Abstract
A rapid molecular-based assay for the detection of the Candida albicans FKS1 gene mutations responsible for resistance to echinocandin drugs was designed and evaluated. The assay consisted of a multiplexed PCR set of 5 tubes able to detect the most commonly described resistance mechanism, including FKS1 hot spot 1 and hot spot 2 mutations. The performance and specificity of the assay was evaluated using a double-blinded panel of 50 C. albicans strains. The assay showed a sensitivity of 96% and was able to detect all homozygous mutants included in the collection of strains, demonstrating that it is a robust, quick, and labor-saving method that is suitable for a routine clinical diagnostic laboratory.
INTRODUCTION
Invasive infections due to Candida albicans are a common and significant clinical problem (1–5), and the echinocandin drugs are becoming the antifungals of choice for the management of these infections (2, 6–8). This expanding use of echinocandins has brought about the emergence of drug resistance (9–15).
Echinocandin in vitro antifungal susceptibility testing (AST) is being performed worldwide to guide therapeutic decisions (16–19). However, testing for antifungal resistance is not routinely performed at many centers. Clinical echinocandin resistance in C. albicans resulting in therapeutic failures is closely linked with amino acid substitutions in the hot spot regions of the Fks1p subunit of the β-d-1,3-glucan synthase complex (10–13, 16, 20, 21). The detection of these mutations has been proposed as the most direct and accurate way to predict echinocandin clinical failure (12, 16, 21, 22).
The aim of this work was to develop a molecular-based method able to quickly and accurately detect the FKS1 mutations linked with clinical echinocandin resistance in C. albicans. The performance of the proposed methodology was evaluated using a blinded collection of clinical echinocandin-susceptible and echinocandin-resistant C. albicans isolates.
MATERIALS AND METHODS
Strains.
Fifty C. albicans strains were used throughout this work. All the strains were isolated from patients with proven invasive fungal disease. Sixteen strains were obtained from the Public Health Research Institute (PHRI) (Rutgers Biomedical and Health Sciences, NJ) and 34 were from the Mycology and Molecular Diagnostics Laboratory (Santa Fe, Argentina). Ten strains showed homozygous FKS1 hot spot region mutations, two strains showed heterozygous mutations at one of the hot spot regions, and one showed a homozygous mutation at the hot spot 1 of the FKS1 gene together with an heterozygous mutation at the hot spot 2 of the FKS1 (Table 1). C. albicans ATCC 90028, ATCC 36082, and SC5314 were used as the wild-type control strains to validate the PCRs. Candida krusei ATCC 6258 and Candida parapsilosis sensu stricto ATCC 22019 were used as AST control strains (17, 18). The isolates were identified by conventional phenotypic methods and by sequencing of the 5.8S RNA gene and adjacent internal transcribed spacer 1 (ITS1) and ITS2 regions (23, 24). The collection of strains was assembled at the PHRI center, and blinded code numbers were assigned. Also, a set of C. albicans strains with known FKS1 mutations were used to develop and test the proposed methodology before confirming its utility with the blind study.
TABLE 1.
Classical PCR set, DNA sequencing, and in vitro susceptibility determinations of the Candida glabrata strains included in this study
| Strain | Classical PCR set result for:a |
DNA sequencing result for Fks1 hot spot:b |
MIC (μg/ml) of:c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F641 | S645 | D648 | P649 | R1361 | 1 | 2 | ANF | CSF | MCF | |
| WT (n = 37) | + | + | + | + | + | WT | WT | 0.05 (S) | 0.06 (S) | 0.03 (S) |
| 2762 | − | + | + | + | + | F641S | WT | 0.50 (I) | 4.00 (R) | 1.00 (R) |
| M119 | − | + | + | + | + | F641L | WT | 0.15 (S) | 2.00 (R) | 0.25 (S) |
| M177 | − | + | + | + | + | F641S | WT | 0.50 (I) | 4.00 (R) | 1.00 (R) |
| M205 | + | − | + | + | + | S645P | WT | 1.26 (R) | 8.00 (R) | 4.00 (R) |
| M89 | + | − | + | + | + | S645Y | WT | 2.00 (R) | 4.00 (R) | 4.00 (R) |
| M85 | + | − | + | + | + | S645F | WT | 1.50 (R) | 2.00 (R) | 2.52 (R) |
| M194 | + | − | + | + | + | S645F | R1361R/H | 4.00 (R) | 4.00 (R) | 2.67 (R) |
| M149 | + | + | − | + | + | D648Y | WT | 4.00 (R) | 4.00 (R) | 2.52 (R) |
| M122 | + | + | + | − | + | P649H | WT | 0.15 (S) | 2.52 (R) | 0.50 (I) |
| A15d | + | + | + | + | + | S645S/P | WT | 0.50 (R) | 4.00 (R) | 0.50 (R) |
| A15-10d | + | − | + | + | + | S645P | WT | 2.00 (R) | 8.00 (R) | 4.00 (R) |
| M121 | + | + | + | + | − | WT | R1361H | 0.25 (S) | 2.00 (R) | 0.25 (S) |
| M90 | + | + | + | + | + | WT | R1361R/H | 0.25 (S) | 1.00 (R) | 0.50 (R) |
Signs represent the presence (+) or absence (−) of the PCR band in an electrophoresis gel.
Wild type (WT) at the corresponding hot spot (hot spot 1, 641-FLTLSLRDP-649; hot spot 2, 1357-DWIRRYTL-1364).
Expressed MICs are geometric means of data obtained on three separate days in micrograms per milliliter (for the 37 wild-type strains, presented is the geometric mean of the MICs of all the strains). ANF, anidulafungin; CSF, caspofungin; MCF, micafungin. Letters in parenthesis indicate that the strain is considered echinocandin susceptible (S), intermediate (I), or resistant (R) using the CLSI M27-S4 interpretative guidelines (18).
Isogenic laboratory-obtained spontaneous echinocandin-resistant mutants isolated by exposing strain Sc5314 to different ANF concentrations (12).
Antifungals and susceptibility testing.
Caspofungin (CSF) (Merck & Co. Inc., Rahway, NJ), anidulafungin (ANF) (Pfizer, New York, NY), and micafungin (MCF) (Astellas Pharma USA Inc., Deerfield, IL) were obtained as standard powder from their respective manufacturers. Echinocandin susceptibility testing was performed in triplicate in accordance with CLSI document M27-A3 and following the interpretive guidelines published in the M27-S4 document (17, 18).
DNA isolation, primer, and PCR design.
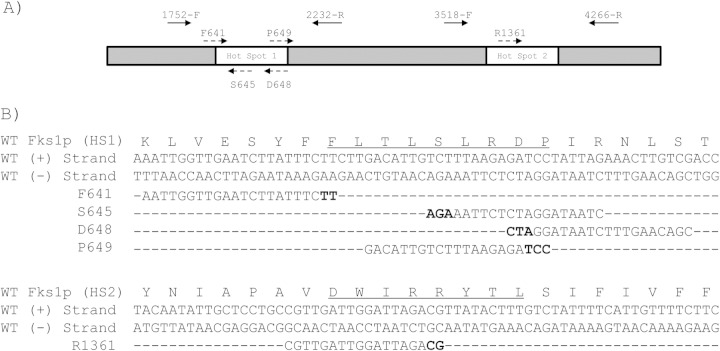
C. albicans genomic DNAs were extracted with the phenol-chloroform method (25) or with a FastDNA kit (QBiogene) following the manufacturer's instructions. The C. albicans FKS1 gene with GenBank accession number XM_446406 was used for primer design. Two groups of primers were used throughout this work. The first primer pair, named PCR control primers, consisted of two primer pairs: 1752-F and 2232-R and 3518-F and 4266-R. These primer pairs were designed to specifically hybridize FKS1 hot spot 1 and hot spot 2 regions, respectively. They were used as amplification control in each of the multiplex PCR tubes. The second group of primers (mutation detection primers) included five oligonucleotides that were designed to detect the 8 most common mutations related with echinocandin resistance in C. albicans (oligonucleotide sequences in Table 2). These mutation detection primers align the FKS1 hot spot 1 (primers F641, S645, D648, and P649) and hot spot 2 regions (primer R1361) and were added to one of the five tubes of the PCR set, which already contained one pair of PCR control primers. The hot spot 1 mutation detection primers were paired with hot spot 1 PCR control primers (1752-F and 2232-R), while the primer R1361 was used together with the hot spot 2 control primers (3518-F and 4266-R) (Fig. 1). Primers were designed using the oligonucleotide design tool of the IDT SciTools (Integrated DNA Technologies, Coralville, IA) and were purchased from Integrated DNA Technologies (IDT-Biodynamics, Buenos Aires, Argentina). Amplifications were carried out in a 25-μl volume of a mixture containing 5 mM (NH4)2SO4, 5 mM KCl, 10 mM Tris-Cl (pH 8.8), 1 Mm MgSO4, 5 ng of bovine serum albumin, 0.1% Triton X-100, 125 μM each of dATP, dGTP, dCTP, and dTTP (Genbiotech, Buenos Aires, Argentina), a 0.5 μM concentration of each of the three primers, 1.25 U of Pegasus DNA polymerase (PBL; EmbioTec, Buenos Aires, Argentina), and 10 to 25 ng of C. albicans genomic DNA. Amplification was performed for one initial step of 2 min at 94°C followed by 25 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C and then a final cycle of 10 min at 72°C in an Applied Biosystems thermocycler (Tecnolab-AB, Buenos Aires, Argentina). The PCR products were analyzed by electrophoresis.
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Target gene | Purposea | Sequence (5′ to 3′) | Primer orientation |
|---|---|---|---|---|
| 1752-F | FKS1 | FKS1 HS1 AfS and AC | CATGCCATTGGGTGGTTTAT | Sense |
| 2232-R | FKS1 | FKS1 HS1 sequencing and AC | GATTTCCATTTCCGTGGTAGC | Antisense |
| 3518-F | FKS1 | FKS1 HS2 AfS and AC | CTGGTGTTTTGGGTGATGTTGC | Sense |
| 4266-R | FKS1 | FKS1 HS2 sequencing and AC | GGTCAAATCAGTGAAAACCG | Antisense |
| F641 | FKS1 | Mutation detection | AATTGGTTGAATCTTATTTCTT | Sense |
| S645 | FKS1 | Mutation detection | CTAATAGGATCTCTTAAAGA | Antisense |
| D648 | FKS1 | Mutation detection | CGACAAGTTTCTAATAGGATC | Antisense |
| P649 | FKS1 | Mutation detection | GACATTGTCTTTAAGAGATCC | Sense |
| R1361 | FKS1 | Mutation detection | CGTTGATTGGATTAGACG | Sense |
| 1892-F | FKS1 | FKS1 HS1 sequencing | CCTTGCCAAATTGGTTGAATC | Sense |
| 3904-F | FKS1 | FKS1 HS2 sequencing | TACTATGGTCATCCAGGTTTCCA | Sense |
| ITS1b | r DNAc | Molecular identification | TCCGTAGGTGAACCTGCGG | Sense |
| ITS4b | r DNA | Molecular identification | TCCTCCGCTTATTGATATGC | Antisense |
AfS, amplification for subsequent sequencing; AC, amplification control; HS1, hot spot 1.
See reference 24.
rDNA, ribosomal DNA.
FIG 1.
(A) Representation of C. albicans FKS1 gene including the hot spot regions (white boxes). Arrows represent the control group primers (filled arrows) and the mutation detection primers (dashed arrows). (B) Alignment of the mutation detection primers with the wild-type (WT) FKS1. Underlined amino acids show the hot spot regions. Nucleotides in bold show the mutations detected by the proposed method.
DNA sequencing.
Two regions of the C. albicans FKS1 gene (flanking the hot spot 1 and hot spot 2 regions between nucleotides [nt] 1892 and 2232 and nt 3904 and 4266, respectively) and 5.8S RNA gene and adjacent internal transcribed spacer 1 (ITS1) and ITS2 regions were amplified and sequenced in each directions using the primers described in Table 2. For sequencing of the FKS1 regions, primer pair 1752-F/2232-R and 3518-F/4266-R were used for PCR amplification. The purified fragments were then subjected to sequencing using primers 1892-F and 2232-R for the hot spot 1 region and 3904-F and 4266-R for the hot spot region 2 (Table 2). In Argentina, DNA sequencing was performed using a BigDye Terminator cycle sequencing ready-reaction system (Applied Biosystems, Buenos Aires, Argentina) according to the manufacturer's instructions. Sequence analysis was performed on an ABI Prism 310 DNA sequencer (Applied Biosystems) using the facilities available at Cromatida S.A. (Buenos Aires, Argentina). In the PHRI Center, DNA sequencing was performed with a CEQ dye terminator cycle sequencing QuickStart kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations. Sequencing analyses were done with CEQ 8000 genetic analysis system software (Beckman Coulter) and with the BioEdit sequence alignment editor (Ibis Therapeutics, Carlsbad, CA).
RESULTS
Multiplex PCR design.
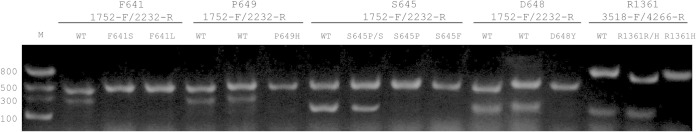
We designed two sets of primers named PCR control primers and mutation primers. These primers were mixed to form the set of 5 multiplex PCR tubes. Mutation primers were designed to hybridize FKS1 hot spot 1 and hot spot 2 regions by considering that a primer with a 3′ mismatch would not hybridize under the appropriate conditions of temperature. The PCR control group of primers was designed to specifically hybridize C. albicans FKS1 gene hot spot flanking regions (nt 1752 to 2232 and nt 3518 to 4266). The 3′ and 5′ ends of these regions share low homology with C. albicans FKS2 and FKS3, giving FKS1 specificity. These primer pairs were used as internal controls for PCR validation (DNA quality and the absence of PCR inhibitors) considering that a hot spot mutation would produce a negative PCR result. The multiplex PCR variables, including annealing temperature were established in order to use the same PCR program regardless of the primer set used. For the detection of amino acid substitutions at the Fks1p hot spot 1, 4 PCR tubes were used. Each of the 4 multiplex PCR tubes contained a pair of control PCR primers (1752-F and 2232-R) and one of the mutation detection primers named F641, S645, D648, or P649. PCRs gave one 481-bp band in all the tubes (corresponding to the amplification with the PCR control primers) and a second band of 332 bp, 200 bp, 211 bp, or 307 bp when the template DNA was wild type at the codon that codified the F641, S645, D648, or P649, respectively. Similarly, for the detection of hot spot 2 substitutions at residue R1361, the last PCR tube of the set included the primers 3518-F, 4266-R, and R1361. PCR with wild-type DNA gave two bands (769 bp and 202 bp corresponding to the control band and wild-type band, respectively). On the other hand, when mutant DNAs were used, a unique PCR band was obtained in one of the tubes corresponding to the control PCR (481 bp or 769 bp for hot spot 1 or hot spot 2 mutants, respectively) (Fig. 2).
FIG 2.
Electrophoresis gel (1.5% agarose). The primers included in each of the PCR tubes are above the photograph. Lane M, molecular marker. Lanes named as WT, wild-type strains (echinocandin susceptible strains). Lane 4, C. albicans strain M177 (Fks1p-F641S). Lane 5, strain M119 (Fks1p-F641L). Lane 8, strain M122 (Fks1p-P649H). Lane 10, strain A15 (Fks1p-S645S/P-heterozygous mutant). Lane 11, strain A15-10 (Fks1p-S645P). Lane 12, strain M85 (Fks1p-S645F). Lane 15, strain M149 (Fks1p-D648Y). Lane 17, strain M90 (Fks1p-R1361R/H heterozygous mutant). Lane 18, strain M121 (Fks1p-R1361H).
Test validation using a blinded collection of strains.
A validation study of the 5-tube multiplex PCR assay was performed using a blinded collection of 50 clinical C. albicans strains that included 13 echinocandin-resistant strains harboring different amino acid substitutions in Fks1p hot spots (Table 1). The resistant strains were mostly homozygous mutants with the exception of three isolates that harbor heterozygous mutations (Table 1). Using the described 5-tube multiplex PCR, 39 C. albicans strains were considered FKS1 wild-type (echinocandin-susceptible) strains. The remaining 11 strain were classified as echinocandin-resistant mutants harboring amino acid substitutions at Fks1p at the following residues: F641 (n = 3), S645 (n = 5), D648 (n = 1), P649 (n = 1), and R1361 (n = 1). When these results were compared with the AST results, 48 strains (96%) were correctly classified as echinocandin susceptible or resistant. Moreover, when our results were matched up with the FKS1 sequencing results, 47 isolates (94%) showed equivalent results. The described false-negative results were all due to the heterozygous mutants. These mutants showed heterozygous mutations at S645S/P (strain A15) and at R1361R/H (strain M90), and the third showed one hot spot 1 homozygous mutation (S645F) together with one hot spot 2 heterozygous mutation at R1361R/H (strain M194). This last isolate was classified as hot spot 1 mutant (S645) and hot spot 2 wild type by our method; hence, it was considered an echinocandin-resistant strain. Similarly, this strain was considered echinocandin resistant by AST. However, by FKS1 sequencing, this strain showed the homozygous S645F and the heterozygous mutation at R1361R/H not detected by the multiplex PCR.
DISCUSSION
It is well understood that rapid initiation of appropriate antifungal therapy reduces candidemia mortality rates (26, 27). Multiple efforts have been evaluated to use biomarkers or patients risk factors to direct early empirical antifungal therapy (2, 28–30). However, it is increasingly clear that the selection of a correct antifungal treatment depends on AST due to the emergence of resistance (31–33). The available AST methods need at least 24 h to obtain confident results for therapy initiation (17–19). In the present work, a molecular-based assay for rapid identification of echinocandin-resistant C. albicans strains was developed and evaluated. The described methodology is able to detect the most common FKS1 mutations linked with clinical echinocandin resistance in 4 h, and it is based in the strict relationship between C. albicans FKS1 mutations and echinocandin treatment failure. The described C. albicans FKS1 mutations comprise substitutions in five amino acid residues, four at the hot spot 1 region (F641, S645, D648, and P649) and one residue at the hot spot 2 region (R1361) (31, 33). Since C. albicans is a diploid organism, the described substitutions can be found in homozygotic and heterozygotic states (12, 22). These mutations are genetically dominant and most of them confer cross-resistance to all echinocandin drugs (12, 21, 22, 31, 33). However, heterozygous mutations confer lower echinocandin MIC values and are uncommonly isolated (10, 12, 31, 33). One possible explanation is that most of the heterozygous mutants would respond in vivo to echinocandin therapy and would act as an intermediate state for a stepwise development of the homozygous mutant displaying complete phenotype, as described for C. albicans in spontaneous laboratory mutants and for clinical C. tropicalis isolates (22, 34).
In the blinded study, we demonstrated that the method described here is able to detect homozygous substitutions in all the described residues, including F641S, F641L, S645P, S645Y, S645F, D648Y, P649H, and R1361H, which accounts for more than 98% of known resistance in C. albicans. There is an important method-inherent limitation that is the difficulty of uncovering heterozygous mutants. These false-negative results would be considered a very major error compared with sequencing or AST given that our PCR method would classify as susceptible a resistant strain. However, as described earlier in this section, heterozygous mutants represent a tiny minority (9–13, 20–22, 33, 35, 36). In view of this limitation, whole-cell antifungal susceptibility testing should be performed coupled with this assay to avoid any potential false results.
In 2006, Balashov et al. reported an allele-specific real-time PCR molecular-beacon assay able to identify heterozygous and homozygous mutations that alters the C. albicans Fks1p S645 residue (22). The method's advantages are that it is based on a more commonly available classical PCR method and that it is able to detect all the described homozygous mutations in the other residues of the hot spot regions. As any molecular-based method designed for the detection of molecular mechanisms of resistance, this method would be unable to detect non-FKS-linked echinocandin resistance mechanisms or to detect newly described mutations. However, the described set of PCRs is suitable to be adapted to detect other resistance mechanisms accompanying epidemiology changes.
In summary, the described PCR method proved to be a quick, simple, and inexpensive tool that is able to detect the main mechanism of echinocandin resistance in C. albicans.
ACKNOWLEDGMENTS
This study was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) grant PIP2011/331 to G.G.-E. and by a Universidad Nacional del Litoral (UNL) grant (CAID-PIRCA) to G.G.-E. and S.G. C.D. and F.L. have a doctoral fellowship from CONICET. The Perlin laboratory was funded by National Institutes of Health (NIH) grant AI109025 to D.S.P. and Astellas.
REFERENCES
- 1.Berdal JE, Haagensen R, Ranheim T, Bjornholt JV. 2014. Nosocomial candidemia; risk factors and prognosis revisited; 11 years experience from a Norwegian secondary hospital. PLoS One 9:e103916. doi: 10.1371/journal.pone.0103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo AL, Guimaraes T, Sukienik T, Pasqualotto AC, Andreotti R, Queiroz-Telles F, Nouer SA, Nucci M. 2014. Prognostic factors and historical trends in the epidemiology of candidemia in critically ill patients: an analysis of five multicenter studies sequentially conducted over a 9-year period. Intensive Care Med 40:1489–1498. doi: 10.1007/s00134-014-3400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez Moral L, Tiraboschi IN, Schijman M, Bianchi M, Guelfand L, Cataldi S. 2012. Fungemia in hospitals of the city of Buenos Aires, Argentina. Rev Iberoam Micol 29:144–149. (In Spanish.) doi: 10.1016/j.riam.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Nucci M, Queiroz-Telles F, Alvarado-Matute T, Tiraboschi IN, Cortes J, Zurita J, Guzman-Blanco M, Santolaya ME, Thompson L, Sifuentes-Osornio J, Echevarria JI, Colombo AL. 2013. Epidemiology of candidemia in Latin America: a laboratory-based survey. PLoS One 8:e59373. doi: 10.1371/journal.pone.0059373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang HJ, Liu WL, Lin HL, Lai CC. 2014. Epidemiology and prognostic factors of candidemia in cancer patients. PLoS One 9:e99103. doi: 10.1371/journal.pone.0099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maertens JA, Madero L, Reilly AF, Lehrnbecher T, Groll AH, Jafri HS, Green M, Nania JJ, Bourque MR, Wise BA, Strohmaier KM, Taylor AF, Kartsonis NA, Chow JW, Arndt CA, DePauw BE, Walsh TJ. 2010. A randomized, double-blind, multicenter study of caspofungin versus liposomal amphotericin B for empiric antifungal therapy in pediatric patients with persistent fever and neutropenia. Pediatr Infect Dis J 29:415–420. doi: 10.1097/INF.0b013e3181da2171. [DOI] [PubMed] [Google Scholar]
- 7.Reboli AC, Rotstein C, Pappas PG, Chapman SW, Kett DH, Kumar D, Betts R, Wible M, Goldstein BP, Schranz J, Krause DS, Walsh TJ. 2007. Anidulafungin versus fluconazole for invasive candidiasis. N Engl J Med 356:2472–2482. doi: 10.1056/NEJMoa066906. [DOI] [PubMed] [Google Scholar]
- 8.Shorr AF, Wu C, Kothari S. 2011. Outcomes with micafungin in patients with candidaemia or invasive candidiasis due to Candida glabrata and Candida krusei. J Antimicrob Chemother 66:375–380. doi: 10.1093/jac/dkq446. [DOI] [PubMed] [Google Scholar]
- 9.Axner-Elings M, Botero-Kleiven S, Jensen RH, Arendrup MC. 2011. Echinocandin susceptibility testing of Candida isolates collected during a 1-year period in Sweden. J Clin Microbiol 49:2516–2521. doi: 10.1128/JCM.00201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dannaoui E, Desnos-Ollivier M, Garcia-Hermoso D, Grenouillet F, Cassaing S, Baixench MT, Bretagne S, Dromer F, Lortholary O. 2012. Candida spp. with acquired echinocandin resistance, France, 2004-2010. Emerg Infect Dis 18:86–90. doi: 10.3201/eid1801.110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desnos-Ollivier M, Dromer F, Dannaoui E. 2008. Detection of caspofungin resistance in Candida spp. by Etest. J Clin Microbiol 46:2389–2392. doi: 10.1128/JCM.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Effron G, Park S, Perlin DS. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob Agents Chemother 53:112–122. doi: 10.1128/AAC.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother 50:2892–2894. doi: 10.1128/AAC.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Tan J, Sun J, Xu Z, Li M, Yang Q, Shao H, Zhang L, Liu W, Wan Z, Cui W, Zang B, Jiang D, Fang Q, Qin B, Qin T, Li W, Guo F, Liu D, Guan X, Yu K, Qiu H, Li R. 2014. Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. J Antimicrob Chemother 69:162–167. doi: 10.1093/jac/dkt330. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, Baughman W, Stein B, Hollick R, Park BJ, Chiller T. 2012. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arendrup MC, Garcia-Effron G, Lass-Florl C, Lopez AG, Rodriguez-Tudela JL, Cuenca-Estrella M, Perlin DS. 2010. Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, disk diffusion, and agar dilution methods with RPMI and isosensitest media. Antimicrob Agents Chemother 54:426–439. doi: 10.1128/AAC.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—3rd ed. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard—4th ed. CLSI document M27-S4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMIDEuropean Committee for Antimicrobial Susceptibility Testing (EUCAST). 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for yeasts. Clin Microbiol Infect 14:398–405. doi: 10.1111/j.1469-0691.2007.01935.x. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Kelly R, Kahn JN, Robles J, Hsu MJ, Register E, Li W, Vyas V, Fan H, Abruzzo G, Flattery A, Gill C, Chrebet G, Parent SA, Kurtz M, Teppler H, Douglas CM, Perlin DS. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother 49:3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat 10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother 50:2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. 2013. Candida Berkhout (1923). p 987–1278. In Kurtzman CP, Fell JW, Boekhout T (ed), The yeasts: a taxonomic study, 5th ed, vol 2 Elsevier, London, United Kingdom. [Google Scholar]
- 24.White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1998. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Garey KW, Rege M, Pai MP, Mingo DE, Suda KJ, Turpin RS, Bearden DT. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis 43:25–31. doi: 10.1086/504810. [DOI] [PubMed] [Google Scholar]
- 27.Morrell M, Fraser VJ, Kollef MH. 2005. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 49:3640–3645. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leon C, Ruiz-Santana S, Saavedra P, Castro C, Ubeda A, Loza A, Martin-Mazuelos E, Blanco A, Jerez V, Ballus J, Alvarez-Rocha L, Utande-Vazquez A, Farinas O. 2012. Value of beta-d-glucan and Candida albicans germ tube antibody for discriminating between Candida colonization and invasive candidiasis in patients with severe abdominal conditions. Intensive Care Med 38:1315–1325. doi: 10.1007/s00134-012-2616-y. [DOI] [PubMed] [Google Scholar]
- 29.Leon C, Ruiz-Santana S, Saavedra P, Galvan B, Blanco A, Castro C, Balasini C, Utande-Vazquez A, Gonzalez de Molina FJ, Blasco-Navalproto MA, Lopez MJ, Charles PE, Martin E, Hernandez-Viera MA. 2009. Usefulness of the “Candida score” for discriminating between Candida colonization and invasive candidiasis in non-neutropenic critically ill patients: a prospective multicenter study. Crit Care Med 37:1624–1633. doi: 10.1097/CCM.0b013e31819daa14. [DOI] [PubMed] [Google Scholar]
- 30.Ostrosky-Zeichner L, Kullberg BJ, Bow EJ, Hadley S, Leon C, Nucci M, Patterson TF, Perfect JR. 2011. Early treatment of candidemia in adults: a review. Med Mycol 49:113–120. doi: 10.3109/13693786.2010.512300. [DOI] [PubMed] [Google Scholar]
- 31.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 10 November 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlin DS. 2014. Echinocandin resistance, susceptibility testing and prophylaxis: implications for patient management. Drugs 74:1573–1585. doi: 10.1007/s40265-014-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen RH, Johansen HK, Arendrup MC. 2013. Stepwise development of a homozygous S80P substitution in Fks1p, conferring echinocandin resistance in Candida tropicalis. Antimicrob Agents Chemother 57:614–617. doi: 10.1128/AAC.01193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeiffer CD, Garcia-Effron G, Zaas AK, Perfect JR, Perlin DS, Alexander BD. 2010. Breakthrough invasive candidiasis in patients on micafungin. J Clin Microbiol 48:2373–2380. doi: 10.1128/JCM.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiederhold NP, Grabinski JL, Garcia-Effron G, Perlin DS, Lee SA. 2008. Pyrosequencing to detect mutations in FKS1 that confer reduced echinocandin susceptibility in Candida albicans. Antimicrob Agents Chemother 52:4145–4148. doi: 10.1128/AAC.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]