Abstract
We describe a case of polymicrobial bloodstream infection with six organisms identified by multiplex PCR that was initially thought to be a monomicrobial infection. Early recognition of specific Gram-positive, Gram-negative, and fungal organisms and resistance elements allowed significantly more rapid optimization of therapy.
CASE REPORT
A 21-year-old female was admitted for a cystic fibrosis exacerbation with increased coughing and shortness of breath. She reported a low-grade fever of 37.7°C accompanied by yellow sputum production for 2 to 3 days. She also had diarrhea, which was chronic, and denied chills, night sweats, vomiting, and abdominal pain. Her medical history was also significant for bronchiectasis, obstructive airway disease, chronic sinusitis, pancreatic insufficiency, malnutrition, and previous infections with Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and methicillin-resistant Staphylococcus aureus (MRSA). In addition, she was colonized by vancomycin-resistant Enterococcus faecium (VRE). Her surgical history included portacath placement.
On presentation, her vital signs included a temperature of 36.9°C, a pulse rate of 91 beats/min, a respiratory rate of 22 breaths/min, and an oxygen saturation level of 94% on 3L oxygen. On physical examination, there were diffuse crackles bilaterally and a murmur near the pulmonic valve region. No erythema, fluctuance, or tenderness was noted at the site of the portacath, and there were no embolic phenomena. Blood cultures were obtained from the portacath and right hand. Radiographic imaging of the chest demonstrated prominent interstitial markings bilaterally with continued patchy airspace opacity in the left lower lung. On admission, her home antibiotic regimen of azithromycin and inhalational tobramycin was continued and intravenous vancomycin, piperacillin-tazobactam, and tobramycin were initiated empirically on the basis of pulmonary consult service recommendations for a cystic fibrosis exacerbation.
On hospitalization day 2, the portacath blood culture obtained on admission became positive by the BacTalert automated blood culture system (bioMérieux Inc., Hazelwood, MO). Trypticase soy agar with 5% sheep blood, chocolate agar, and MacConkey agar plates (BD Diagnostics, Sparks, MD) were inoculated from the positive standard anaerobic blood culture bottle. Gram staining displayed Gram-positive cocci in clusters. Multiplex PCR with the FilmArray blood culture identification (BCID) panel (BioFire Diagnostics, Salt Lake City, UT) identified Candida tropicalis, Candida glabrata, Pseudomonas aeruginosa, Staphylococcus aureus, an Enterococcus species, and a Streptococcus species. Additionally, the mecA gene and the vanA and vanB genes were detected, which encode methicillin and vancomycin resistance in staphylococci and enterococci, respectively. In accordance with hospital policy, Gram staining and multiplex PCR results were telephoned to the patient's nurse for communication to the on-call physician. Results entered into the laboratory information system were immediately available in the electronic medical record and Premier SafetySurveillor (Premier Healthcare Alliance, Charlotte, NC) antimicrobial utilization tool. The portacath standard aerobic blood culture bottle became positive the same day, showing only Gram-positive cocci in clusters on Gram staining. In accordance with lab protocol, a multiplex PCR was not performed on this aerobic bottle. The venipuncture pediatric fastidious antibiotic neutralization (FAN) blood culture obtained on admission also revealed Gram-positive cocci in clusters on Gram staining that were later confirmed to be MRSA by conventional microbiology procedures. FAN bottles are not tested by multiplex PCR, in accordance with the BCID package insert, because of potential false-positive results from nonviable organisms and/or nucleic acid that may be present at detectable levels in these bottles. In accordance with lab protocol, additional media were not used for the subculture of the portacath aerobic bottle or the venipuncture pediatric FAN bottle.
Further extensive review of the original portacath Gram stain showed rare Gram-positive cocci in chains and one budding yeast. On the basis of these Gram stain findings and multiplex PCR results, in accordance with lab protocol, selective media (Trypticase soy agar with 5% sheep blood with colistin and nalidixic acid [CNA], inhibitory mold agar [IMA; BD Diagnostics, Sparks, MD), and chromogenic MRSA [Hardy Diagnostics, Santa Maria, CA]) were immediately inoculated to enhance recovery. After overnight incubation, S. aureus and an alpha-hemolytic streptococcus grew on blood agar, chocolate agar, and CNA plates and large (C. tropicalis) and small (C. glabrata) colony types of yeast grew only on the IMA plate. In addition, S. aureus grew on the chromogenic MRSA agar plate, supporting the multiplex PCR identification of MRSA. P. aeruginosa grew only on MacConkey agar after 48 h of incubation. Because no enterococci were recovered after 48 h of incubation, the original positive anaerobic blood culture bottle was subcultured to a chromogenic VRE agar plate (Hardy Diagnostics, Santa Maria, CA); colonies consistent with VRE were recovered, supporting the multiplex PCR identification of VRE. Each organism was identified by standard microbiological techniques. The alpha-hemolytic streptococcus was not further characterized beyond ruling out pneumococcus and enterococcus because of the large number of organisms present in this culture. The S. aureus and E. faecium bacteria were confirmed by standard susceptibility testing as MRSA and VRE, respectively. For organisms other than the alpha-hemolytic streptococcus, susceptibility testing was performed by an automated method (MicroScan Walkaway; Siemens Healthcare, Washington, DC) or disk diffusion. No additional media were used for the venipuncture blood culture subculture, as there were no multiplex PCR results to suggest that those media were needed.
On the basis of the multiplex PCR results, the antimicrobial stewardship program recommended therapy changes. Vancomycin therapy was changed to daptomycin for VRE and MRSA bacteremia. Linezolid was initiated for pulmonary coverage of cystic fibrosis-related chronic MRSA infection, micafungin was started for Candida species, and piperacillin-tazobactam therapy was changed to intravenous ciprofloxacin for P. aeruginosa on the basis of prior susceptibility data from a cystic fibrosis respiratory culture. The time from reporting of multiplex PCR results to the administration of an active agent was 1.75 h for daptomycin and linezolid, 6 h for micafungin, and 18.5 h for ciprofloxacin. An infectious disease consult was obtained. On hospital day 3, one of two blood cultures obtained from the portacath on hospital day 2 revealed yeast by Gram staining; the organisms were identified by multiplex PCR and conventional microbiology as C. glabrata and Candida albicans. All cultures thereafter were negative. The patient completed 14 days of therapy with micafungin, linezolid, daptomycin, and ciprofloxacin and was subsequently discharged. The portacath was retained because of complicated access issues but was ultimately removed 1 month later during a subsequent admission for a cystic fibrosis exacerbation, which included two sets of negative blood cultures, a negative catheter tip culture, and a sputum culture yielding P. aeruginosa and MRSA, known cystic fibrosis-related chronic bacteria.
The recent addition of rapid molecular testing to the clinical armamentarium has begun to shift the paradigm in the standard of care for clinical microbiology. For bloodstream infections in particular, rapid diagnostic methods have been shown to impact patient care by rapidly identifying pathogens within hours of a culture becoming positive—24 to 48 h before conventional microbiological methods. This has aided empirical therapy decisions and improved patient outcomes (1–10). The time advantage of early identification of organisms by rapid diagnostics versus conventional microbiological methods such that optimization of therapies can occur can be seen in this patient's case (Fig. 1).
FIG 1.
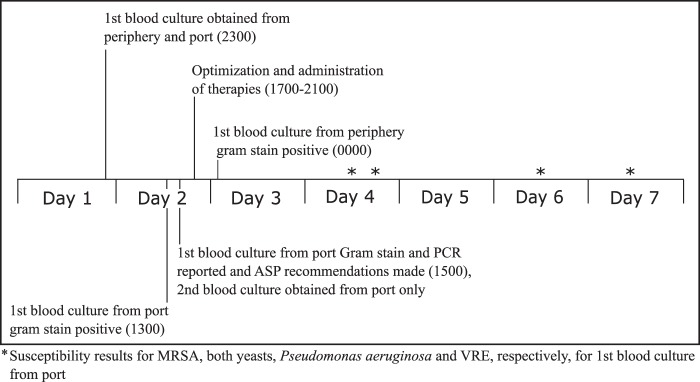
Blood culture timeline.
Bloodstream infections have historically been reported as polymicrobial in 5 to 15% of cases. Regrettably, blood cultures may detect only 20 to 40% of pathogens (11, 12). Therefore, many polymicrobial infections may go underreported. Multiplex PCR technology increases polymicrobial bloodstream infection detection sensitivity by allowing the identification of a multitude of common clinically relevant organisms from positive cultures. Within our own institution, after the initiation of multiplex PCR testing, 11% of the first 215 consecutive positive cultures were polymicrobial, with 5 detecting three or more organisms (13). The case reported here offers a potentially unique perspective on the utility of rapid molecular testing methods beyond the speed with which results are obtained. Implementation of multiplex PCR unexpectedly impacted therapy because of the identification of a polymicrobial infection with six organisms originally thought to be monomicrobial by Gram staining. The multiplex PCR results enabled the lab to subsequently confirm all six pathogens, several of which are likely to have gone unrecognized without longer incubation of MacConkey agar or the use of selective media. However, line colonization cannot be excluded in this case, as MRSA alone was isolated from the single positive venipuncture sample that was not subjected to the multiplex PCR assay.
Our institution uses the FilmArray BCID multiplex PCR panel, which detects 19 bacterial targets, 5 Candida targets, and three resistance mechanisms. In contrast, the Verigene system (Nanosphere, Northbrook, IL) splits microbial detection into Gram-positive, Gram-negative, and yeast panels. A recent study evaluated the performance of the two multiplex PCR systems, but clinical comparisons of outcomes have not been published to date (14). As the Verigene system uses Gram staining to direct panel selection, this patient would likely have had a different clinical course with that system, as the original Gram stain reading showed only Gram-positive cocci in clusters. Yeast and Gram-negative Verigene panels would not have been performed, and those organisms would likely have been missed. The outcome of our case suggests that these methods may offer clinical utility in the diagnosis of polymicrobial bloodstream infections, as demonstrated by the significant change in the clinical and microbiologic management of this patient on the basis of the multiplex PCR results. Our case report offers a unique observation of the clinical impact of multiplex PCR testing and suggests that more inquiry into the best utility of this technology is needed, specifically, for the diagnosis of polymicrobial infections.
ACKNOWLEDGMENT
We have no conflict of interest related to this study to declare.
REFERENCES
- 1.Alexander BD, Ashley ED, Reller LB, Reed SD. 2006. Cost savings with implementation of PNA FISH testing for identification of Candida albicans in blood cultures. Diagn Microbiol Infect Dis 54:277–282. doi: 10.1016/j.diagmicrobio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Bauer KA, West JE, Balada-Llasat J, Pancholi P, Stevenson KB, Goff DA. 2010. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis 51:1074–1080. doi: 10.1086/656623. [DOI] [PubMed] [Google Scholar]
- 3.Carver PL, Lin S-W, DePestel DD, Newton DW. 2008. Impact of mecA gene testing and intervention by infectious disease clinical pharmacists on time to optimal antimicrobial therapy for Staphylococcus aureus bacteremia at a university hospital. J Clin Microbiol 46:2381–2383. doi: 10.1128/JCM.00801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dierkes C, Ehrenstein B, Siebig S, Linde H-J, Reischl U, Salzberger B. 2009. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis 9:126. doi: 10.1186/1471-2334-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forrest GN, Mankes K, Jabra-Rizk MA, Weekes E, Johnson JK, Lincalis DP, Venezia RA. 2006. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol 44:3381–3383. doi: 10.1128/JCM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forrest GN, Roghmann M-C, Toombs LS, Johnson JK, Weekes E, Lincalis DP, Venezia RA. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother 52:3558–3563. doi: 10.1128/AAC.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heil EL, Daniels LM, Long DM, Rodino KG, Weber DJ, Miller MB. 2012. Impact of a rapid peptide nucleic acid fluorescence in situ hybridization assay on treatment of Candida infections. Am J Health Syst Pharm 69:1910–1914. doi: 10.2146/ajhp110604. [DOI] [PubMed] [Google Scholar]
- 8.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. 2006. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother 58:154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 9.Holtzman C, Whitney D, Barlam T, Miller NS. 2011. Assessment of impact of peptide nucleic acid fluorescence in situ hybridization for rapid identification of coagulase-negative staphylococci in the absence of antimicrobial stewardship intervention. J Clin Microbiol 49:1581–1582. doi: 10.1128/JCM.02461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ly T, Gulia J, Pyrgos V, Waga M, Shoham S. 2008. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag 4:637–640. doi: 10.2147/TCRM.S2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler AP, Bernard GR. 1999. Treating patients with severe sepsis. N Engl J Med 340:207–214. doi: 10.1056/NEJM199901213400307. [DOI] [PubMed] [Google Scholar]
- 12.Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, Knoblich B, Peterson E, Tomlanovich M. 2001. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med 345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 13.Nolte FS, Gullett JC, Youngberg LA, Steed LL. 2014. Clinical evaluation of a rapid multiplex polymerase chain reaction blood culture identification panel, abstr D-914. 54th ICAAC, Washington, DC. [Google Scholar]
- 14.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. 2014. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J Clin Microbiol 52:3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


