Abstract
mariPOC is a novel point-of-care test system for rapid detection of respiratory tract infections. We compared the performance of the mariPOC test to that of bacterial culture for detecting group A streptococcus (GAS) in 219 pharyngitis patients (ages 1–64 years) and 109 healthy asymptomatic controls (ages 19–69 years). In addition, 42 patient samples were analyzed by quantitative PCR (qPCR). Of the 219 pharyngeal patient samples, 32 were positive in a GAS bacterial culture (prevalence 15%) and 65 (30%) in the mariPOC test. The amount of GAS in samples reported positive by the mariPOC test and negative by culture was, on average, 10-fold less than that of those positive in both methods. This indicated that the negative results in bacterial cultures were due to lower sensitivity. The qPCR results were positive and in line with the mariPOC results in 43% of the discordant samples studied. Two GAS culture-positive samples were negative by the mariPOC test. The prevalences of GAS in the control subjects were 2% and 6% by culture and mariPOC results, respectively. We conclude that the mariPOC antigen detection test is more sensitive than the conventional bacterial culture for the detection of GAS among symptomatic pharyngitis patients. The higher prevalence of GAS by the mariPOC test among symptomatic patients was probably not due to carriership, since among the control patients, the difference in the prevalence of GAS by the mariPOC test and culture was not nearly as high, 15% versus 4%, respectively. Clinical trials are needed to show the clinical importance of our findings.
INTRODUCTION
Lancefield group A beta-hemolytic streptococcus (GAS) is a common causative agent for pharyngitis and skin infections such as erysipelas, impetigo, and perianal cellulitis. In addition, it is found less commonly as a pathogen in otitis, pneumonia, pericarditis, arthritis, and osteomyelitis. Conditions like scarlet fever, necrotizing fasciitis, toxic shock syndrome, and rheumatoid fever have also been linked with GAS infections. Although the majority of GAS infections are easy to treat and not life threatening, significant effort and cost are used for the diagnosis and treatment of GAS infections worldwide. Thus, it is important to evaluate and improve methods used in the diagnosis of GAS infections.
In typical cases of GAS infections, like streptococcal pharyngitis, the symptoms and signs are readily recognized by an experienced clinician, and decisions are in line with the laboratory diagnoses in 70 to 80% of cases (1). The frequency of misdiagnosed patients, however, is still much too high to be acceptable as it can result in 20 to 30% of patients being unnecessarily treated with antibiotics. In practice, however, the proportion of patients receiving unnecessary antibiotic treatment is closer to 50%, even in industrialized countries (2). This is due to the fact that atypical cases are common, and the experience levels of clinicians are varied. Thus, a GAS diagnosis has been advised to be confirmed either by a rapid antigen test or bacterial culture. Although rapid tests are available, a significant proportion of patients remain without laboratory confirmation for various reasons, including time constraints and the price of the test.
A prerequisite for accurate diagnosis and therapy for GAS is a sensitive and specific in vitro diagnostic test by which the presence or absence of GAS can be proven. In particular, the faster that GAS can be ruled out, the higher the value of the testing is for clinicians. Accordingly, the test should provide sensitivity which allows reliance on negative test results. For decades, the gold standard in GAS diagnostics has been bacterial culture. The method has properties which are potentially advantageous over other laboratory tests. In culture, only live bacteria are detected, and further investigations for subtyping, strain identification, and antimicrobial susceptibility testing are possible. However, the biggest disadvantage from the clinician and the patient's point of view is related to the delay in obtaining the results; i.e., a minimum of 24 but typically 48 to 72 h are required for the result to be ready. Physicians are usually reluctant to defer the treatment decisions that long.
Qualitative antigen detection tests to provide rapid results have been developed. The fastest immunoassay tests can be read within 5 min from the addition of the reagents and the sample. The disadvantages include the relatively long hands-on time and the requirement to read the test lines subjectively by the human eye during a limited time window. The specificity of rapid antigen tests for GAS is usually good. However, the sensitivity of the assays in clinical practice may be inferior to that of cultures (53% to 97%), although there are also reports of similar sensitivities (3). It has even been recommended that negative test results should be confirmed with bacterial cultures (4). In practice, this recommendation is complex and uneconomical. Despite extensive efforts, we have not been able to identify commercial PCR-based methods for the diagnostic testing of GAS directly from throat swab samples that could potentially take the diagnostic sensitivity to the next level. Irrespective of the test method, careful sampling is pivotal and a necessity for an accurate diagnostic test result. It, however, is still being overlooked in too many clinical units.
The recently developed rapid and automated test system, mariPOC (ArcDia International Ltd., Turku, Finland), is based on two-photon excitation fluorometry and the concentrations of antigens and fluorescent tracer on microspheres by antigen-antibody reactions (5). The technology utilizes microvolume reaction chambers and separation-free fluorescence measurement. The technology allows real-time follow-up of reaction kinetics, and in this application, the test results are read at approximately 20 min and 2 h from the beginning of the reactions. Accordingly, strong positive samples can be revealed very rapidly and even the lowest positive samples can be detected at the point of care. The mariPOC test has been validated in other studies for eight respiratory viruses from nasopharyngeal aspirate and swab samples (6, 7). We tested the applicability of the mariPOC GAS test and compared its performance with that of bacterial culture both for symptomatic cases and asymptomatic controls. Because there is little knowledge about what level of bacterial load can be detected from throat samples, our further aim was to compare the sensitivity of bacterial culture to that of the mariPOC antigen detection test.
MATERIALS AND METHODS
Study samples.
Pharyngeal swabs for the patient cohort were collected from patients with clinical suspicion of streptococcal throat infections visiting the Mehiläinen Laboratories in Helsinki and Turku, Finland. Tests were ordered by clinicians who were not aware of the study. Samples from the Helsinki unit in Töölö consisted of 121 outpatients between March and June 2012 (ages: mean, 24.9 years; median, 9.3 years). Samples from the Turku unit consisted of 98 outpatients between February and May 2013 (mean, 9.9 years; median, 7.0 years). The study samples were collected during an internal laboratory method validation study which does not require ethical permission and was not linked with recruitment or treatment of patients. The asymptomatic cohort (control group) consisted of 109 healthy university students without clinical suspicion of streptococcal infection. These samples were collected as a part of microbiological laboratory courses included in the students' academic studies. The ages of the control subjects were not recorded, but the vast majority of the university students are 20 to 30 years old.
Sample collection.
Two swab samples were collected simultaneously from each control and patient, one for the mariPOC test using Copan flocked swabs for throat sampling (Copan Ltd., Brescia, Italy) and one for culture using regular cotton swabs.
Culture.
Throat swab cultures were performed using selective sheep blood agar plates (Tammer-Tutka Oy, Tampere, Finland) or Columbia CNA agar with 5% sheep blood (bioMérieux, France). The plates were incubated at +35°C for 18 to 24 h for primary inspection and for an additional 24 h if the first readout was negative. Beta-hemolytic colonies were picked, and possible GAS was identified using an Oxoid Strep plus DR 575 kit (Thermo Fisher Scientific).
mariPOC.
The mariPOC tests were performed according to the manufacturer's instructions (ArcDia International Ltd., Turku, Finland). The throat swab head was cut into a sample tube, 6 drops of extraction solutions A and B were added, the tube was vortexed, and the extraction mixture was incubated at room temperature for 2 to 10 min. The tube was labeled with a barcode printed from the user interface. One push of sample buffer (1.3 ml) was added from a bottle-top dispenser, and the tube was vortexed and inserted into an analyzer for automated analysis. The following steps have been fully automated in the test system: the sample is aspirated into the dispensing tubing and needle, the sample is dispensed (20 μl per well) into the assay reaction wells (1 well per tested pathogen on a 384-microtiter well plate) containing microbe-specific immunoassay reagents in the dry state (22 to 66 multianalyte tests per plate), the reaction wells are shaken and incubated, separation-free fluorescence measurements are taken at 20 min and 2 h, data analysis, and the result is report onto the user interface.
The mariPOC immunoassay tests are quantitative in nature with an average of approximately 10% coefficients of variation (5). The nature of the swab sampling, however, is qualitative or semiquantitative at best. Therefore, the results are reported qualitatively in the user interface, i.e., positive or negative. In this study, we analyzed the quantitative fluorescence signals to estimate the bacterial loads in the samples.
qPCR.
The development and validation of a quantitative PCR (qPCR)-based method for detection of GAS directly from patient samples is described in the supplemental material. The lowest limit of detection (analytical sensitivity) of the GAS qPCR test has been determined to be 10,000 bacteria/ml (approximately 500 CFU/ml) using a dilution series of a standard GAS control sample. There were 42 throat swab patient samples, stored in mariPOC buffers, available for qPCR. All control group (n = 109) samples were available for qPCR.
RESULTS
Prevalence of GAS in the symptomatic patient population is higher by mariPOC test than by conventional culture.
In the asymptomatic cohort (n = 109), the prevalences of GAS were 2% by bacterial culture and 6% by the mariPOC test. The 4% unit difference indicated that the set reporting level (see Fig. 2) in the mariPOC assay is clinically relevant, keeping the number of asymptomatic carriers relatively low.
FIG 2.
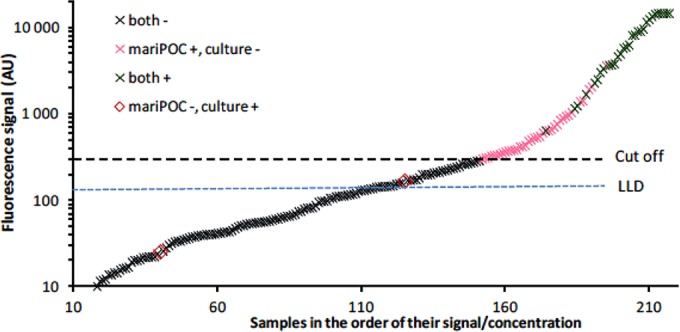
Relation of samples with coinciding or contradictory GAS results in culture and in the mariPOC test to the lowest analytical detection limit (blue dotted line, lowest limit of detection [LLD]) and to the clinical reporting level (black dashed line, Cut off) of the mariPOC test. One arbitrary unit (AU) corresponds to 1 CFU/ml.
In the symptomatic patient population (n = 219), however, the prevalence of GAS was 15% units higher with the mariPOC test (30%, n = 65) than with conventional culture (15%, n = 32) (Table 1). In the Helsinki unit, the prevalences of GAS by the mariPOC test and culture were 26% (31/121) and 14% (17/121), respectively. In the Turku unit, the corresponding figures were 35% (34/98) and 15% (15/98).
TABLE 1.
Symptomatic pharyngitis patients were tested for group A streptococcus using culture and mariPOC methodsa
| Culture result | mariPOC test result (no.) |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 30 | 2 | 32 |
| Negative | 35 | 152 | 187 |
| Total | 65 | 154 | |
Prevalence of group A streptococcus by culture was 15% and by the mariPOC test was 30%. n = 219.
Discrepancy between mariPOC test and conventional culture correlates with low bacterial load in swab samples.
In order to study whether differences in the GAS loads in the samples might explain the differences in the prevalences between the mariPOC test and culture in symptomatic patients, we took advantage of the quantitative nature of the mariPOC test methodology (see Materials and Methods). The samples that were positive in both the mariPOC test and culture had average GAS concentrations of 7,490 (range, 637 to 14,700) CFU/ml, while the samples that were positive in the mariPOC test but negative in culture had an average GAS bacterial load of 745 (302 to 3,623) CFU/ml (Fig. 1). Thus, the samples that were negative in bacterial culture had, on average, 10-fold lower bacterial levels. The samples that were negative in bacterial culture and were also negative in the mariPOC method gave bacterial loads between 0 and 287 CFU/ml. Combined, these results suggest that throat samples with low bacterial loads could easily remain undetected by bacterial culture, but not by the mariPOC test.
FIG 1.
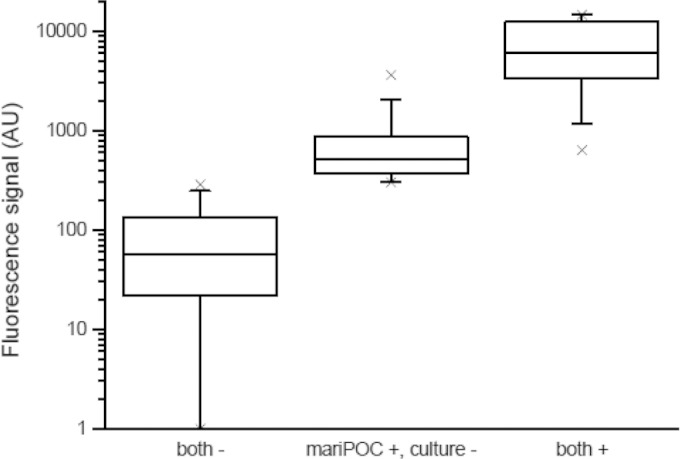
Box (25th to 75th percentiles) and whiskers (5th to 95th percentile) plot with medians showing the differences in group A streptococcus (GAS) concentrations between the samples negative or positive by both culture and the mariPOC method, and the samples negative in culture but positive in the mariPOC test. × represents minimum and maximum values. One arbitrary unit (AU) corresponds to 1 CFU/ml.
The quantitative data were also plotted as an increase in the fluorescent signal level of the mariPOC test (Fig. 2). The cutoff is the present level of signal that determines whether the sample is reported as positive or negative. The cutoff is significantly higher than the analytical detection limit (0 + 3 SD in Fig. 2). The difference between the cutoff and lowest limit of detection (above which the sample can be said to be analytically positive for the target antigen) indicates that if a reasonable compromise is accepted in the form of increased frequency of carriers found, the test can be made even more sensitive by adjusting the cutoff.
qPCR results are in line with the results obtained by mariPOC assay.
Of the 42 patient samples available for qPCR testing, 10 samples positive by both bacterial culture and the mariPOC test were also positive in qPCR. All 18 samples that were negative in bacterial culture and the mariPOC test were also negative in qPCR. Of the 14 samples negative in the bacterial culture but positive in the mariPOC test, 6 were positive in qPCR. The average load of bacteria as measured by the mariPOC test for these 6 samples was 1,750 CFU/ml. Of the samples positive in the mariPOC test but negative in the culture and in qPCR, the bacterial load was, on average, 520 CFU/ml. The average bacterial load in samples positive by all methods was 10,700 CFU/ml.
In the samples from healthy controls (n = 109), qPCR was positive for the same three samples as the mariPOC test. Three additional samples reported positive by the mariPOC test were not detected as positive by qPCR.
DISCUSSION
In this study, we show the superior sensitivity of the novel mariPOC test to detect group A streptococcus from pharyngeal swabs compared to that of the traditional bacterial culture. Of the 219 clinical pharyngeal samples, 32 were positive in bacterial culture whereas a total of 65 samples were positive in the mariPOC test. To verify this finding, we analyzed the quantitative data of the bacterial load in the samples as determined by the mariPOC test. The average bacterial load measured by the mariPOC fluorescence for samples positive in bacterial culture was, on average, 7,490 (range, 637 to 14,700) CFU/ml, whereas samples negative in culture but positive in the mariPOC test had an average bacterial load of 745 (range, 302 to 3,623) CFU/ml. Furthermore, bacterial loads of samples that were positive only in the mariPOC test but not in culture or qPCR were even lower (520 CFU/ml). Although the difference between the last two subgroups is small and the number of samples is limited, this level of difference might be expected from the slightly greater sensitivity of the mariPOC test compared to that of qPCR.
In order to study the methodological sensitivity of a suspension culture (plating of a solution instead of a swab to make the results more quantitative) with 50-μl plating of sample (suspended in 2,000 μl of ESwab medium) and mariPOC antigen detection, we also analyzed a dilution series from the Streptococcus pyogenes ATCC 9615 strain using both methods (data not shown). In this particular experiment, the culture sensitivity was 50 CFU/ml (a few colonies per plate), and the mariPOC clinical reporting level was 700 CFU/ml and analytical detection sensitivity was 200 CFU/ml. This study reflects theoretical laboratory conditions and samples that have been in a growth phase in culture medium. In clinical samples, the situation is evidently different. The bacteria may not be able to grow and multiply in the throat or in the culture plate due to ongoing antibiotic regimens, competition by other commensal flora such as Staphylococcus aureus (3), or other factors. Our results suggest, however, that in clinical practice, the sensitivity of the mariPOC fluorescent antigen detection is far better than that of culture.
The potential cross-reactivities of the mariPOC GAS test have been excluded extensively in earlier studies (8, 9). The studies have covered a significant proportion of Streptococcus species other than pyogenes, such as Streptococcus milleri/Streptococcus anginosus group strains, and group C and G streptococci. According to the literature, about 5% of isolates of Streptococcus anginosus (10, 11, 12) and about 3% of isolates of Streptococcus dysgalactiae subsp. equisimilis, species which are usually type C or G (13), express the Lancefield group A surface antigen measured in the antigen detection tests for Streptococcus pyogenes. For this reason, a sample containing S. anginosus or S. dysgalactie subsp. equisimilis bacteria may give false-positive results in a test for group A streptococci. The frequency of S. anginosus and S. dysgalactie subsp. equisimilis carriership in respiratory epithelia is not well known, although false clinical diagnoses due to a patient carrying a non-S. pyogenes bacteria expressing Lancefield type A antigen appear rare. In the Antikainen et al. study (9), all S. anginosus group strains from the national strain collection of Finland (n = 18) were tested with the mariPOC test, but none reacted with the GAS test. In our study, group C or G streptococci findings in culture did not correlate with positive results in the mariPOC test (data not shown). Accordingly, our results seem unlikely to be explained by cross-reactions.
The limitations of our study include at least the discrepancies that may have been caused by sample collection. The two swabs taken from each patient and control for the two test methods may have had different amounts of sample collected. However, the samples were collected at the same occasion so we feel that in a data set as large as ≥200 samples, such discrepancies are diluted. It is also of note that the symptomatic patients and the asymptomatic controls were not matched according to age. More than 50% of the symptomatic patients were children, whereas in asymptomatic group their proportion was 0%. Since it is well known that the prevalence of GAS is higher in children than in adults, this may interfere with the results. However, the symptomatic cohort was in fact composed of two separate subgroups (Turku and Helsinki) that were significantly different (P < 0.001) according to age. Yet the results measured both by the mariPOC test and GAS culture did not differ between these groups. Moreover, to our knowledge, there are no data supporting the assumption that either the sensitivity of the GAS culture or antigen tests might be age dependent.
The data presented in this study suggest that the results are indeed explained by the differences in the analytical sensitivities of the tests in routine use. Similar results have been recently reported by Cohen et al. (3), who showed that the sensitivity of standard culture suffers from competing microbiota and that most patients negative in culture but positive in conventional lateral flow rapid tests may actually be false negatives in culture. Cohen et al. (3) also reported that they found confirmation with PCR in 76% of lateral flow rapid test-positive but culture-negative GAS results and suggested that the rapid test specificity could be considered close to 100% in practice. Interestingly, our results and the results by Cohen et al. (3) are in line with those presented in the marketing material of another new test with an automated readout, the FDA-cleared illumigene DNA test for GAS from Meridian Bioscience, Inc. (14). This test is not based on PCR but on an isothermal amplification technique. According to the manufacturer, the analytical sensitivity of the test is 400 CFU/test (note that this result is not per milliliter units), and it finds 50% more positive cases than culture. Based on a theoretical calculation, the sensitivity of a PCR method using typical protocols cannot be much better than that of the mariPOC test because there have to be at least 2 to 3 gene copies on average per PCR to be able to determine if a sample is positive. Antigen detection has, in this sense, an advantage over PCR. There may be up to 100,000 antigens per bacterial genome in the sample. Thus, although PCR is more sensitive in terms of molecules needed to detect, there are so many more antigens compared to genes, more than compensating for the lower analytical sensitivity in molecules of mariPOC antigen testing. The situation is significantly different in viral diagnostics, where the proportion of genes to antigens is on the order of 1 to 100 to 1,000, more preferable to PCR.
In the process of verifying the high sensitivity of the mariPOC GAS test in clinical practice compared to that of culture, we performed detailed retrospective analysis of the medical records and other supporting evidence for the laboratory findings. For example, there were several patients who visited the Helsinki clinic for the same symptoms twice within a few days. During the first visit, the patients were diagnosed as GAS negative based on culture, but, in contrast, the mariPOC test gave positive results for these patients at the first visit. At the second visit, both culture and the mariPOC test gave positive results for group A streptococcus. The data allowing us to confirm the GAS diagnosis varied between different patients and included, e.g., clinical signs and symptoms typically found in acute and complicated GAS diseases, a positive result with another rapid antigen test, a recent GAS-positive bacterial culture of another family member, or conversion of a patient's negative bacterial culture into a positive result within 4 weeks from the mariPOC testing date. Importantly, a sample from the first visit was available for qPCR in one patient and confirmed the mariPOC finding in this case. Our findings encourage pursuit of prospective clinical studies to evaluate the clinical importance and impact of the mariPOC GAS test.
In conclusion, for decades the gold standard for GAS detection has been bacterial culture but in spite of being affordable and specific, the results are delayed and therefore more rapid point-of-care (POC) tests are required. This study in which we compared the fully automated mariPOC GAS test with bacterial culture provided data suggesting that the new rapid POC test is more sensitive than bacterial culture. The high analytical sensitivity of the mariPOC GAS test enabled the detection of symptomatic patients that harbor only a low amount of group A streptococcal bacteria in their throat swab samples. This may also result in better understanding of symptomatic GAS pharyngitis and other GAS-related disorders.
Supplementary Material
ACKNOWLEDGMENTS
The work of Jukka Vakkila is supported, in part, by the Gyllenberg Foundation, Finland.
Janne O. Koskinen is employed by Arcdia Ltd., the manufacturer of the mariPOC antigen test. None of the other authors is involved with or has any financial connections with Arcadia.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00018-15.
REFERENCES
- 1.Breese BB, Disney FA. 1954. The accuracy of diagnosis of beta streptococcal infections on clinical grounds. J Pediatr 44:670–673. doi: 10.1016/S0022-3476(54)80008-4. [DOI] [PubMed] [Google Scholar]
- 2.Rautakorpi UM, Lumio J, Huovinen P, Klaukka T. 1999. Indication-based use of antimicrobials in Finnish primary health care. Description of a method for data collection and results of its application. Scand J Prim Health Care 17:93–99. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JF, Cohen R, Bidet P, Levy C, Deberdt P, d'Humières C, Liguori S, Corrard F, Thollot F, Mariani-Kurkdjian P, Chalumeau M, Bingen E. 2013. Rapid-antigen detection tests for group a streptococcal pharyngitis: revisiting false-positive results using polymerase chain reaction testing. J Pediatr 162:1282–1284. doi: 10.1016/j.jpeds.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 4.Gerber MA, Shulman ST. 2004. Rapid diagnosis of pharyngitis caused by group A streptococci. Clin Microbiol Rev 17:571–580. doi: 10.1128/CMR.17.3.571-580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koskinen JO, Vainionpää R, Meltola NJ, Soukka J, Hänninen PE, Soini AE. 2007. Rapid method for detection of influenza A and B virus antigens by use of a two-photon excitation assay technique and dry-chemistry reagents. J Clin Microbiol 45:3581–3588. doi: 10.1128/JCM.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivaska L, Niemelä J, Heikkinen T, Vuorinen T, Peltola V. 2013. Identification of respiratory viruses with a novel point-of-care multianalyte antigen detection test in children with acute respiratory tract infection. J Clin Virol 57:136–140. doi: 10.1016/j.jcv.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuuminen T, Suomala P, Koskinen JO. 2013. Evaluation of automated multianalyte point-of-care mariPOC test for the detection of influenza A virus and respiratory syncytial virus. J Med Virol 85:1598–1601. doi: 10.1002/jmv.23660. [DOI] [PubMed] [Google Scholar]
- 8.Koskinen JO, Järvinen E, Saukkoriipi A, Kaijalainen T, Soini AE. 2009. Sensitivity and specificity of group A streptococci and pneumococcus tests in a new rapid multianalyte respiratory infection test system, abstr 292, poster 561 27th Annu Meet of the European Society for Paediatric Infectious Diseases, ESPID 2009, Brussels, Belgium, 9–13 June, 2009. [Google Scholar]
- 9.Antikainen P, Toivola H, Haanperä-Heikkinen M, Hakanen AJ, Koskinen JM, Koskinen JO. 2013. No cross-reactions detected by mariPOC group A streptococci antigen test with Streptococcus anginosus, abstr p 40, poster 9 GAS Infections, Rome, Italy, 21–23 March 2013. [Google Scholar]
- 10.Lawrence J, Yajko DM, Hadley WK. 1985. Incidence and characterization of beta-hemolytic Streptococcus milleri and differentiation from S. pyogenes (group A), S. equisimilis (group C), and large-colony group G streptococci. J Clin Microbiol 22:772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruoff KL. 1988. Streptococcus anginosus (“Streptoccus milleri”): the unrecognized pathogen. Clin Microbiol Rev 1:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DR, Kaplan EL. 2001. False-positive rapid antigen detection test results: reduced specificity in the absence of group A streptococci in the upper respiratory tract. J Infect Dis 183:1135–1137. doi: 10.1086/319286. [DOI] [PubMed] [Google Scholar]
- 13.Jensen A, Kilian M. 2012. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol 50:113–126. doi: 10.1128/JCM.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meridian Bioscience, Inc. illumigene group A streptococcus (group A strep) DNA amplification assay package insert. Meridian Bioscience, Inc., Cincinnati, OH: http://www.meridianbioscience.com/Content/Assets/Files/Group%20A%20Strep/illumigeneGroupAStrepPackageInsert.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


