-17–positive CD4+T cells in PBMCs and of IL-17 and IL-23 synovial fluid levels, the significance of the differences between patients and controls was determined using the unpaired Student’s t-test.
Results
Gene-array analysis
a) DEGs in synovial biopsies
In order to identify genes potentially involved in the pathogenesis of PsA, we first compared the gene expression profiles of 10 synovial biopsies obtained from patients with PsA with synovial specimens obtained from normal healthy subjects undergoing post-traumatic surgery.
When both a Bonferroni–corrected P-value criterion (p≤0.01) and a fold change criterion (FC≥2) were applied to the signal variation of every single gene to detect robust and statistically significant changes between baseline and experimental arrays [25,27], we obtained 196 modulated genes that were further analyzed. 135 and 61 transcripts resulted to be up- and downregulated respectively; in many cases the fold changes were very high. In particular three genes showed a very high level of induction: osteopontin, SPP1/OPN (FC 450), fibronectin 1, FN1 (FC 405) and osteoactivin, GPNMB (FC 147).
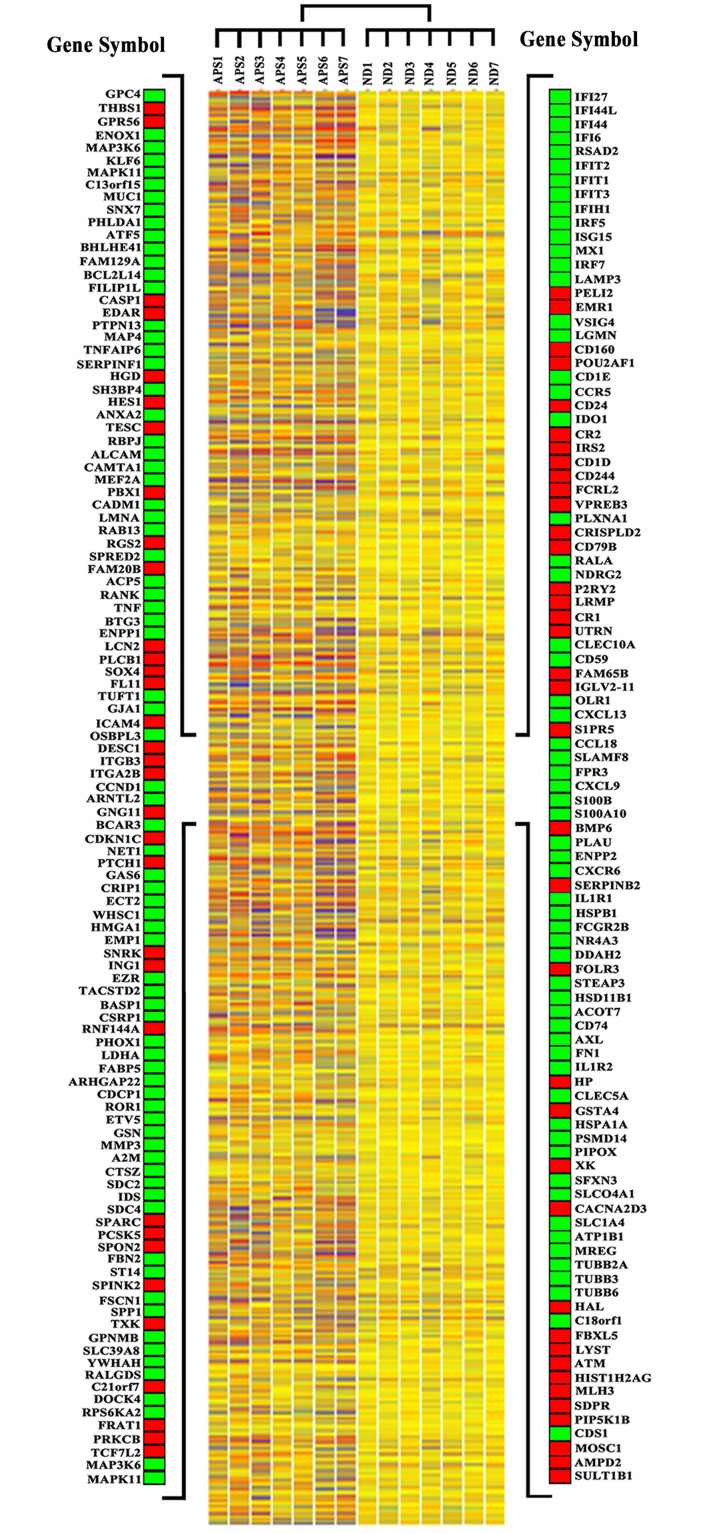
Fig 1 shows the heat map representing some Affymetrix arrays with a selection of modulated genes.
Fig 1. Gene expression profiles of synovial membranes obtained from 10 PsA patients and from 10 healthy subjects.
The heat map shows the expression levels of all the modulated genes. Blue-violet indicates genes that are expressed in synovial biopsies of PsA patients at lower levels when compared with the mean values of the control subjects, orange-red indicates genes that are expressed at higher levels when compared to the control means and yellow indicates genes that are not differently expressed in the patients group versus the control group. Each row represents a gene; each column shows the expression of selected genes in each individual (see S1 Table for complete list of genes expression data). On both sides of the heat map a selection of genes is shown. Green and red squares indicate upregulated and downregulated genes respectively.
The Gene Ontology analysis of the regulated transcripts showed that the vast majority of them are involved in several biological processes that may play a role in PsA including: inflammation, immune response, apoptosis, cell cycle regulation and proliferation, cell migration and invasion, extra-cellular matrix (ECM) and ECM remodeling, bone remodeling, angiogenesis, signal transduction.
Table 2 shows a detailed representation of selected genes within the above-mentioned clusters. The table also includes GeneBank accession numbers and fold changes. The complete list of modulated genes can be found in S1 Table.
Table 2. Annotated genes differentially expressed in PsA synovial membranes versus healthy synovial membranes grouped according to their function.
| Probe Set ID | Gene Title | Gene Symbol | FC | Representative Public ID |
|---|---|---|---|---|
| inflammation | ||||
| 207113_s_at | tumor necrosis factor (TNF superfamily, member 2) | TNF | 3.55 | NM_000594 |
| 210004_at | oxidized low density lipoprotein (lectin-like) receptor 1 | OLR1 | 83.10 | AF035776 |
| 205242_at | chemokine (C-X-C motif) ligand 13 | CXCL13 | 79.22 | NM_006419 |
| 32128_at | chemokine (C-C motif) ligand 18 | CCL18 | 20.32 | NM_002988 |
| 203915_at | chemokine (C-X-C motif) ligand 9 | CXCL9/MIG | 11.16 | NM_002416 |
| 209686_at | S100 calcium binding protein B | S100B | 7.80 | NM_006272 |
| 200872_at | S100 calcium binding protein A10 | S100A10 | 2.21 | NM_002966 |
| 206974_at | chemokine (C-X-C motif) receptor 6 | CXCR6 | 6.28 | NM_006564 |
| 202948_at | interleukin 1 receptor, type I | IL1R1 | 5.67 | NM_000877 |
| 211719_x_at | fibronectin 1 | FN1 | 405.39 | NM_212482 |
| immune response | ||||
| a) adaptive | ||||
| 205242_at | chemokine (C-X-C motif) ligand 13 | CXCL13 | 79.22 | NM_006419 |
| 216876_s_at | interleukin 17A | IL17A | 2.30 | U32659 |
| 221165_s_at | interleukin 22 | IL22 | 2.04 | AF279437 |
| 221111_at | interleukin 26 | IL26 | 3.67 | NM_018402 |
| 220054_at | interleukin 23, alpha subunit p19 | IL23A | 2.00 | NM_016584 |
| 208991_at | signal transducer and activator of transcription 3 | STAT3 | 2.26 | NM_139276.2 |
| 206983_at | chemokine (C-C motif) receptor 6 | CCR6 | 2.01 | NM_004367 |
| 205476_at | chemokine (C-C motif) ligand 20 | CCL20 | 3.61 | NM_004591 |
| 206991_s_at | chemokine (C-C motif) receptor 5 | CCR5 | 8.42 | NM_000579 |
| 208488_s_at | complement component (3b/4b) receptor 1 | CR1 | -2.36 | NM_000651 |
| 205544_s_at | complement component receptor 2 | CR2 | -5.05 | NM_001877 |
| 221239_s_at | Fc receptor-like 2 | FCRL2 | -3.90 | NM_030764 |
| 220068_at | pre-B lymphocyte 3 | VPREB3 | -3.59 | NM_013378 |
| b) innate | ||||
| 205569_at | lysosomal-associated membrane protein 3 | LAMP3 | 36.99 | NM_014398 |
| 204787_at | V-set and immunoglobulin domain containing 4 | VSIG4/Z39IG | 26.76 | NM_007268 |
| 221538_s_at | plexin A1 | PLXNA1 | 3.36 | NM_032242 |
| 210889_s_at | Fc fragment of IgG, low affinity IIb, receptor (CD32) | FCGR2B | 5.25 | M31933 |
| 209959_at | nuclear receptor subfamily 4, group A, member 3 | NR4A3 | 5.20 | U12767 |
| 219890_at | C-type lectin domain family 5, member A | CLEC5A | 16.91 | NM_013252 |
| 206682_at | C-type lectin domain family 10, member A | CLEC10A | 4.01 | NM_006344 |
| apoptosis | ||||
| 205573_s_at | sorting nexin 7 | SNX7 | 7.76 | NM_015976 |
| 217996_at | pleckstrin homology-like domain, family A, member 1 | PHLDA1 | 6.09 | NM_007350 |
| 204998_s_at | activating transcription factor 5 | ATF5 | 5.79 | NM_012068 |
| 221530_s_at | basic helix-loop-helix family, member e41 | BHLHE41 | 5.03 | AB044088 |
| 217966_s_at | family with sequence similarity 129, member A | FAM129A | 3.54 | AF288391 |
| 211367_s_at | caspase 1, apoptosis-related cysteine peptidase | CASP1 | -2.83 | NM_033292 |
| 220048_at | ectodysplasin A receptor | EDAR | -2.22 | NM_022336 |
| 204201_s_at | protein tyrosine phosphatase, non-receptor type 13 | PTPN13 | 2.18 | NM_006264 |
| 243_g_at | microtubule-associated protein 4 | MAP4 | 2.40 | M64571 |
| 204614_at | serpin peptidase inhibitor, clade B, member 2 | SERPINB2 | -5.90 | NM_002575 |
| cell cycle regulation | ||||
| 208712_at | cyclin D1 | CCND1 | 9.94 | NM_053056.2 |
| 220658_s_at | aryl hydrocarbon receptor nuclear translocator-like 2 | ARNTL2 | 4.03 | NM_020183 |
| 204115_at | guanine nucleotide binding protein (G protein), gamma 11 | GNG11 | -42.28 | NM_004126 |
| 204032_at | breast cancer anti-estrogen resistance 3 | BCAR3 | 3.94 | NM_003567 |
| cell proliferation | ||||
| 213348_at | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | -17.49 | NM_001122631 |
| 201830_s_at | neuroepithelial cell transforming 1 | NET1 | 4.27 | NM_005863 |
| 209815_at | patched homolog 1 (Drosophila) | PTCH1 | -7.22 | NM_000264 |
| 202177_at | growth arrest-specific 6 | GAS6 | 4.12 | NM_000820 |
| 205081_at | cysteine-rich protein 1 | CRIP1 | 3.11 | NM_001311 |
| 219787_s_at | epithelial cell transforming sequence 2 oncogene | ECT2 | 3.04 | NM_018098 |
| 201324_at | epithelial membrane protein 1 | EMP1 | 45.31 | NM_001423 |
| 209481_at | SNF related kinase | SNRK | -2.15 | AF226044 |
| 202286_s_at | tumor-associated calcium signal transducer 2 | TACSTD2 | 56.82 | NM_002353 |
| cell migration and/or invasion | ||||
| 202345_s_at | fatty acid binding protein 5 (psoriasis-associated) | FABP5 | 12.73 | NM_001444 |
| 218451_at | CUB domain containing protein 1 | CDCP1 | 3.61 | NM_022842 |
| 203349_s_at | ets variant 5 | ETV5 | 7.03 | NM_004454 |
| 200696_s_at | gelsolin (amyloidosis, Finnish type) | GSN | 3.65 | NM_000177 |
| ECM/ECM remodeling | ||||
| 205828_at | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 | 26.05 | NM_002422 |
| 217757_at | alpha-2-macroglobulin | A2M | 25.43 | NM_000014 |
| 210042_s_at | cathepsin Z | CTSZ | 7.14 | NM_001336 |
| 212158_at | syndecan 2 | SDC2 | 15.68 | NM_002998 |
| 202439_s_at | iduronate 2-sulfatase | IDS | 2.31 | NM_000202 |
| 202071_at | syndecan 4 | SDC4 | 5.98 | NM_002999 |
| 205559_s_at | proprotein convertase subtilisin/kexin type 5 | PCSK5 | -4.52 | NM_006200 |
| 206310_at | serine peptidase inhibitor, Kazal type 2 | SPINK2 | -2.30 | NM_021114 |
| 214768_x_at | anti-thyroid peroxidase monoclonal autoantibody IgK chain, V region | FAM20B | -4.03 | NM_014864 |
| angiogenesis | ||||
| 204984_at | glypican 4 | GPC4 | 2.24 | NM_001448 |
| 201110_s_at | thrombospondin 1 | THBS1 | -8.29 | NM_003246 |
| 212070_at | G protein-coupled receptor 56 | GPR56 | -7.31 | NM_001145774 |
| 219501_at | ecto-NOX disulfide-thiol exchanger 1 | ENOX1 | 3.47 | NM_017993 |
| 219278_at | mitogen-activated protein kinase kinase kinase 6 | MAP3K6 | 2.41 | NM_004672 |
| 211499_s_at | mitogen-activated protein kinase 11 | MAPK11 | 2.24 | NM_002751 |
| 213693_s_at | mucin 1, cell surface associated | MUC1 | 2.46 | X80761 |
| bone remodeling | ||||
| 209875_s_at | secreted phosphoprotein 1 | SPP1/OPN | 448.78 | J04765 |
| 201141_at | glycoprotein (transmembrane) nmb | GPNMB | 147.53 | NM_001005340 |
| a) bone growth | ||||
| 209031_at | cell adhesion molecule 1 | CADM1 | 14.85 | NM_014333 |
| 203411_s_at | lamin A/C | LMNA | 13.75 | NM_005572 |
| 212458_at | sprouty-related, EVH1 domain containing 2 | SPRED2 | 4.33 | AY299090 |
| 205066_s_at | ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | 7.20 | NM_006208 |
| 206026_s_at | tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 4.84 | NM_007115 |
| 202283_at | serpin peptidase inhibitor, clade F, member 1 | SERPINF1 | 4.77 | NM_002615 |
| 201951_at | activated leukocyte cell adhesion molecule | ALCAM | 5.95 | L38608 |
| b) bone erosion | ||||
| 207113_s_at | tumor necrosis factor (TNF superfamily, member 2) | TNF | 3.55 | NM_000594 |
| 204638_at | acid phosphatase 5, tartrate resistant | ACP5 | 3.91 | NM_001611 |
| 207037_at | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | TNFRSF11A | 3.77 | NM_003839 |
| 202252_at | RAB13, member RAS oncogene family | RAB13 | 5.37 | NM_002870 |
| 212151_at | pre-B-cell leukemia homeobox 1 | PBX1 | -2.36 | NM_002585 |
| 202388_at | regulator of G-protein signaling 2, 24kDa | RGS2 | -4.34 | NM_002923 |
| 212531_at | lipocalin 2 | LCN2 | -3.26 | NM_005564 |
| 201417_at | SRY (sex determining region Y)-box 4 | SOX4 | -2.78 | NM_003107 |
| 203395_s_at | hairy and enhancer of split 1 | HES1 | -2.18 | NM_005524 |
| signaling pathways | ||||
| a) Wnt and beta-catenin pathway | ||||
| 205003_at | dedicator of cytokinesis 4 | DOCK4 | 13.81 | AY233380 |
| 213693_s_at | mucin 1, cell surface associated | MUC1 | 2.46 | X80761 |
| 219889_at | frequently rearranged in advanced T-cell lymphomas | FRAT1 | -2.59 | NM_005479 |
| 205805_s_at | receptor tyrosine kinase-like orphan receptor 1 | ROR1 | 2.02 | NM_005012 |
| 208712_at | cyclin D1 | CCND1 | 9.94 | NM_053056.2 |
| 201667_at | gap junction protein, alpha 1, 43kDa | GJA1 | 9.03 | NM_000165 |
| 216511_s_at | transcription factor 4 | TCF4/TCF7L2 | -7.27 | NM_001146274 |
| b) MAP-kinase pathway | ||||
| 219278_at | mitogen-activated protein kinase kinase kinase 6 | MAP3K6 | 2.41 | NM_004672 |
| 211499_s_at | mitogen-activated protein kinase 11 | MAPK11 | 2.24 | NM_002751 |
| 212912_at | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | RPS6KA2 | 2.65 | NM_021135 |
| 202581_at | heat shock 70kDa protein 1A | HSPA1A | 6.94 | NM_005345 |
| d) Insulin pathway | ||||
| 209185_s_at | insulin receptor substrate 2 | IRS2 | -4.83 | AF073310 |
| 205066_s_at | ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | 7.20 | NM_006208 |
| 206020_at | suppressor of cytokine signaling 6 | SOCS6 | 2.26 | NM_004232 |
| b) Notch pathway | ||||
| 211974_x_at | recombination signal binding protein for immunoglobulin kappa J region | RBPJ | 2.92 | NM_203284 |
| 203395_s_at | hairy and enhancer of split 1 | HES1 | -2.18 | NM_005524 |
| 207113_s_at | tumor necrosis factor (TNF superfamily, member 2) | TNF | 3.55 | NM_000594 |
| e) Type I interferon pathway | ||||
| 202411_at | interferon, alpha-inducible protein 27 | IFI27 | 13.74 | NM_005532 |
| 204439_at | interferon-induced protein 44-like | IFI44L | 2.23 | NM_006820 |
| 214453_s_at | interferon-induced protein 44 | IFI44 | 2.32 | NM_006417 |
| 204415_at | interferon, alpha-inducible protein 6 | IFI6 | 2.20 | NM_022873 |
| 213797_at | radical S-adenosyl methionine domain containing 2 | RSAD2 | 3.68 | NM_080657 |
| 217502_at | interferon-induced protein with tetratricopeptide repeats 2 | IFIT2 | 2.03 | NM_001547 |
| 203153_at | interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 3.17 | NM_001548 |
| 204747_at | interferon-induced protein with tetratricopeptide repeats 3 | IFIT3 | 2.58 | NM_001549 |
| 219209_at | interferon induced with helicase C domain 1 | IFIH1 | 2.15 | NM_022168 |
| 205469_s_at | interferon regulatory factor 5 | IRF5 | 2.03 | NM_001098629 |
| 205483_s_at | ISG15 ubiquitin-like modifier | ISG15 | 2.03 | NM_005101 |
| 202086_at | myxovirus resistance 1, interferon-inducible protein p78 | MX1 | 3.05 | NM_002462 |
| 208436_s_at | interferon regulatory factor 7 | IRF7 | 2.15 | NM_001572 |
| f) others | ||||
| 201020_at | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta | YWHAH | 3.27 | NM_003405 |
| 209050_s_at | ral guanine nucleotide dissociation stimulator | RALGDS | 2.43 | NM_006266 |
| 221211_s_at | chromosome 21 open reading frame 7 | C21orf7 | -8.00 | NM_020152 |
A large number (29/196) of modulated genes have a role in the inflammatory process.
The upregulated transcripts comprise chemokine (C-X-C motif) ligand 13, CXCL13 (FC 79.22), CXCL9/MIG (FC 11.16), chemokine (C-C motif) ligand 18, CCL18 (FC 20.32), interleukin 1 receptor, type I, IL1R1 [30,31] (FC 5.67), oxidized low density lipoprotein receptor 1, OLR1/LOX1 (FC 83.1), S100 calcium binding protein namely S100B [32] (FC 7.8) and S100A102 [33] (FC 2.21), fibronectin1, FN1 [34] (FC 405.4), tumour necrosis factor alpha TNF (FC 3.5).
Among genes involved in the immune response, many Th17-lymphocytes related genes were up-regulated including CCR6 (FC 2.01), CCL20 (FC 3.6), CXCL13 (FC 79.2) [35] interleukin 17A, IL17A [36] (FC 2.3), IL22 [37] (FC 2.04), IL26 [38] (FC 3.67); IL23A [39] (FC 2.2) and signal transducer and activator of transcription 3, STAT3 (FC 2.26) [40].
Other genes involved in B cell activity are downregulated in PsA samples [40,41].
Several upregulated genes play a role in innate immunity and are expressed in dendritic cells [42–45] and in macrophages [46].
Many genes coding for protein involved in apoptosis and/or in apoptosis regulation resulted modulated in pathological samples [47–54].
Genes that positively regulate cell cycle resulted overexpressed in PsA synovial samples [55–57]. Moreover there was a strong downregulation of the guanine nucleotide binding protein, gamma 11, GNG11 (FC -42.3), an inducer of cellular senescence in human cells [58]. Genes that control cell proliferation were also modulated in PsA biopsies even with very high fold change [59–66].
Another group of differentially expressed genes comprises transcripts involved in cell migration and invasion. Interestingly genes that promote these processes resulted up-regulated, i.e. the CUB domain containing protein 1, CDCP1 (FC 3.6) [67]; gelsolin, GSN (FC 3.65) [68]; the fatty acid binding protein 5, FABP5 (FC 12.73) [69], and the ets variant 5, ETV5 (FC 7.03), a pro-invasive transcription factor tought to be involved in bone invasion [70].
Several genes encoding for ECM components or involved in ECM remodeling were modulated in PsA samples. We found a strong overexpression of syndecan 2, SDC2 (FC 15.68) and syndecan 4, SDC4 (FC 5.98) and of several proteases-encoding genes, including matrix metallopeptidase 3, MMP3 (FC 26.05), suppression of tumorigenicity 14, ST14/matriptase (FC 2.78); carboxy-peptidase cathepsin Z, CTSZ (FC 7.14) [71] and iduronate 2-sulfatase, IDS (FC 2.31) [72]. On the contrary, inhibitors of metalloproteases, proprotein convertase subtilisin/kexin type 5, PCSK5/PC5/6 (FC -4.52) [73], and serine peptidase inhibitor, Kazal type 2, SPINK2 (FC -2.30) [74] were underexpressed.
Differentially expressed genes also comprise transcripts which regulate the angiogenesis process [75–78].
A large number (28/196) of genes differentially expressed in PsA biopsies belong to the bone remodeling cluster which comprises transcripts involved in bone-growth and bone resorption. Upregulated genes promote bone growth with different mechanisms such as osteoblast differentiation i.e: lamin A, LMNA (FC 13.75) [79]; bone-matrix deposition, i.e: cell adhesion molecule 1, CADM1 (FC 14.85) [80]; bone development, i.e: sprouty-related, EVH1 domain containing 2, SPRED2 (FC 4.33) [81]; bone mineralization i.e: ectonucleotide pyrophosphatase/phosphodiesterase 1, ENPP1/NPP1 (FC 7.20), [82] and bone morphogenesis, i.e: activated leukocyte cell adhesion molecule, ALCAM/CD166 (FC 5.95), [83].
Genes inhibiting osteoblast proliferation [84,85]and osteoblast differentiation [86–88] were expressed in lower levels in PsA samples.
In addition the following upregulated genes may support bone growth by interfering with osteoclast activity: tumor necrosis factor, alpha-induced protein 6, TNFAIP6/TSG6 (FC 4.84) [89], and serpin peptidase inhibitor, clade F member 1, SERPINF1/PEDF (FC 4.77).
In the bone resorption cluster, the following upregulated genes promote osteoclast activity and/or differentiation: the acid phosphatase 5, tartrate resistant, ACP5/TRAP (FC 3.91) [90], the tumor necrosis factor a, TNFa (FC 3.55); member RAS oncogene family, RAB13 (FC 5.37) [91] and tumor necrosis factor receptor superfamily, member 11a, NFKB activator, TNFRSF11A/RANK (FC 3.77) [92].
Noteworthy the bone remodeling category contains two genes, osteopontin and osteoactivin, which show a very high level of induction (FC 450 and 147 respectively).
Finally we analysed DEGs which belong to /or are connected to the gene cluster signal transduction using a pathway analysis tool to evaluate if they contribute to known signaling networks that may be relevant in the pathogenesis of PsA and found that a conspicuous number of genes (also including genes originally ascribed to different functional classes) is involved in well characterized signaling networks already associated to human diseases, including immune-mediated diseases, bone diseases and inflammatory diseases.
These signal cascades include: 1) the Interferon-alpha (IFN-A) pathway also named “Type I Interferon signature” [93], 2) the Wnt/beta-catenin signaling pathway, 3) the Notch signaling pathway, 4) the mitogen activated protein (MAP) kinase pathway, and 5) the insulin pathway.
In particular 13 type I interferon inducible genes (IFIG) were upregulated, thus indicating the presence of an IFN type I signature, typically associated with autoimmune disease such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), Crohn’s disease and Sjogren syndrome [94–100].
Seven modulated genes belong to the WNT signalling pathway which has been associated with the pathogenesis of RA, and in particular with synovial inflammation and bone remodeling [101–107].
Three genes belong to the Notch signalling pathway that is crucial for bone homeostasis and is involved in the pathogenesis of several immune-mediated diseases, including RA [108].
Four genes of the MAP kinase pathway are upregulated including: mitogen-activated protein kinase 11, MAPK11 (FC 2.24), mitogen-activated protein kinase 6,MAP3K6 (FC 2.41), ribosomal protein S6 kinase, 90 kDa, polypeptide 2, RPS6KA2 (FC 2.65), heat shock 70 kDa protein 1 A, HSPA1A (FC 6.94). The p38 MAPK signalling pathway has been associated with psoriasis and psoriatic arthritis [109].
Two genes belong to the insulin pathway: insulin receptor substrate 2, IRS2 (FC-4.83) and ectonucleotyde pyrophosphatase/phosphodiesterase 1, ENPP1 (FC 7.2). The insulin pathway is involved in the pathogenesis of the metabolic syndrome [110] and an increased body mass index increases the risk of PsA development in patients with cutaneous psoriasis, supporting a link between fat-mediated inflammation and joint involvement [111].
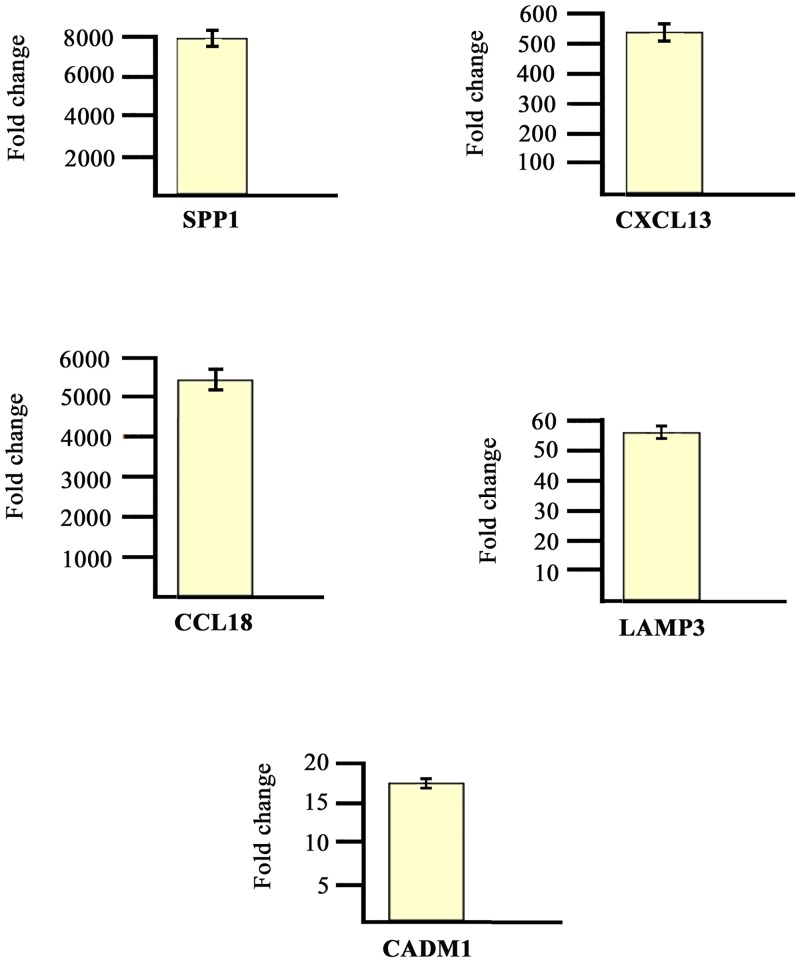
The modulation of some genes observed by gene array analysis was validated by Q-PCR (Fig 2).
Fig 2. Real time RT-PCR of some modulated genes.
Genes selected for validation were SPP1, CXCL13, CCL18, LAMP3 and CADM1. The transcripts of the selected genes were increased in PsA samples when compared to healthy donors. Relative expression levels were calculated for each sample after normalization against the housekeeping gene GAPDH. Experiments have been conducted in triplicates. Similar results were obtained using the housekeeping genes18s rRNA and beta-actin (data not shown).
B) DEGs in PBC
We next compared the gene expression profiles of 10 PBC samples obtained from the same PsA patients used for the biopsy collection with 10 PBC samples obtained from healthy age and sex matched donors. When the same statistical criteria and fold change criteria were applied to the modulated genes, we obtained 187 genes that were further analysed. 118 and 69 genes resulted to be up- or downregulated respectively. While the number of DEGs in PBC is similar to that obtained in synovial biopsies, the average level of gene induction (FC) is much lower in PBC when compared to the synovium. DEGs are distributed in several functional classes which partially overlap with the ones identified by genes modulated in PsA synovium. The gene categories comprise: inflammation, immune response, apoptosis, cell cycle regulation and proliferation, cell migration and invasion, ECM/ECM remodeling, bone remodelling, angiogenesis, signal transduction.
A detailed representation of modulated genes within the above-mentioned clusters vcan be found in S2 Table.
Interestingly modulated genes are distributed in various gene categories that regulate different biological processes (ie proliferation, apoptosis, immune response), however the functional classes which show the highest enrichment in modulated trascripts are the immune response (36/187), the signal transduction (25/187) the inflammation (25/187), and the bone remodeling (17/187) gene clusters.
Remarkably, in the immune response group, we observed increased expression of several Th17 related genes (CCR6 [112]; CCL20 [113]; lymphocyte antigen 9, LY9 [114,115]; interleukin 6 signal transducer, IL6ST [116–118], interleukin 12 receptor, beta 2, IL12RB [119]).
Noteworthy 10 IFIG were modulated also in PBC samples, thus confirming the presence of an INFA signature observed within the synovium.
Consistently with the presence of a strong inflammatory response typical of PsA we also observed upregulation of several proinflammatory genes (IL8; CCL18; chemokine (C-X-C motif) ligand 1, CXCL1; IL1A; OLR1; HSPA1A; CXCL9; see S2 Table). New bone formation is a typical feature of PsA and consistently with this aspect of the disease several genes involved in the bone growth process are modulated also in PBC.
The analysis of the expression profiles of the PBC samples revealed that 90 out of 187 transcripts differently regulated in PBC were also modulated in PsA synovium (Table 3). These genes belong to different functional classes including: immune response and inflammation; angiogenesis, apoptosis; bone remodeling; cell proliferation; extracellular matrix remodeling and ECM components; signal transduction.
Table 3. Annotated genes differentially expressed in PsA synovial membranes and in PsA PBC versus healthy controls grouped according to their function.
| Probe Set ID | Gene Title | Gene Symbol | FC SM | FC PBC | Representative Public ID |
|---|---|---|---|---|---|
| inflammation | |||||
| 210004_at | oxidized low density lipoprotein (lectin-like) receptor 1 | OLR1 | 83.10 | 7.16 | AF035776 |
| 32128_at | chemokine (C-C motif) ligand 18 | CCL18 | 20.32 | 2.03 | NM_002988 |
| 219386_s_at | SLAM family member 8 | SLAMF8 | 14.24 | 4.52 | NM_020125 |
| 202948_at | interleukin 1 receptor, type I | IL1R1 | 5.67 | 3.31 | NM_000877 |
| 209959_at | nuclear receptor subfamily 4, group A, member 3 | NR4A3 | 5.20 | 2.48 | U12767 |
| 211372_s_at | interleukin 1 receptor, type II | IL1R2 | 6.57 | 4.27 | U64094 |
| 219890_at | C-type lectin domain family 5, member A | CLEC5A | 16.91 | 5.64 | NM_013252 |
| 202581_at | heat shock 70kDa protein 1A | HSPA1A | 6.94 | 10.43 | NM_005345 |
| 206697_s_at | haptoglobin | HP | -2.71 | -2.34 | NM_005143 |
| 203915_at | chemokine (C-X-C motif) ligand 9 | CXCL9/MIG | 11.16 | 8.48 | NM_002416 |
| 209686_at | S100 calcium binding protein B | S100B | 7.80 | 7.82 | NM_006272 |
| immune response | |||||
| 209771_x_at | CD24 molecule | CD24 | -8.30 | -2.06 | NM_013230 |
| 205789_at | CD1d molecule | CD1D | -4.49 | -2.62 | NM_001766 |
| 206277_at | purinergic receptor P2Y, G-protein coupled, 2 | P2RY2 | -2.90 | -2.2 | NM_002564 |
| 216984_x_at | immunoglobulin lambda variable 2–11 | IGLV2-11 | -3.66 | -3.31 | D84143 |
| 206983_at | chemokine (C-C motif) receptor 6 | CCR6 | 2.01 | 2.67 | NM_004367 |
| 205476_at | chemokine (C-C motif) ligand 20 | CCL20 | 3.61 | 2.14 | NM_004591 |
| 220307_at | CD244 molecule, natural killer cell receptor 2B4 | CD244/2B4 | -4.05 | -2.73 | AF242540 |
| 210029_at | indoleamine 2,3-dioxygenase 1 | IDO1 | 5.87 | 2.34 | M34455 |
| 205544_s_at | complement component receptor 2 | CR2 | -5.05 | -3.12 | NM_001877 |
| 220068_at | pre-B lymphocyte 3 | VPREB3 | -3.59 | -3.43 | NM_013378 |
| 207840_at | CD160 molecule | CD160 | -14.75 | -2.51 | NM_007053 |
| apoptosis | |||||
| 217996_at | pleckstrin homology-like domain, family A, member 1 | PHLDA1 | 6.09 | 4.32 | NM_007350 |
| 204998_s_at | activating transcription factor 5 | ATF5 | 5.79 | 2.79 | NM_012068 |
| 204201_s_at | protein tyrosine phosphatase, non-receptor type 13 | PTPN13 | 2.18 | 2.16 | NM_006264 |
| 204614_at | serpin peptidase inhibitor, clade B, member 2 | SERPINB2/PAI2 | -5.90 | -2.46 | NM_002575 |
| cell cycle regulation | |||||
| 204115_at | guanine nucleotide binding protein (G protein), gamma 11 | GNG11 | -42.28 | -2.65 | NM_004126 |
| cell proliferation | |||||
| 201830_s_at | neuroepithelial cell transforming 1 | NET1 | 4.27 | 3.05 | NM_005863 |
| 201324_at | epithelial membrane protein 1 | EMP1 | 45.31 | 2.06 | NM_001423 |
| 208623_s_at | ezrin | EZR | 2.41 | 3.04 | J05021 |
| 209481_at | SNF related kinase | SNRK | -2.15 | -2.54 | AF226044 |
| 209815_at | patched homolog 1 (Drosophila) | PTCH1 | -7.22 | -2.65 | NM_000264 |
| 209808_x_at | inhibitor of growth family, member 1 | ING1 | -2.08 | -2.68 | AF149723 |
| 202286_s_at | tumor-associated calcium signal transducer 2 | TACSTD2 | 56.82 | 3.76 | NM_002353 |
| ECM/ECM remodeling | |||||
| 210042_s_at | cathepsin Z | CTSZ | 7.14 | 2.67 | NM_001336 |
| 212158_at | syndecan 2 | SDC2 | 15.68 | 2.84 | NM_002998 |
| 202071_at | syndecan 4 | SDC4 | 5.98 | 2.71 | NM_002999 |
| 200665_s_at | secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | -5.60 | -2.11 | NM_003118 |
| 206310_at | serine peptidase inhibitor, Kazal type 2 | SPINK2 | -2.30 | -2.32 | NM_021114 |
| 214768_x_at | anti-thyroid peroxidase monoclonal autoantibody IgK chain, V region | FAM20B | -4.03 | -2.52 | NM_014864 |
| angiogenesis | |||||
| 212070_at | G protein-coupled receptor 56 | GPR56 | -7.31 | -3.25 | NM_001145774 |
| 218723_s_at | chromosome 13 open reading frame 15 | C13orf15 | 3.11 | 2.74 | NM_014059 |
| 201110_s_at | thrombospondin 1 | THBS1 | -8.29 | -3.48 | NM_003246 |
| bone remodeling | |||||
| 206026_s_at | tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 4.84 | 4.23 | NM_007115 |
| 202283_at | serpin peptidase inhibitor, clade F, member 1 | SERPINF1 | 4.77 | 2.77 | NM_002615 |
| 222258_s_at | SH3-domain binding protein 4 | SH3BP4 | 3.73 | 3.11 | AF015043 |
| 203395_s_at | hairy and enhancer of split 1 | HES1 | -2.18 | -2.23 | NM_005524 |
| 208328_s_at | myocyte enhancer factor 2A | MEF2A | 2.51 | 2.45 | NM_005587 |
| 212151_at | pre-B-cell leukemia homeobox 1 | PBX1 | -2.36 | -2.2 | NM_002585 |
| 205548_s_at | BTG family, member 3 | BTG3/ANA | 2.29 | 4.76 | NM_006806 |
| 212531_at | lipocalin 2 | LCN2 | -3.26 | -3.43 | NM_005564 |
| 213222_at | phospholipase C, beta 1 (phosphoinositide-specific) | PLCB1 | -3.68 | -2.73 | NM_182734 |
| 201417_at | SRY (sex determining region Y)-box 4 | SOX4 | -2.78 | -3.03 | NM_003107 |
| 210786_s_at | Friend leukemia virus integration 1 | FLI1/ETS1 | -2.25 | -2.16 | M93255 |
| 209875_s_at | secreted phosphoprotein 1 | SPP1/OPN | 448.78 | 4.32 | J04765 |
| 201141_at | glycoprotein (transmembrane) nmb | GPNMB | 147.53 | 3.27 | NM_001005340 |
| 203411_s_at | lamin A/C | LMNA | 13.75 | 2.4 | NM_005572 |
| 202252_at | RAB13, member RAS oncogene family | RAB13 | 5.37 | 2.62 | NM_002870 |
| signaling pathways | |||||
| 201020_at | tyrosine 3-monooxygenase | YWHAH | 3.27 | 2.37 | NM_003405 |
| 221211_s_at | chromosome 21 open reading frame 7 | C21orf7 | -8.00 | -2.81 | NM_020152 |
| 206020_at | suppressor of cytokine signaling 6 | SOCS6 | 2.26 | 3.15 | NM_004232 |
| 202581_at | heat shock 70kDa protein 1A | HSPA1A | 6.94 | 10.43 | NM_005345 |
| 209185_s_at | insulin receptor substrate 2 | IRS2 | -4.83 | -2.05 | AF073310 |
| 203395_s_at | hairy and enhancer of split 1 | HES1 | -2.18 | -2.23 | NM_005524 |
| 214453_s_at | interferon-induced protein 44 | IFI44 | 2.32 | 2.06 | NM_006417 |
| 204439_at | interferon-induced protein 44-like | IFI44L | 2.23 | 2.34 | NM_006820 |
| 204415_at | interferon, alpha-inducible protein 6 | IFI6 | 2.20 | 3.04 | NM_022873 |
| 213797_at | radical S-adenosyl methionine domain containing 2 | RSAD2 | 3.68 | 2.65 | NM_080657 |
| 203153_at | interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 3.17 | 2.21 | NM_001548 |
| 204747_at | interferon-induced protein with tetratricopeptide repeats 3 | IFIT3 | 2.58 | 3.06 | NM_001549 |
| 205469_s_at | interferon regulatory factor 5 | IRF5 | 2.03 | 2.78 | NM_001098629 |
| 205483_s_at | ISG15 ubiquitin-like modifier | ISG15 | 2.03 | 2.23 | NM_005101 |
| 202086_at | myxovirus resistance 1, interferon-inducible protein p78 | MX1 | 3.05 | 2.66 | NM_002462 |
| 205003_at | dedicator of cytokinesis 4 | DOCK4 | 13.81 | 2.04 | AY233380 |
| 216511_s_at | transcription factor 4 | TCF7L2/TCF4 | -7.27 | -2.25 | NM_001146274 |
| cell junctions | |||||
| 204627_s_at | integrin, beta 3 | ITGB3 | -9.21 | -2.46 | NM_000212 |
| 206493_at | integrin, alpha 2b | ITGA2B | -5.36 | -2.33 | NM_000419 |
| microtubule-based process | |||||
| 204141_at | tubulin, beta 2A | TUBB2A | 5.21 | 3.06 | NM_001069 |
| transport | |||||
| 209267_s_at | solute carrier family 39 (zinc transporter), member 8 | SLC39A8 | 6.68 | 2.96 | AB040120 |
| 209610_s_at | solute carrier family 1, member 4 | SLC1A4 | 2.58 | 2.27 | NM_003038 |
| 220974_x_at | sideroflexin 3 | SFXN3 | 2.93 | 2.14 | NM_030971 |
| 201243_s_at | ATPase, Na+/K+ transporting, beta 1 polypeptide | ATP1B1 | 3.86 | 2.97 | NM_001677 |
| others | |||||
| 221541_at | cysteine-rich secretory protein LCCL domain containing 2 | CRISPLD2 | -6.35 | -2.52 | NM_031476 |
| 206643_at | histidine ammonia-lyase | HAL | -4.98 | -3.37 | NM_002108 |
| 209574_s_at | chromosome 18 open reading frame 1 | C18orf1/LDLRAD4 | 2.27 | 2.13 | NM_181481 |
| 203518_at | lysosomal trafficking regulator | LYST | -2.31 | -2.17 | NM_000081 |
| 208442_s_at | ataxia telangiectasia mutated | ATM | -3.32 | -3.2 | NM_000051 |
| 207156_at | histone cluster 1, H2ag | HIST1H2AG | -3.58 | -2.04 | NM_021064 |
| 204838_s_at | mutL homolog 3 | MLH3 | -4.14 | -2.33 | NM_014381 |
| 218711_s_at | serum deprivation response | SDPR | -19.65 | -2.04 | NM_004657 |
| 218865_at | MOCO sulphurase C-terminal domain containing 1 | MOSC1/MARC1 | -4.86 | -2.45 | NM_022746 |
| 213022_s_at | utrophin | UTRN/DRP1 | -2.15 | -2.59 | NM_007124 |
Frequency of IL-17–positive CD4+T cells in PBMCs from patients with PsA
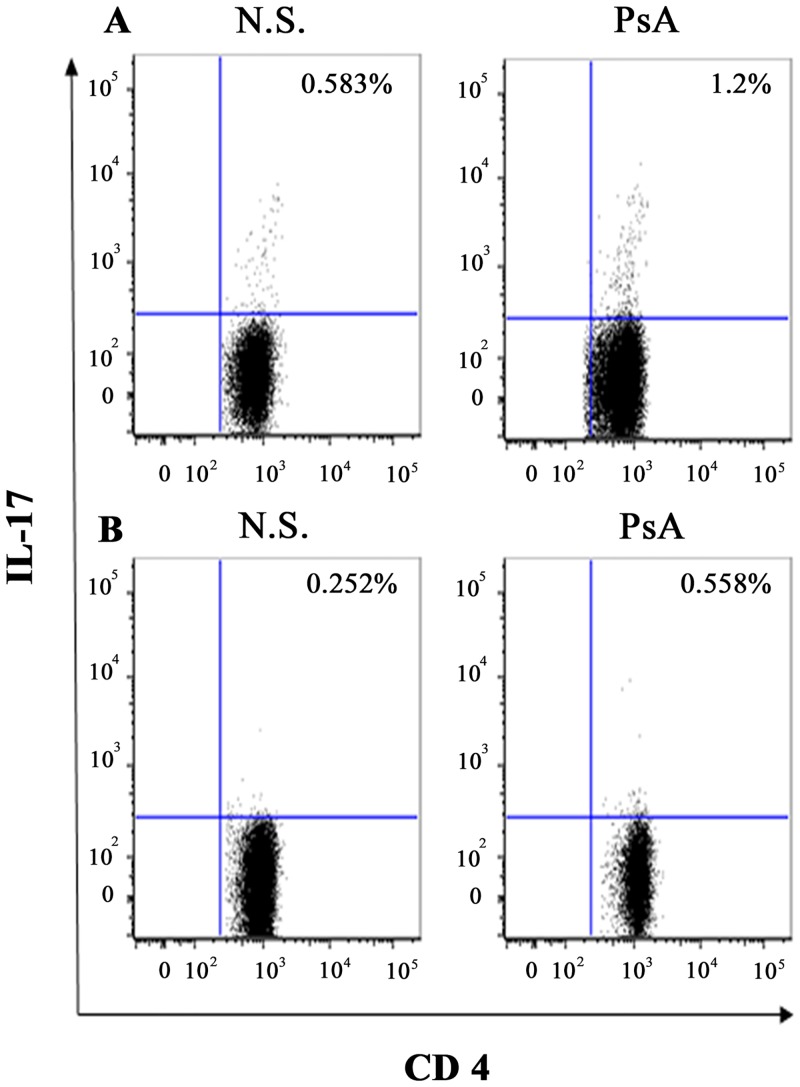
Using flow cytometry, we evaluated the intracellular expression of the cytokine IL-17, by PBMCs from 30 patients with PsA and from 20 healthy control subjects, following stimulation with microbeads coated with anti-CD3 and CD28 or with Candida Albicans. We observed a higher proportion of IL-17–producing CD4+ T cells within the PBMCs of patients with PsA compared with healthy control subjects.
The mean values obtained in 30 PsA PBMC following stimulation with anti-CD3 and CD28 were 1.3% +/- 0.28 versus 0.65% +/- 0.2 (p = 0.005), while the mean values obtained in PSA PBMC following stimulation with Candida Albicans were 0.63% +/- 0.18 versus 0.33%+/- 0.07 (p = 0.004). Representative FACS experiments are shown in Fig (3A and 3B).
Fig 3. Flow cytometric analysis of IL-17 producing CD4+ lymphocytes in patients with PsA.
Data are representative of all the 30 subjects studied. A and B panels show the percentages of IL-17 producing CD4+ lymphocytes in PBMC of healthy donors (N.S) and patients affected by PsA (PsA) after stimulation with Dynabeads Human T-Activator CD3/CD28 (panel a) or with heat-inactivated Candida albicans (panel b).
Detection of IL-17 and IL-23 in synovial fluids from patients with PsA
Il-17 and IL-23 levels were investigated in 20 SFs of patients with PsA and in 20 SF of patients with OA used as controls. SF IL-17 levels were higher in PSA patients (17.87±11.13 pg/mL) than in controls (5.12±1.30 pg/mL) (p<0.01). SF IL-23 levels were higher in PSA patients (37.17±18.13 pg/mL) than in controls (14.12±7.23 pg/mL) (p<0.01)
Detection of soluble mediators in PsA sera
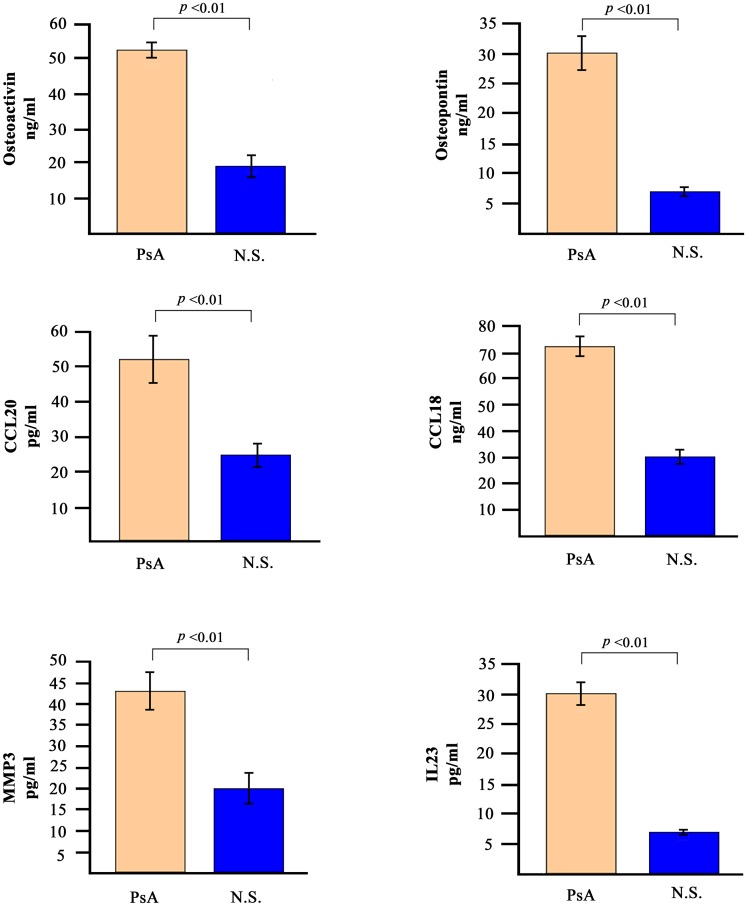
The analysis of gene expression profiles was paralleled by the detection of some of the corresponding soluble mediators in the sera of patients with PsA. We decided to analyse the levels of osteopontin, osteoactivin, CCL20, CCL18, MMP-3 and IL-23. Fig 4 shows the concentration of these molecules in the sera of the 30 PsA patients. The serum levels of all the molecules tested was significantly higher in PsA patients when compared to 30 normal healthy donors.
Fig 4. Serum levels of selected soluble mediators in PsA patients and in normal subjects.
The histograms represent the mean of the results obtained in 30 healthy donors and in 30 PsA patients. p values were calculated using the non-parametric Mann-Whitney test.
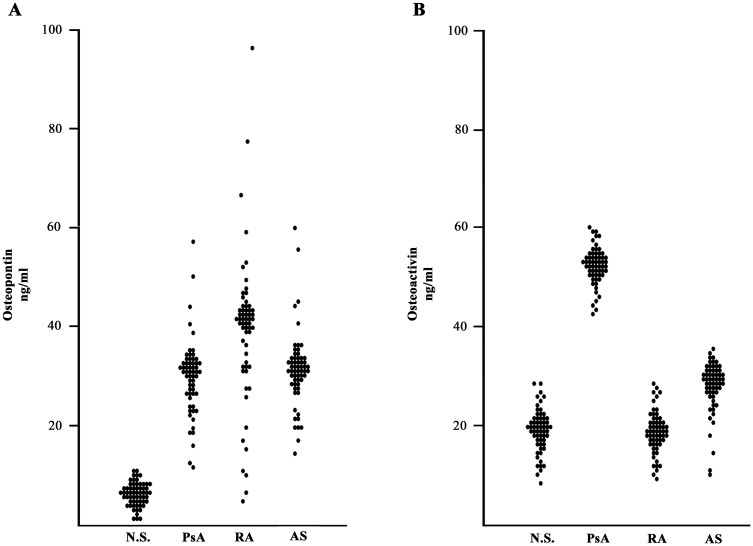
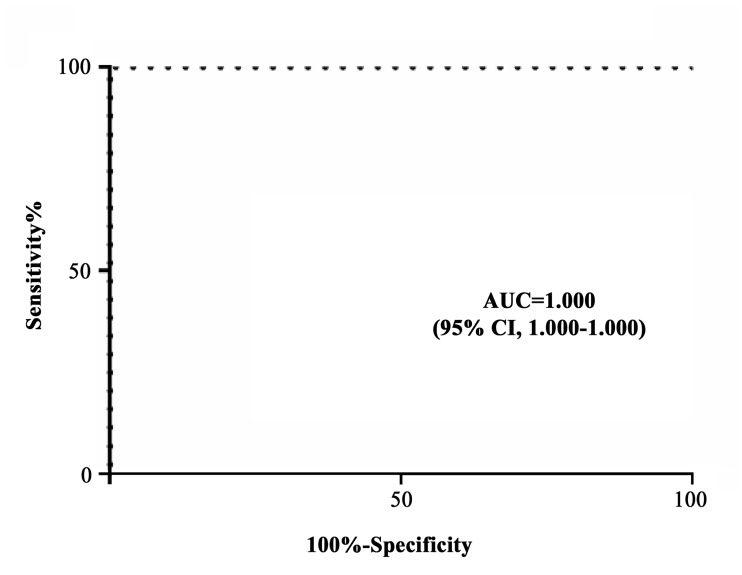
When osteopontin and osteoactivin were evaluated in the entire cohort of 60 patients with PsA and compared with 60 normal subjects and with 60 patients affected by RA and AS, osteoactivin was found to be statistically higher in subjects with PsA (Fig 5) with a high sensitivity and specificity (Fig 6).
Fig 5. Serum levels of osteopontin and osteoactivin in PsA patients and in control subjects.
Graphical representation of the distributions of osteopontin (A) and osteoactivin (B) serum levels in: 60 psoriatic arthritis (PsA) patients, 60 rheumatoid arthritis (RA) patients, 60 ankylosing spondylitis (AS) patients and in 60 normal subjects (NS). p values were calculated using the non-parametric Mann-Whitney test: Osteopontin: PsA vs NS: p <0.01; PsA vs SA: p <0.01, PsA vs AR: p not significant. Osteoactivin: PsA vs NS: p <0.01; PsA vs SA: p <0.05; PsA vs AR: p <0.01.
Fig 6. Sensitivity and specificity of the assay of osteoactivin levels between PsA patients and control subjects.
AUC: area under the curve; CI: confidence interval.
Discussion
In this paper we provide for the first time a comprehensive analysis of the transcriptome within SM and PBC obtained from the same patients affected by PsA. We believe that the combined analysis of the gene expression profiles of paired synovial biopsies and PBC of patients with PsA is likely to better dissect the complex molecular pathways that regulate the different clinical and histopatological aspects of the disease. One contribution of this work is therefore a detailed compilation of genes relevant to the pathogenesis of PsA. Indeed the vast majority of DEGs is involved in biological processes closely connected to the main features of the disease. Noteworthy we observed that, when comparing DEGs in synovial biopsies with DEGs in PBC, a large number of genes were similarly modulated in the two settings, indicating that PBC may be a significant promise for gene expression studies as substitute of tissues that are not easily accessible.
A feature of PsA histopathology is the synovial membrane lining layer hyperplasia, which in RA has been at least in part attributed to an impairement of fibroblast-like-synoviocyte (FLS) apoptosis [120]. Our analysis of apoptosis-related genes indicate a predominant overexpression of anti-apoptotic transcripts which may contribute to the cell accumulation phenomena typical of synovial hyperplasia.
The synovial hyperplastic growth may also be sustained by the global upregulation of genes with mitogenic effect and by the repression of anti-proliferative transcripts observed in PsA samples.
Interestingly, the overexpression of the GAS6 and EZR genes has been already associated with FLS proliferation in inflammatory arthitides [60,121]. FLSs are indeed the predominant cell type in synovial membrane and, especially when switching their phenotype to macrophage-like cells, they can destroy extracellular matrix, deeply penetrating into bone and cartilage [122]. It is worthwhile mentioning that EZR has already been reported as differentially expressed in PsA by Pollock et al. [123]
In PsA synovium, genes involved in cell migration and tissue invasion are strongly upregulated (ie ETV5, and FABP5) consistently with the acquisition of a proinvasive phenotype by PsA synoviocytes. This behaviour may also depend on overexpression of extracellular matrix degrading enzymes.
In this regard we found a strong upregulation of transcripts encoding for proteases (i.e. MMP3, IDS, and CTSZ) and for the metalloproteases inducers ETV5 and FSCN1 [124], whereas proteinases inhibitors (i.e. PCSK5 and SPINK2) showed a decreased expression.
The upregulation of the MMP3 gene is particularly interesting, since its overexpression has been reported in PsA synovium [125] and increased MMP3 levels have been detected in sera of PsA patients by several investigators [126] including ourselves (Fig 4).
Among genes coding for ECM components, the upregulation of the two syndecans transcribing genes (SDC2 and SDC4) is remarkable since these molecules are involved in the retention and activation of leukocytes in inflamed synovium [127,128] and can induce synovial fibroblasts to produce cartilage matrix degrading enzymes such as ADAMTS5 [129].
Another aspect associated to synovial hyperplasia is neoangiogenesis, considered a typical feature of the early phase of PsA [130,131]. Synovial angiogenesis is mediated by several factors produced by both synovial tissue and infiltrating inflammatory cells [131] and promotes the synovial infiltration into the intraarticular cartilage [132,133].
Our gene array data show an overall up-regulation of proangiogenic genes involved in different steps of neoangiogenesis, such as GPC4, abundantly expressed in PsA synovium [75], and MAPK11, also known as p38β, a downstream target of VEGF signaling during angiogenesis [78]. An increased expression for MUC1, an activator of multiple pro-angiogenic factors during hypoxia-driven angiogenesis [134], typical of inflammatory joint diseases [135], was also observed.
Moreover we found a decreased expression of two anti-angiogenic genes, GPR56, a potent inhibitor of vessel formation [136] and THBS1, which inhibits vasculogenesis through the triggering of CD36 on endothelial cells, leading to apoptosis of endothelial cells [137]. Interestingly, using a rat model of osteoarthritis (OA) Hsie et al. [138] demonstrated that THBS1 gene transfer significantly reduced microvessel density and inflammation, thus controlling the progression of the disease.
A very peculiar aspect of PsA pathology is the presence of new bone deposition.
In this context our DEGs indicate that the process of bone formation may be due to genes which regulate osteoblast differentiation (LMNA, CAMTA1 and LCN2) [79,86,139] and proliferation (PBX1, RGS2 and SOX4) [84,85,87,140] or correlate with the bone-forming capacity of mesenchymal stem cells (MSCs) (CADM1). As a matter of fact the synovial membrane has been recently considered a reservoir of MSCs able to differentiate in osteoblasts and chondroblasts [141,142].
Moreover some upregulated genes have specific roles in particular aspects of bone development, such as bone matrix mineralization (ENPP1 and TUFT1) [82], bone morphogenesis (ALCAM) and endochondral ossification (SPRED2). Noteworthy endochondral ossification plays an important role in the pathogenesis of spondyloarthropathies, including PsA [143].
DEGs are therefore consistent with the new bone formation process typical of PsA, and mostly characterized by ankylosis, periostitis, and syndesmophytes [143].
PsA is characterized by bone erosions mediated by osteoclasts at the synovial-bone junction. It is well known that bone erosion is sustained by the osteoclast proteolytic activity and indeed a great increase of the osteoclast-specific marker ACP5 gene, was present in our arrays [90]. Accordingly, we also noticed a remarkable induction for the SH3BP4 gene (FC 3.73), a positive regulator of autophagy, which is a TNFα-dependent process thought to play an important role in joint destruction [144,145]. Two important mediators of bone remodeling typically associated with PsA are RANK and TNF both upregulated in PsA samples. RANK plays a pivotal role in osteoclastogenesis since its gene product is a macrophage-colony stimulating factor (M-CSF)-inducible molecule located on the osteoclast precursor surface that, upon engagement by its ligand RANKL, induces these cells to differentiate [146].
Interestingly the RANKL expression on osteoblasts and other effector cell is primarily regulated by TNF and by Annexin II [147], the product of the gene ANXA2, also upregulated in our PsA samples (see S2 Table).
TNF is involved in a number of autoimmune /inflammatory diseases and is one of the major proinflammatory factors in arthritis causing joint inflammation and cartilage destruction [148]. TNF level is increased both in the synovium and in the synovial fluid of PSA patients [149].
Besides these wellknown effects on inflammation and on bone resorption by inducing osteoclastogenesis and osteoclast recruitment, TNF is also able to induce the expression of the glycoprotein dickkopf-1 (DKK-1), the Wnt-signaling antagonist that suppresses the bone-formation process and the production of the osteoclastogenesis inhibitor, osteoprotegerin [92].
However several phase-3 studies showed that TNF inhibitors significantly ameliorated radiographic progression of the disease, but failed to control new bone formation [92]. It has been suggested that continued suppression of inflammation via anti-TNF agents may accelerate new bone formation and ankylosis [92] possibly through upregulation of DKK-1.
As expected, a large number of modulated genes encode for proinflammatory transcripts and the highest levels of induction were observed for FN1 (F.C. 405) followed by OLR1 (F.C. 83) and CXCL13 (F.C. 79).
Fibronectin is a component of the cartilage matrix but is also locally produced in the synovial fluid where its level positively correlates with joint destruction [150]. Moreover citrullinated forms of fibronectin (cFn) are commonly present in inflammed synovium [150] and notably, fibronectin is a target of autoantibodies in RA [151]. Furthermore cFn inhibits apoptosis and stimulates the secretion of proinflammatory cytokines in FLS isolated from RA patients [150] and induces osteoblast differentiation from human mesenchymal stem cells [152].
OLR1/LOX-1, the receptor for LDL oxidized expressed by endothelial cells and macrophages in the atherosclerotic lesions, is also produced by FLS in the RA synovium where it drives inflammation and cartilage degradation by inducing MMP-1 and MMP-3 expression [153].
CXCL13 is a B cell chemoattractant that has been related to the ectopic lymphoid neogenesis and T/B cells aggregates seen in PsA synovial tissues [130,154].
Other proinflammatory mediators highly induced in PsA samples include CXCL9/MIG, CCL18, CXCR6. CCL18 has been found increased in synovial tissue from RA patients [155] whereas MIG has been involved in regulating leucocyte traffic in both RA and PsA synovium [156].
CXCR6 is highly expressed by skin CD8+ T cells of psoriatic skin lesions [157] and in PsA synovium [158] where it has been suggested to play a role in T-cell homing [157].
We also found increased expression for the IL1R1 gene, an important regulator in both inflammation and autoimmunity. Indeed anti-IL-1 strategies have had a huge impact in autoimmune and inflammatory diseases [159].
Our results indicate a conspicuous participation of macrophages in the PSA synovium inflammatory infiltrate as showed by the up-regulation of several macrophage related genes, such as: FCGR2B, NR4A3, and CLEC5A/MDL1. Indeed macrophages are the principal source of TNFα [160] and in the inflammatory setting, TNF-α in synergy with RANKL, can induce the acquisition of the osteoclast phenotype by macrophages [161] thus contributing to promote osteolysis.
Among the above mentioned genes, NR4A3 and CLEC5A/MDL1 are abundantly expressed by activated macrophages [162,163]. Interestingly, during joint inflammation, overexpression of CLEC5A/MDL-1 recruits inflammatory cells and induces the production of IL-1, IL-6, IL-17A, and TNF, contributing to cartilage damage and bone erosion. FCGR2B, present on several cell types including synovial mononuclear phagocytes, monocytes, neutrophils and myeloid dendritic cells (DCs) [164] is an inhibitory receptor with important regulatory functions on Fc receptor activation, which has been found overexpressed in RA synovium samples [165]. Noteworthy variation in the gene encoding FCGR2B has been associated with susceptibility to autoimmune disease such as RA and SLE [166].
Another important gene cluster is the signal transduction gene category which includes several modulated transcripts particularly important for PsA pathogenesis. Indeed we noticed overexpression of 13 IFIG in PsA synovium, thus indicating the presence of an IFN type I signature, typically associated with autoimmune disease such as SLE, RA, Crohn’s disease and Sjogren syndrome [94–100]. Noteworthy a type I interferon signature is present also in DEGs from PBC of patients with PsA, indicating that indeed peripheral blood cells reflects, although at lower level, the gene modulation present in the synovium.
Finally, DEGs indicate a T cell immune response with prevalent upregulation of several Th17 related genes both in PsA synovium (8 upregulated transcripts) and, to a lesser extent, in PBC (5 upregulated genes). Th17 cells have been implicated in autoimmune diseases including SLE, RA and psoriasis. In these last two disease conditions, experimental evidence indicates that Th17 cells and the related IL-17/IL-23 cytokine axis, play a role in disease pathogenesis in animal models and in the human disease suggesting that targeting the IL-17/IL23 cytokine axis may represent a promising therapeutic strategy. In this regard it is now clear that Th17 cells and related cytokines play a crucial role in sustaining autoimmunity especially when associated to type I IFN-driven inflammation. A recent report [20] shows that CCR6+ memory T-helper cells producing IL-17A, IL-17F, IL-21, and/or IL-22 are increased in SLE patients and that this increase is related to the presence of IFN type I signature thus providing evidence that IFN type I signature co-acts with Th17 cells and related cytokines in the pathogenesis of systemic autoimmune diseases such as SLE.
In order to further confirm our gene expression data on overexpression of IFIG and Th17 pathways, we analysed the presence of IL-17 producing CD4+ T cells and found a significantly increased percentage of these cells in PBMC of patients with PsA compared with normal subjects. Moreover the levels of IL-17 and IL-23 in synovial fluid were higher than in control synovial fluids further confirming the findings of the gene array analysis. Taken together these data suggest an autoimmune origin of PsA, probably through the activation of the IL-23/IL-17 cytokine network. Indeed PsA has always been considered of autoimmune origin, driven by autoreactive T cells directed against autoantigens present in the skin and in the joints. This view has been recently questioned by McGonagle et al. [167] who have proposed that PsA may be considered an autoinflammatory rather than an autoimmune disease. One of the reasons adduced is that the autoimmune model would fail to explain diffuse enthesitis, and that the proposed autoantigens common to skin and joints have not been identified so far. However a recent work by Sherlock et al. [168,169]shows that IL-23 is able to induce enthesitis in animal models of spondyloarthropathy acting on resident T cells within the enthesis; once activated these entheseal T cells can promote local inflammation and bone remodeling through a variety of effector molecules such as IL-17 and IL-22. Therefore these findings suggest that mechanisms other then local injury (microtrauma, microdamage, altered vascularity and repair) [170] may be responsible for the onset of enthesitis and that enthesitis may be sustained by effector mediators of the Th17 cell subset. As far as the absence of autoantigens common to skin and joints adduced as evidence for an autoinflammatory origin of the disease we have recently found that PsA is characterized by the presence of serum autoantibodies crossreacting with an epitope shared by skin and joint antigens [171].
Finally our work shows that the two genes, SSP1 and GPNMB, encoding for osteopontin and osteoactivin are among the most expressed genes found in the gene array analysis (FC 450 and 147). Osteopontin (OPN), also called bone sialoprotein I (BSP-1 or BNSP), is encoded by the SPP1 gene (secreted phosphoprotein 1), first identified in osteoblasts. OPN is biosynthesized by a variety of cells including fibroblasts, osteoblasts, osteocytes, chondrocytes, some bone marrow cells, dendritic cells, endothelial cells. OPN plays a role in bone metabolism (bone mineralization and remodeling) but has also a role in immunity. OPN is a cytokine with pleiotropic effect produced by activated T cells, dendritic cells and macrophages. It is released at increased levels during inflammation and the secreted form (s-OPN) has been shown to act as a chemoattractant for many cells through integrin receptors and CD44. Recently, it has been shown that OPN is able to regulate the expression of Toll like receptor-9 (TLR-9), of TLR-7-dependent interferon-alpha (IFN-alpha) in plasmacytoid dendritic cells. Moreover it seems to be implicated in Th17 developmenent [172]. Indeed OPN seems to play a role in the pathogenesis of several autoimmune diseases including RA, MS and SLE [173]. Osteoactivin (OA), also known as transmembrane glycoprotein GPNMB, is a type I glycoprotein expressed in osteoblasts, osteoclasts, melanocytes and other cell types. In osteoblast progenitors OA is a positive regulator of osteoblast differentiation in the final phases of matrix maturation and mineralization [174]. In addition, as observed in bone fracture in animal models, OA is able to accelerate bone repair [175].
We have found that osteopontin is elevated also in the sera of subjects with RA and AS, whereas high levels of osteoactivin are present only in the sera of PsA patients.
In conclusion, we report here the gene array analysis of paired SM and PBC in patients with PsA; the analysis has identified the modulation of cluster of genes encoding for molecules involved in the pathogenesis of the disease. The findings have been validated with different methods and further support that PsA is of autoimmune origin.
Moreover, one of the more expressed genes encodes for a molecule, osteoactivin, that is selectively elevated in the sera of patients with PsA suggesting that this molecule may be used as a marker of the disease.
Supporting Information
(DOC)
(DOC)
Data Availability
All the relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Eder L, Gladman DD (2013) Psoriatic arthritis: phenotypic variance and nosology. Curr Rheumatol Rep 15: 316 10.1007/s11926-013-0316-4 [DOI] [PubMed] [Google Scholar]
- 2. Rahman P, Elder JT (2005) Genetic epidemiology of psoriasis and psoriatic arthritis. Ann Rheum Dis 64 Suppl 2: ii37–39; discussion ii40-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ritchlin CT (2005) Pathogenesis of psoriatic arthritis. Curr Opin Rheumatol 17: 406–412. [DOI] [PubMed] [Google Scholar]
- 4. Korendowych E, McHugh N (2005) Genetic factors in psoriatic arthritis. Curr Rheumatol Rep 7: 306–312. [DOI] [PubMed] [Google Scholar]
- 5. Moll JM, Wright V (1973) Psoriatic arthritis. Semin Arthritis Rheum 3: 55–78. [DOI] [PubMed] [Google Scholar]
- 6. Mease P (2013) Psoriatic arthritis and spondyloarthritis assessment and management update. Curr Opin Rheumatol 25: 287–296. 10.1097/BOR.0b013e32835fd8d5 [DOI] [PubMed] [Google Scholar]
- 7. McGonagle DG, Helliwell P, Veale D (2012) Enthesitis in psoriatic disease. Dermatology 225: 100–109. 10.1159/000341536 [DOI] [PubMed] [Google Scholar]
- 8. McQueen F, Lassere M, Ostergaard M (2006) Magnetic resonance imaging in psoriatic arthritis: a review of the literature. Arthritis Res Ther 8: 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anandarajah A (2013) Imaging in psoriatic arthritis. Clin Rev Allergy Immunol 44: 157–165. [DOI] [PubMed] [Google Scholar]
- 10. Veale DJ, Ritchlin C, FitzGerald O (2005) Immunopathology of psoriasis and psoriatic arthritis. Ann Rheum Dis 64 Suppl 2: ii26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kruithof E, Baeten D, De Rycke L, Vandooren B, Foell D, Roth J, et al. (2005) Synovial histopathology of psoriatic arthritis, both oligo- and polyarticular, resembles spondyloarthropathy more than it does rheumatoid arthritis. Arthritis Res Ther 7: R569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivieri I, D'Angelo S, Palazzi C, Padula A (2014) Advances in the management of psoriatic arthritis. Nat Rev Rheumatol. [DOI] [PubMed]
- 13. Staudt LM (2002) Gene expression profiling of lymphoid malignancies. Annu Rev Med 53: 303–318. [DOI] [PubMed] [Google Scholar]
- 14. Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100: 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batliwalla FM, Baechler EC, Xiao X, Li W, Balasubramanian S, Khalili H, et al. (2005) Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun 6: 388–397. [DOI] [PubMed] [Google Scholar]
- 16. Bomprezzi R, Ringner M, Kim S, Bittner ML, Khan J, Chen Y, et al. (2003) Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum Mol Genet 12: 2191–2199. [DOI] [PubMed] [Google Scholar]
- 17. Crow MK (2010) Type I interferon in organ-targeted autoimmune and inflammatory diseases. Arthritis Res Ther 12 Suppl 1: S5 10.1186/ar2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ambrosi A, Espinosa A, Wahren-Herlenius M (2012) IL-17: a new actor in IFN-driven systemic autoimmune diseases. Eur J Immunol 42: 2274–2284. 10.1002/eji.201242653 [DOI] [PubMed] [Google Scholar]
- 19. Axtell RC, de Jong BA, Boniface K, van der Voort LF, Bhat R, De Sarno P, et al. (2010) T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat Med 16: 406–412. 10.1038/nm.2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brkic Z, Corneth OB, van Helden-Meeuwsen CG, Dolhain RJ, Maria NI, Paulissen SM, et al. (2014) T helper 17 cell cytokines and interferon type I: partners in crime in systemic lupus erythematosus? Arthritis Res Ther 16: R62 10.1186/ar4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dolcino M, Lunardi C, Ottria A, Tinazzi E, Patuzzo G, Puccetti A (2014) Crossreactive autoantibodies directed against cutaneous and joint antigens are present in psoriatic arthritis. PLoS One. 9(12):e115424 10.1371/journal.pone.0115424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duarte GV, Faillace C, Freire de Carvalho J (2012) Psoriatic arthritis. Best Pract Res Clin Rheumatol 26: 147–156. 10.1016/j.berh.2012.01.003 [DOI] [PubMed] [Google Scholar]
- 23. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd et al. (2010) 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62: 2569–2581. 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 24. Raychaudhuri SP, Deodhar A (2014) The classification and diagnostic criteria of ankylosing spondylitis. J Autoimmun 48–49: 128–133. [DOI] [PubMed] [Google Scholar]
- 25. Dolcino M, Cozzani E, Riva S, Parodi A, Tinazzi E, Lunardi C, et al. (2012) Gene expression profiling in dermatitis herpetiformis skin lesions. Clin Dev Immunol 2012: 198956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tinazzi E, Dolcino M, Puccetti A, Rigo A, Beri R, Valenti MT, et al. (2010) Gene expression profiling in circulating endothelial cells from systemic sclerosis patients shows an altered control of apoptosis and angiogenesis that is modified by iloprost infusion. Arthritis Res Ther. 2010;12(4):R131 10.1186/ar3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dolcino M, Patuzzo G, Barbieri A, Tinazzi E, Rizzi M, Beri R, et al. (2014) Gene expression profiling in peripheral blood mononuclear cells of patients with common variable immunodeficiency: modulation of adaptive immune response following intravenous immunoglobulin therapy. PLoS One. 9(5):e97571 10.1371/journal.pone.0097571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Abdelmagid SM, Belcher JY, Moussa FM, Lababidi SL, Sondag GR, Novak KM, et al. (2014) Mutation in osteoactivin decreases bone formation in vivo and osteoblast differentiation in vitro. Am J Pathol 184: 697–713. 10.1016/j.ajpath.2013.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakki A, Kouhia ST, Saarela J, Harilainen A, Tallroth K, Videman T, et al. (2010) Allelic variants of IL1R1 gene associate with severe hand osteoarthritis. BMC Med Genet 11: 50 10.1186/1471-2350-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rahman P, Sun S, Peddle L, Snelgrove T, Melay W, Greenwood C, et al. (2006) Association between the interleukin-1 family gene cluster and psoriatic arthritis. Arthritis Rheum 54: 2321–2325. [DOI] [PubMed] [Google Scholar]
- 32. Yammani RR (2012) S100 proteins in cartilage: role in arthritis. Biochim Biophys Acta 1822: 600–606. 10.1016/j.bbadis.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song C, Zhou X, Dong Q, Fan R, Wu G, Biao Ji, et al. (2012) Regulation of inflammatory response in human chondrocytes by lentiviral mediated RNA interference against S100A10. Inflamm Res 61: 1219–1227. 10.1007/s00011-012-0519-6 [DOI] [PubMed] [Google Scholar]
- 34. La Fleur M, Beaulieu AD, Kreis C, Poubelle P (1987) Fibronectin gene expression in polymorphonuclear leukocytes. Accumulation of mRNA in inflammatory cells. J Biol Chem 262: 2111–2115. [PubMed] [Google Scholar]
- 35. Takagi R, Higashi T, Hashimoto K, Nakano K, Mizuno Y, Yosuke Mizuno, et al. (2008) B cell chemoattractant CXCL13 is preferentially expressed by human Th17 cell clones. J Immunol 181: 186–189. [DOI] [PubMed] [Google Scholar]
- 36. Qu N, Xu M, Mizoguchi I, Furusawa J, Kaneko K, Watanabe K, et al. (2013) Pivotal roles of T-helper 17-related cytokines, IL-17, IL-22, and IL-23, in inflammatory diseases. Clin Dev Immunol 2013: 968549 10.1155/2013/968549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rutz S, Noubade R, Eidenschenk C, Ota N, Zeng W, Zheng Y, et al. (2011) Transcription factor c-Maf mediates the TGF-beta-dependent suppression of IL-22 production in T(H)17 cells. Nat Immunol 12: 1238–1245. 10.1038/ni.2134 [DOI] [PubMed] [Google Scholar]
- 38. Braum O, Pirzer H, Fickenscher H (2012) Interleukin-26, a highly cationic T-cell cytokine targeting epithelial cells. Antiinflamm Antiallergy Agents Med Chem 11: 221–229. [DOI] [PubMed] [Google Scholar]
- 39. Suzuki E, Mellins ED, Gershwin ME, Nestle FO, Adamopoulos IE (2014) The IL-23/IL-17 axis in psoriatic arthritis. Autoimmun Rev 13: 496–502. 10.1016/j.autrev.2014.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hemdan NY, Birkenmeier G, Wichmann G, Abu El-Saad AM, Krieger T, Thorsten Krieger, et al. (2010) Interleukin-17-producing T helper cells in autoimmunity. Autoimmun Rev 9: 785–792. 10.1016/j.autrev.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 41. Rodig SJ, Kutok JL, Paterson JC, Nitta H, Zhang W, Chapuy B, et al. (2010) The pre-B-cell receptor associated protein VpreB3 is a useful diagnostic marker for identifying c-MYC translocated lymphomas. Haematologica 95: 2056–2062. 10.3324/haematol.2010.025767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson TA, Haga CL, Ehrhardt GR, Davis RS, Cooper MD (2010) FcR-like 2 Inhibition of B cell receptor-mediated activation of B cells. J Immunol 185: 7405–7412. 10.4049/jimmunol.1002305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. E XQ, Meng HX, Cao Y, Zhang SQ, Bi ZG, Yamakawa M (2012) Distribution of regulatory T cells and interaction with dendritic cells in the synovium of rheumatoid arthritis. Scand J Rheumatol 41: 413–420. 10.3109/03009742.2012.696135 [DOI] [PubMed] [Google Scholar]
- 44. O'Connor BP, Ting JP (2008) The evolving role of semaphorins and plexins in the immune system: Plexin-A1 regulation of dendritic cell function. Immunol Res 41: 217–222. 10.1007/s12026-008-8026-0 [DOI] [PubMed] [Google Scholar]
- 45. van Vliet SJ, Saeland E, van Kooyk Y (2008) Sweet preferences of MGL: carbohydrate specificity and function. Trends Immunol 29: 83–90. 10.1016/j.it.2007.10.010 [DOI] [PubMed] [Google Scholar]
- 46. Tanaka M, Nagai T, Tsuneyoshi Y, Sunahara N, Matsuda T, Nakamura T, et al. (2008) Expansion of a unique macrophage subset in rheumatoid arthritis synovial lining layer. Clin Exp Immunol 154: 38–47. 10.1111/j.1365-2249.2008.03733.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu L, Yin W, Xia J, Peng M, Li S, Lin S, et al. (2012) An antiapoptotic role of sorting nexin 7 is required for liver development in zebrafish. Hepatology 55: 1985–1993. 10.1002/hep.25560 [DOI] [PubMed] [Google Scholar]
- 48. Toyoshima Y, Karas M, Yakar S, Dupont J, Lee H, LeRoith D (2004) TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J Biol Chem 279: 25898–25904. [DOI] [PubMed] [Google Scholar]
- 49. Li W, Yeo LS, Vidal C, McCorquodale T, Herrmann M, LeRoith D (2011) Decreased bone formation and osteopenia in lamin a/c-deficient mice. PLoS One 6: e19313 10.1371/journal.pone.0019313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu XS, Masedunskas A, Weigert R, Copeland NG, Jenkins NA, Hammer JA (2012) Melanoregulin regulates a shedding mechanism that drives melanosome transfer from melanocytes to keratinocytes. Proc Natl Acad Sci U S A 109: E2101–2109. 10.1073/pnas.1209397109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujikawa K, Shiraki K, Sugimoto K, Ito T, Yamanaka T, Takase K, et al. (2000) Reduced expression of ICE/caspase1 and CPP32/caspase3 in human hepatocellular carcinoma. Anticancer Res 20: 1927–1932. [PubMed] [Google Scholar]
- 52. Wajant H (2003) Death receptors. Essays Biochem 39: 53–71. [DOI] [PubMed] [Google Scholar]
- 53. Lu Y, Stinnette TW, Westrick E, Klein PJ, Gehrke MA, Cross VA, et al. (2011) Treatment of experimental adjuvant arthritis with a novel folate receptor-targeted folic acid-aminopterin conjugate. Arthritis Res Ther 13: R56 10.1186/ar3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boncela J, Przygodzka P, Papiewska-Pajak I, Wyroba E, Cierniewski CS (2011) Association of plasminogen activator inhibitor type 2 (PAI-2) with proteasome within endothelial cells activated with inflammatory stimuli. J Biol Chem 286: 43164–43171. 10.1074/jbc.M111.245647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cai CK, Zhao GY, Tian LY, Liu L, Yan K, Ma YL, et al. (2012) miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep 28: 1764–1770. 10.3892/or.2012.1995 [DOI] [PubMed] [Google Scholar]
- 56. Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, et al. (2000) The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J Neurosci 20: RC83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oh MJ, van Agthoven T, Choi JE, Jeong YJ, Chung YH, Kim CM, et al. (2008) BCAR3 regulates EGF-induced DNA synthesis in normal human breast MCF-12A cells. Biochem Biophys Res Commun 375: 430–434. 10.1016/j.bbrc.2008.08.040 [DOI] [PubMed] [Google Scholar]
- 58. Hossain MN, Sakemura R, Fujii M, Ayusawa D (2006) G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun 351: 645–650. [DOI] [PubMed] [Google Scholar]
- 59. Vessichelli M, Ferravante A, Zotti T, Reale C, Scudiero I, Picariello G, et al. (2012) Neuroepithelial transforming gene 1 (Net1) binds to caspase activation and recruitment domain (CARD)- and membrane-associated guanylate kinase-like domain-containing (CARMA) proteins and regulates nuclear factor kappaB activation. J Biol Chem 287: 13722–13730. 10.1074/jbc.M111.304436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ruiz-Heiland G, Zhao Y, Derer A, Braun T, Engelke K, Neumann E, et al. (2013) Deletion of the receptor tyrosine kinase Tyro3 inhibits synovial hyperplasia and bone damage in arthritis. Ann Rheum Dis. [DOI] [PubMed] [Google Scholar]
- 61. Rotival M, Zeller T, Wild PS, Maouche S, Szymczak S, Schillert A, et al. (2011) Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans. PLoS Genet 7: e1002367 10.1371/journal.pgen.1002367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu K, Xu P, Yao JF, Zhang YG, Hou WK, Lu SM (2013) Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis. Inflamm Res 62: 229–237. 10.1007/s00011-012-0572-1 [DOI] [PubMed] [Google Scholar]
- 63. Jain A, Tindell CA, Laux I, Hunter JB, Curran J, Galkin A, et al. (2005) Epithelial membrane protein-1 is a biomarker of gefitinib resistance. Proc Natl Acad Sci U S A 102: 11858–11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stoyanova T, Goldstein AS, Cai H, Drake JM, Huang J, Witte ON (2012) Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via beta-catenin signaling. Genes Dev 26: 2271–2285. 10.1101/gad.196451.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, et al. (1995) p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev 9: 650–662. [DOI] [PubMed] [Google Scholar]
- 66. Barnes EA, Kong M, Ollendorff V, Donoghue DJ (2001) Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J 20: 2214–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miyazawa Y, Uekita T, Hiraoka N, Fujii S, Kosuge T, Kanai Y, et al. (2010) CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res 70: 5136–5146. 10.1158/0008-5472.CAN-10-0220 [DOI] [PubMed] [Google Scholar]
- 68. Zhuo J, Tan EH, Yan B, Tochhawng L, Jayapal M, Koh S, et al. (2012) Gelsolin induces colorectal tumor cell invasion via modulation of the urokinase-type plasminogen activator cascade. PLoS One 7: e43594 10.1371/journal.pone.0043594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jeong CY, Hah YS, Cho BI, Lee SM, Joo YT, Jung EJ, et al. (2012) Fatty acid-binding protein 5 promotes cell proliferation and invasion in human intrahepatic cholangiocarcinoma. Oncol Rep 28: 1283–1292. 10.3892/or.2012.1922 [DOI] [PubMed] [Google Scholar]
- 70. Power PF, Mak IW, Singh S, Popovic S, Gladdy R, Ghert M (2013) ETV5 as a regulator of matrix metalloproteinase 2 in human chondrosarcoma. J Orthop Res 31: 493–501. 10.1002/jor.22227 [DOI] [PubMed] [Google Scholar]
- 71. Wang J, Chen L, Li Y, Guan XY (2011) Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PLoS One 6: e24967 10.1371/journal.pone.0024967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Charoenwattanasatien R, Cairns JR, Keeratichamroen S, Sawangareetrakul P, Tanpaiboon P, Wattanasirichaigoon D, et al. (2012) Decreasing activity and altered protein processing of human iduronate-2-sulfatase mutations demonstrated by expression in COS7 cells. Biochem Genet 50: 990–997. 10.1007/s10528-012-9538-9 [DOI] [PubMed] [Google Scholar]
- 73. Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, et al. (2008) The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol 40: 1111–1125. 10.1016/j.biocel.2008.01.030 [DOI] [PubMed] [Google Scholar]
- 74. Chen T, Lee TR, Liang WG, Chang WS, Lyu PC (2009) Identification of trypsin-inhibitory site and structure determination of human SPINK2 serine proteinase inhibitor. Proteins 77: 209–219. 10.1002/prot.22432 [DOI] [PubMed] [Google Scholar]
- 75. Patterson EI, Swarbrick CM, Roman N, Forwood JK, Raidal SR (2013) Differential expression of two isolates of beak and feather disease virus capsid protein in Escherichia coli. J Virol Methods 189: 118–124. 10.1016/j.jviromet.2013.01.020 [DOI] [PubMed] [Google Scholar]
- 76. Venkateswaran A, Friedman DB, Walsh AJ, Skala MC, Sasi S, Rachakonda G, et al. (2013) The novel antiangiogenic VJ115 inhibits the NADH oxidase ENOX1 and cytoskeleton-remodeling proteins. Invest New Drugs 31: 535–544. 10.1007/s10637-012-9884-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Eto N, Miyagishi M, Inagi R, Fujita T, Nangaku M (2009) Mitogen-activated protein 3 kinase 6 mediates angiogenic and tumorigenic effects via vascular endothelial growth factor expression. Am J Pathol 174: 1553–1563. 10.2353/ajpath.2009.080190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han SW, Jung YK, Lee EJ, Park HR, Kim GW, Jeong JH, et al. (2013) DICAM inhibits angiogenesis via suppression of AKT and p38 MAP kinase signalling. Cardiovasc Res 98: 73–82. 10.1093/cvr/cvt019 [DOI] [PubMed] [Google Scholar]
- 79. Alves RD, Eijken M, Swagemakers S, Chiba H, Titulaer MK, Burgers PC, et al. (2010) Proteomic analysis of human osteoblastic cells: relevant proteins and functional categories for differentiation. J Proteome Res 9: 4688–4700. 10.1021/pr100400d [DOI] [PubMed] [Google Scholar]
- 80. Mentink A, Hulsman M, Groen N, Licht R, Dechering KJ, Alves HA, et al. (2013) Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials 34: 4592–4601. 10.1016/j.biomaterials.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 81. Bundschu K, Knobeloch KP, Ullrich M, Schinke T, Amling M, Engelhardt CM et al. (2005) Gene disruption of Spred-2 causes dwarfism. J Biol Chem 280: 28572–28580. [DOI] [PubMed] [Google Scholar]
- 82. Goding JW, Grobben B, Slegers H (2003) Physiological and pathophysiological functions of the ecto-nucleotide pyrophosphatase/phosphodiesterase family. Biochim Biophys Acta 1638: 1–19. [DOI] [PubMed] [Google Scholar]
- 83. Alsalameh S, Amin R, Gemba T, Lotz M (2004) Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum 50: 1522–1532. [DOI] [PubMed] [Google Scholar]
- 84. Gordon JA, Hassan MQ, Saini S, Montecino M, van Wijnen AJ, Stein GS, et al. (2010) Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol 30: 3531–3541. 10.1128/MCB.00889-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, et al. (2008) Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem 283: 27585–27597. 10.1074/jbc.M802453200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Costa D, Lazzarini E, Canciani B, Giuliani A, Spano R, Marozzi K, et al. (2013) Altered bone development and turnover in transgenic mice over-expressing Lipocalin-2 in bone. J Cell Physiol 228: 2210–2221. 10.1002/jcp.24391 [DOI] [PubMed] [Google Scholar]
- 87. Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, et al. (2007) Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci 120: 2785–2795. [DOI] [PubMed] [Google Scholar]
- 88. Zanotti S, Smerdel-Ramoya A, Canalis E (2011) HES1 (hairy and enhancer of split 1) is a determinant of bone mass. J Biol Chem 286: 2648–2657. 10.1074/jbc.M110.183038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mahoney DJ, Swales C, Athanasou NA, Bombardieri M, Pitzalis C, Kliskey K, et al. (2011) TSG-6 inhibits osteoclast activity via an autocrine mechanism and is functionally synergistic with osteoprotegerin. Arthritis Rheum 63: 1034–1043. 10.1002/art.30201 [DOI] [PubMed] [Google Scholar]
- 90. Logar DB, Komadina R, Prezelj J, Ostanek B, Trost Z, Marc J (2007) Expression of bone resorption genes in osteoarthritis and in osteoporosis. J Bone Miner Metab 25: 219–225. [DOI] [PubMed] [Google Scholar]
- 91. Hirvonen MJ, Mulari MT, Buki KG, Vihko P, Harkonen PL, Vaananen HK (2012) Rab13 is upregulated during osteoclast differentiation and associates with small vesicles revealing polarized distribution in resorbing cells. J Histochem Cytochem 60: 537–549. 10.1369/0022155412448069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mensah KA, Schwarz EM, Ritchlin CT (2008) Altered bone remodeling in psoriatic arthritis. Curr Rheumatol Rep 10: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ronnblom L, Eloranta ML (2013) The interferon signature in autoimmune diseases. Curr Opin Rheumatol 25: 248–253. 10.1097/BOR.0b013e32835c7e32 [DOI] [PubMed] [Google Scholar]
- 94. Gordon RA, Grigoriev G, Lee A, Kalliolias GD, Ivashkiv LB (2012) The interferon signature and STAT1 expression in rheumatoid arthritis synovial fluid macrophages are induced by tumor necrosis factor alpha and counter-regulated by the synovial fluid microenvironment. Arthritis Rheum 64: 3119–3128. 10.1002/art.34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nzeusseu Toukap A, Galant C, Theate I, Maudoux AL, Lories RJ, Houssiau FA, et al. (2007) Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum 56: 1579–1588. [DOI] [PubMed] [Google Scholar]
- 96. Thurlings RM, Boumans M, Tekstra J, van Roon JA, Vos K, van Westingdm, et al. (2010) Relationship between the type I interferon signature and the response to rituximab in rheumatoid arthritis patients. Arthritis Rheum 62: 3607–3614. 10.1002/art.27702 [DOI] [PubMed] [Google Scholar]
- 97. Raterman HG, Vosslamber S, de Ridder S, Nurmohamed MT, Lems WF, Boers M, et al. (2012) The interferon type I signature towards prediction of non-response to rituximab in rheumatoid arthritis patients. Arthritis Res Ther 14: R95 10.1186/ar3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Moschella F, Torelli GF, Valentini M, Urbani F, Buccione C, Petrucci MT, et al. (2013) Cyclophosphamide induces a type I interferon-associated sterile inflammatory response signature in cancer patients' blood cells: implications for cancer chemoimmunotherapy. Clin Cancer Res 19: 4249–4261. 10.1158/1078-0432.CCR-12-3666 [DOI] [PubMed] [Google Scholar]
- 99. Maria NI, Brkic Z, Waris M, van Helden-Meeuwsen CG, Heezen K, van de Merwe JP, et al. (2013) MxA as a clinically applicable biomarker for identifying systemic interferon type I in primary Sjogren's syndrome. Ann Rheum Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Caignard G, Lucas-Hourani M, Dhondt KP, Labernardiere JL, Petit T, Vidalain JP (2013) The V protein of Tioman virus is incapable of blocking type I interferon signaling in human cells. PLoS One 8: e53881 10.1371/journal.pone.0053881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miao CG, Yang YY, He X, Li XF, Huang C, Huang Y, et al. (2013) Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal 25: 2069–2078. 10.1016/j.cellsig.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 102. Upadhyay G, Goessling W, North TE, Xavier R, Zon LI, Yajnik V (2008) Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene 27: 5845–5855. 10.1038/onc.2008.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Guang W, Twaddell WS, Lillehoj EP (2012) Molecular Interactions between MUC1 Epithelial Mucin, beta-Catenin, and CagA Proteins. Front Immunol 3: 105 10.3389/fimmu.2012.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hino S, Michiue T, Asashima M, Kikuchi A (2003) Casein kinase I epsilon enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of beta-catenin. J Biol Chem 278: 14066–14073. [DOI] [PubMed] [Google Scholar]
- 105. Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L, et al. (2012) Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A 109: 4044–4051. 10.1073/pnas.1200421109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Q, Symes AJ, Kane CA, Freeman A, Nariculam J, Munson P, et al. (2010) A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer. PLoS One 5: e10456 10.1371/journal.pone.0010456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Talhouk RS, Fares MB, Rahme GJ, Hariri HH, Rayess T, Dbuok HA, et al. (2013) Context dependent reversion of tumor phenotype by connexin-43 expression in MDA-MB231 cells and MCF-7 cells: Role of beta-catenin/connexin43 association. Exp Cell Res 319: 3065–3080. 10.1016/j.yexcr.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 108. Park JS, Kim SH, Kim K, Jin CH, Choi KY (2013) Inhibition of Notch signalling ameliorates experimental inflammatory arthritis. Ann Rheum Dis. [DOI] [PubMed] [Google Scholar]
- 109. Mavropoulos A, Orfanidou T, Liaskos C, Smyk DS, Billinis C, Billinis C, et al. (2013) p38 mitogen-activated protein kinase (p38 MAPK)-mediated autoimmunity: lessons to learn from ANCA vasculitis and pemphigus vulgaris. Autoimmun Rev 12: 580–590. 10.1016/j.autrev.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 110. Guo S (2014) Insulin signaling, resistance, and the metabolic syndrome: insights from mouse models into disease mechanisms. J Endocrinol 220: T1–T23. 10.1530/JOE-13-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Russolillo A, Iervolino S, Peluso R, Lupoli R, Di Minno A, Pappone N, et al. (2013) Obesity and psoriatic arthritis: from pathogenesis to clinical outcome and management. Rheumatology (Oxford) 52: 62–67. 10.1093/rheumatology/kes242 [DOI] [PubMed] [Google Scholar]
- 112. Kim CH (2009) Migration and function of Th17 cells. Inflamm Allergy Drug Targets 8: 221–228. [DOI] [PubMed] [Google Scholar]
- 113. Maddur MS, Miossec P, Kaveri SV, Bayry J (2012) Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am J Pathol 181: 8–18. 10.1016/j.ajpath.2012.03.044 [DOI] [PubMed] [Google Scholar]
- 114. Chatterjee M, Hedrich CM, Rauen T, Ioannidis C, Terhorst C, Tsokos GC (2012) CD3-T cell receptor co-stimulation through SLAMF3 and SLAMF6 receptors enhances RORgammat recruitment to the IL17A promoter in human T lymphocytes. J Biol Chem 287: 38168–38177. 10.1074/jbc.M112.415067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhors C, et al. (2012) Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol 188: 1206–1212. 10.4049/jimmunol.1102773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ogura H, Murakami M, Okuyama Y, Tsuruoka M, Kitabayashi C, Kanamoto M, et al. (2008) Interleukin-17 promotes autoimmunity by triggering a positive-feedback loop via interleukin-6 induction. Immunity 29: 628–636. 10.1016/j.immuni.2008.07.018 [DOI] [PubMed] [Google Scholar]
- 117. Li Y, Yu C, Zhu WM, Xie Y, Qi X, Li N, et al. (2010) Triptolide ameliorates IL-10-deficient mice colitis by mechanisms involving suppression of IL-6/STAT3 signaling pathway and down-regulation of IL-17. Mol Immunol 47: 2467–2474. 10.1016/j.molimm.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 118. Jones GW, Greenhill CJ, Williams JO, Nowell MA, Williams AS, Jenkings BJ, et al. (2013) Exacerbated inflammatory arthritis in response to hyperactive gp130 signalling is independent of IL-17A. Ann Rheum Dis 72: 1738–1742. 10.1136/annrheumdis-2013-203771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang RX, Yu CR, Mahdi RM, Egwuagu CE (2012) Novel IL27p28/IL12p40 cytokine suppressed experimental autoimmune uveitis by inhibiting autoreactive Th1/Th17 cells and promoting expansion of regulatory T cells. J Biol Chem 287: 36012–36021. 10.1074/jbc.M112.390625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li H, Wan A (2013) Apoptosis of rheumatoid arthritis fibroblast-like synoviocytes: possible roles of nitric oxide and the thioredoxin 1. Mediators Inflamm 2013: 953462 10.1155/2013/953462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Huang H, Xiao Y, Lin H, Fu D, Zhan Z, Liang L, et al. (2011) Increased phosphorylation of ezrin/radixin/moesin proteins contributes to proliferation of rheumatoid fibroblast-like synoviocytes. Rheumatology (Oxford) 50: 1045–1053. 10.1093/rheumatology/keq440 [DOI] [PubMed] [Google Scholar]
- 122. Chang SK, Gu Z, Brenner MB (2010) Fibroblast-like synoviocytes in inflammatory arthritis pathology: the emerging role of cadherin-11. Immunol Rev 233: 256–266. 10.1111/j.0105-2896.2009.00854.x [DOI] [PubMed] [Google Scholar]
- 123. Pollock RA, Abji F, Liang K, Chandran V, Pellett FJ, Virtanen C, et al. (2015) Gene Expression Differences between Psoriasis Patients with and without Inflammatory Arthritis. J Invest Dermatol. 135:620–3. 10.1038/jid.2014.414 [DOI] [PubMed] [Google Scholar]
- 124. Al-Alwan M, Olabi S, Ghebeh H, Barhoush E, Tulbah A, Al-Tweigeri T, et al. (2011) Fascin is a key regulator of breast cancer invasion that acts via the modification of metastasis-associated molecules. PLoS One 6: e27339 10.1371/journal.pone.0027339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kane D, Jensen LE, Grehan S, Whitehead AS, Bresnihan B, Fitzgerald O (2004) Quantitation of metalloproteinase gene expression in rheumatoid and psoriatic arthritis synovial tissue distal and proximal to the cartilage-pannus junction. J Rheumatol 31: 1274–1280. [PubMed] [Google Scholar]
- 126. Ramonda R, Modesti V, Ortolan A, Scanu A, Bassi N, Oliviero F, et al. (2013) Serological markers in psoriatic arthritis: promising tools. Exp Biol Med (Maywood) 238: 1431–1436. 10.1177/1535370213506435 [DOI] [PubMed] [Google Scholar]
- 127. Patterson AM, Cartwright A, David G, Fitzgerald O, Bresnihan B, Ashton BA, et al. (2008) Differential expression of syndecans and glypicans in chronically inflamed synovium. Ann Rheum Dis 67: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Korb-Pap A, Stratis A, Muhlenberg K, Niederreiter B, Hayer S, Stange R, et al. (2012) Early structural changes in cartilage and bone are required for the attachment and invasion of inflamed synovial tissue during destructive inflammatory arthritis. Ann Rheum Dis 71: 1004–1011. 10.1136/annrheumdis-2011-200386 [DOI] [PubMed] [Google Scholar]
- 129. Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, et al. (2009) Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat Med 15: 1072–1076. 10.1038/nm.1998 [DOI] [PubMed] [Google Scholar]
- 130. Chimenti MS, Ballanti E, Perricone C, Cipriani P, Giacomelli R, Perricone R (2013) Immunomodulation in psoriatic arthritis: focus on cellular and molecular pathways. Autoimmun Rev 12: 599–606. 10.1016/j.autrev.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 131. Yamamoto T (2013) Angiogenic and inflammatory properties of psoriatic arthritis. ISRN Dermatol 2013: 630620 10.1155/2013/630620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Fearon U, Veale DJ (2007) Angiogenesis in arthritis: methodological and analytical details. Methods Mol Med 135: 343–357. [DOI] [PubMed] [Google Scholar]
- 133. Weng LH, Ko JY, Wang CJ, Sun YC, Wang FS (2012) Dkk-1 promotes angiogenic responses and cartilage matrix proteinase secretion in synovial fibroblasts from osteoarthritic joints. Arthritis Rheum 64: 3267–3277. 10.1002/art.34602 [DOI] [PubMed] [Google Scholar]
- 134. Kitamoto S, Yokoyama S, Higashi M, Yamada N, Takao S, Yonezawa S (2012) MUC1 enhances hypoxia-driven angiogenesis through the regulation of multiple proangiogenic factors. Oncogene. [DOI] [PubMed] [Google Scholar]
- 135. Strehl C, Fangradt M, Fearon U, Gaber T, Buttgereit F, Veale DJ (2014) Hypoxia: how does the monocyte-macrophage system respond to changes in oxygen availability? J Leukoc Biol 95: 233–241. 10.1189/jlb.1212627 [DOI] [PubMed] [Google Scholar]
- 136. Yang L, Chen G, Mohanty S, Scott G, Fazal F, Rahman A, et al. (2011) GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res 71: 5558–5568. 10.1158/0008-5472.CAN-10-4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bornstein P (2009) Thrombospondins function as regulators of angiogenesis. J Cell Commun Signal 3: 189–200. 10.1007/s12079-009-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hsieh JL, Shen PC, Shiau AL, Jou IM, Lee CH, Wang C, et al. (2010) Intraarticular gene transfer of thrombospondin-1 suppresses the disease progression of experimental osteoarthritis. J Orthop Res 28: 1300–1306. 10.1002/jor.21134 [DOI] [PubMed] [Google Scholar]
- 139. Gao J, Yang T, Han J, Yan K, Qiu X, Zhou J, et al. (2011) MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem 112: 1844–1856. 10.1002/jcb.23106 [DOI] [PubMed] [Google Scholar]
- 140. Cheung CL, Chan BY, Chan V, Ikegawa S, Kou I, Ngai H, et al. (2009) Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Hum Mol Genet 18: 679–687. 10.1093/hmg/ddn397 [DOI] [PubMed] [Google Scholar]
- 141. Fan J, Varshney RR, Ren L, Cai D, Wang DA (2009) Synovium-derived mesenchymal stem cells: a new cell source for musculoskeletal regeneration. Tissue Eng Part B Rev 15: 75–86. 10.1089/ten.teb.2008.0586 [DOI] [PubMed] [Google Scholar]
- 142. Scanzello CR, Goldring SR (2012) The role of synovitis in osteoarthritis pathogenesis. Bone 51: 249–257. 10.1016/j.bone.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rahimi H, Ritchlin CT (2012) Altered bone biology in psoriatic arthritis. Curr Rheumatol Rep 14: 349–357. 10.1007/s11926-012-0259-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Lin NY, Stefanica A, Distler JH (2013) Autophagy: a key pathway of TNF-induced inflammatory bone loss. Autophagy 9: 1253–1255. 10.4161/auto.25467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, et al. (2013) Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Ann Rheum Dis 72: 761–768. 10.1136/annrheumdis-2012-201671 [DOI] [PubMed] [Google Scholar]
- 146. Kikuta J, Ishii M (2013) Osteoclast migration, differentiation and function: novel therapeutic targets for rheumatic diseases. Rheumatology (Oxford) 52: 226–234. 10.1093/rheumatology/kes259 [DOI] [PubMed] [Google Scholar]
- 147. Li F, Chung H, Reddy SV, Lu G, Kurihara N, Zhao AZ, et al. (2005) Annexin II stimulates RANKL expression through MAPK. J Bone Miner Res 20: 1161–1167. [DOI] [PubMed] [Google Scholar]
- 148. Lee A, Qiao Y, Grigoriev G, Chen J, Park-Min KH, Park SK, et al. (2013) Tumor necrosis factor alpha induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum 65: 928–938. 10.1002/art.37853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Robinson H, Kelly S, Pitzalis C (2009) Basic Synovial Biology and Immunopathology in Psoriatic Arthritis. Journal of Rheumatology 36: 14–16. [DOI] [PubMed] [Google Scholar]
- 150. Fan L, Wang Q, Liu R, Zong M, He D, Zhang H, et al. (2012) Citrullinated fibronectin inhibits apoptosis and promotes the secretion of pro-inflammatory cytokines in fibroblast-like synoviocytes in rheumatoid arthritis. Arthritis Res Ther 14: R266 10.1186/ar4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Trouw LA, Mahler M (2012) Closing the serological gap: promising novel biomarkers for the early diagnosis of rheumatoid arthritis. Autoimmun Rev 12: 318–322. 10.1016/j.autrev.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 152. Mathews S, Bhonde R, Gupta PK, Totey S (2012) Extracellular matrix protein mediated regulation of the osteoblast differentiation of bone marrow derived human mesenchymal stem cells. Differentiation 84: 185–192. 10.1016/j.diff.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 153. Ishikawa M, Ito H, Akiyoshi M, Kume N, Yoshitomi H, Mitzuoka H, et al. (2012) Lectin-like oxidized low-density lipoprotein receptor 1 signal is a potent biomarker and therapeutic target for human rheumatoid arthritis. Arthritis Rheum 64: 1024–1034. 10.1002/art.33452 [DOI] [PubMed] [Google Scholar]
- 154. Canete JD, Santiago B, Cantaert T, Sanmarti R, Palacin A, Celis R, et al. (2007) Ectopic lymphoid neogenesis in psoriatic arthritis. Ann Rheum Dis 66: 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Iwamoto T, Okamoto H, Toyama Y, Momohara S (2008) Molecular aspects of rheumatoid arthritis: chemokines in the joints of patients. FEBS J 275: 4448–4455. 10.1111/j.1742-4658.2008.06580.x [DOI] [PubMed] [Google Scholar]
- 156. Konig A, Krenn V, Toksoy A, Gerhard N, Gillitzer R (2000) Mig, GRO alpha and RANTES messenger RNA expression in lining layer, infiltrates and different leucocyte populations of synovial tissue from patients with rheumatoid arthritis, psoriatic arthritis and osteoarthritis. Virchows Arch 436: 449–458. [DOI] [PubMed] [Google Scholar]
- 157. Gunther C, Carballido-Perrig N, Kaesler S, Carballido JM, Biedermann T (2012) CXCL16 and CXCR6 are upregulated in psoriasis and mediate cutaneous recruitment of human CD8+ T cells. J Invest Dermatol 132: 626–634. 10.1038/jid.2011.371 [DOI] [PubMed] [Google Scholar]
- 158. Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, et al. (2001) Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest 107: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Garlanda C, Mantovani A (2013) Ligands and receptors of the interleukin-1 family in immunity and disease. Front Immunol 4: 396 10.3389/fimmu.2013.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. McGonagle D, Marzo-Ortega H, O'Connor P, Gibbon W, Hawkey P, Henshaw K, et al. (2002) Histological assessment of the early enthesitis lesion in spondyloarthropathy. Ann Rheum Dis 61: 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL (2000) TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest 106: 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. McMorrow JP, Murphy EP (2011) Inflammation: a role for NR4A orphan nuclear receptors? Biochem Soc Trans 39: 688–693. 10.1042/BST0390688 [DOI] [PubMed] [Google Scholar]
- 163. Joyce-Shaikh B, Bigler ME, Chao CC, Murphy EE, Blumenschein WM, Adamopoulos IE, et al. (2010) Myeloid DAP12-associating lectin (MDL)-1 regulates synovial inflammation and bone erosion associated with autoimmune arthritis. J Exp Med 207: 579–589. 10.1084/jem.20090516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Su K, Yang H, Li X, Li X, Gibson AW, Cafardi JM, et al. (2007) Expression profile of FcgammaRIIb on leukocytes and its dysregulation in systemic lupus erythematosus. J Immunol 178: 3272–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Magnusson SE, Engstrom M, Jacob U, Ulfgren AK, Kleinau S (2007) High synovial expression of the inhibitory FcgammaRIIb in rheumatoid arthritis. Arthritis Res Ther 9: R51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Smith KG, Clatworthy MR (2010) FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol 10: 328–343. 10.1038/nri2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. McGonagle D, Ash Z, Dickie L, McDermott M, Aydin SZ (2011) The early phase of psoriatic arthritis. Ann Rheum Dis 70 Suppl 1: i71–76. 10.1136/ard.2010.144097 [DOI] [PubMed] [Google Scholar]
- 168. Sherlock JP, Joyce-Shaikh B, Turner SP, Chao CC, Sathe M, Grein J, et al. (2012) IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nat Med 18: 1069–1076. 10.1038/nm.2817 [DOI] [PubMed] [Google Scholar]
- 169. Sherlock JP, Buckley CD, Cua DJ (2014) The critical role of interleukin-23 in spondyloarthropathy. Mol Immunol 57: 38–43. 10.1016/j.molimm.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 170. Benjamin M, Toumi H, Suzuki D, Redman S, Emery P, Emery P, et al. (2007) Microdamage and altered vascularity at the enthesis-bone interface provides an anatomic explanation for bone involvement in the HLA-B27-associated spondylarthritides and allied disorders. Arthritis Rheum 56: 224–233. [DOI] [PubMed] [Google Scholar]
- 171. Dolcino M, Lunardi C, Ottria A, Tinazzi E, Patuzzo G, Puccetti A (2014) Crossreactive Autoantibodies Directed against Cutaneous and Joint Antigens Are Present in Psoriatic Arthritis. PLoS One 16;9:e115424 10.1371/journal.pone.0115424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Morimoto J, Kon S, Matsui Y, Uede T (2010) Osteopontin; as a target molecule for the treatment of inflammatory diseases. Curr Drug Targets 11: 494–505. [DOI] [PubMed] [Google Scholar]
- 173. Kaleta B (2014) Role of Osteopontin in Systemic Lupus Erythematosus. Arch Immunol Ther Exp (Warsz). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Abdelmagid SM, Barbe MF, Rico MC, Salihoglu S, Arango-Hisijara I, Selim AH, et al. (2008) Osteoactivin, an anabolic factor that regulates osteoblast differentiation and function. Exp Cell Res 314: 2334–2351. 10.1016/j.yexcr.2008.02.006 [DOI] [PubMed] [Google Scholar]
- 175. Abdelmagid SM, Barbe MF, Hadjiargyrou M, Owen TA, Razmpour R, Rehman S, et al. (2010) Temporal and spatial expression of osteoactivin during fracture repair. J Cell Biochem 111: 295–309. 10.1002/jcb.22702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Data Availability Statement
All the relevant data are within the paper and its Supporting Information files.