Abstract
Respiratory infection in cystic fibrosis (CF) is polymicrobial, but standard sputum microbiology does not account for the lung microbiome or detect changes in microbial diversity associated with disease. As a clinically applicable CF microbiome surveillance scheme, total sputum nucleic acids isolated by a standard high-throughput robotic method for accredited viral diagnosis were profiled for bacterial diversity using ribosomal intergenic spacer analysis (RISA) PCR. Conventional culture and RISA were performed on 200 paired sputum samples from 93 CF adults; pyrosequencing of the 16S rRNA gene was applied to 59 patients to systematically determine bacterial diversity. Compared to the microbiology data, RISA profiles clustered into two groups: the emerging nonfermenting Gram-negative organisms (eNFGN) and Pseudomonas groups. Patients who were culture positive for Burkholderia, Achromobacter, Stenotrophomonas, and Ralstonia clustered within the eNFGN group. Pseudomonas group RISA profiles were associated with Pseudomonas aeruginosa culture-positive patients. Sequence analysis confirmed the abundance of eNFGN genera and Pseudomonas within these respective groups. Low bacterial diversity was associated with severe lung disease (P < 0.001) and the presence of Burkholderia (P < 0.001). An absence of Streptococcus (P < 0.05) occurred in individuals with lung function in the lowest quartile. In summary, nucleic acids isolated from CF sputum can serve as a single template for both molecular virology and bacteriology, with a RISA PCR rapidly detecting the presence of dominant eNFGN pathogens or P. aeruginosa missed by culture (11% of cases). We provide guidance for how this straightforward CF microbiota profiling scheme may be adopted by clinical laboratories.
INTRODUCTION
Cystic fibrosis (CF) is an inherited condition characterized by chronic endobronchial infection leading eventually to respiratory failure (1). Traditional culture-based microbiological techniques readily identify pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus, which are common and typical of CF infection (2). Other CF pathogens, such as Burkholderia cepacia complex and emerging pathogens such as Achromobacter, Stenotrophomonas, Ralstonia, and Pandoraea species, are a challenge for conventional microbiology due to their taxonomic complexity (2). These emerging non-Pseudomonas, nonfermenting Gram-negative (eNFGN) species present multiple problems for people with CF, including innate antibiotic resistance, uncertain pathogenic outcome, and potential for transmission, and are possible contraindicators for subsequent lung transplantation (3).
CF therapy has evolved to optimize the prevention and suppression of bacterial infections known to be associated with clinical deterioration, such as P. aeruginosa (4, 5) or B. cepacia complex (6). Recent culture-independent analysis of CF infection revealed the presence of considerable bacterial diversity (collectively known as the CF microbiota) that is not captured by standard culture (7). Facultative and obligately anaerobic bacterial genera such as Streptococcus and Prevotella, respectively, frequently go undetected unless dedicated anaerobic culture or molecular analyses are used (8, 9). It has also been suggested that different CF lung microbial communities might act as pathogenic entities in their own right (10). Studies demonstrating loss of bacterial diversity as patients age and their lung function declines provide evidence for a wider role of the respiratory microbiome in disease progression (11–13). A lack of microbial diversity in patients with severe lung disease has also been observed by direct sampling of explanted lungs from CF individuals with end-stage disease, where pathogens such as P. aeruginosa, B. cepacia complex, and Achromobacter xylosoxidans were found to be dominant in the limited number of cases studied (14).
Understanding the role of the CF lung microbiota and detecting dysbiosis associated with severe infection are key challenges for the delivery of future CF therapy. Although next-generation sequencing of the 16S rRNA gene is the gold standard in bacterial diversity analysis (7, 15), its widespread implementation in routine diagnostic microbiology is not yet possible. However, CF bacteriology laboratories increasingly use PCR for bacterial identification, especially to confirm infections with CF pathogens that are difficult to identify using phenotypic methods such as B. cepacia complex species (2). Ribosomal intergenic spacer analysis (RISA) is a simple, single-step PCR-based method for profiling microbial diversity that detects the variation in size of the intergenic transcribed spacer (ITS) region between the bacterial 16S and 23S rRNA genes (16). RISA has been extensively used to profile microbial diversity in a range of environmental settings (16, 17). A basic RISA PCR was employed to detect bacterial diversity changes in 6 patients in a study demonstrating that antipseudomonal therapy reduced the presence of Aspergillus in sputum (18); during that study, RISA appeared useful as an indicator of diverse or pathogen-dominated infections. To evaluate if RISA could be employed as a routine PCR diagnostic to detect microbial diversity in CF, it was applied to a large observational survey of adult CF patients. RISA was compared to conventional culture and microbiota profiling by16S rRNA gene pyrosequencing, with the sequence-based diversity also being compared to patient clinical data. To facilitate routine uptake of this microbial diversity profiling scheme, total DNA extracted from sputum using an automated, clinically accredited procedure employed in viral diagnosis was used. The performance of the PCR profiling diagnostic scheme compared to culture and its correlation to sequence-based microbial diversity analysis and clinical outcome data are described.
MATERIALS AND METHODS
Clinical samples and conventional microbiology.
Sputum samples (n = 200) were collected from 93 adult CF patients attending the Manchester Adult Cystic Fibrosis Centre, Wythenshawe Hospital, United Kingdom, with consent and full ethical permission (Greater Manchester West NHS Research Ethics Committee study reference number 10/H1014/71). In all cases, CF had been diagnosed prior to study entry on the basis of clinical, biochemical, and genetic results in line with standard clinical practice. At each visit, patients provided paired sputum samples for conventional culture and culture-independent analysis (RISA and 16S rRNA gene pyrosequencing; see below). Samples were expectorated spontaneously by the patient into sterile containers, and a single sample was split for each analysis. Sputum samples were transported on the same day to the on-site microbiology laboratory and processed according to established guidelines published by the United Kingdom Cystic Fibrosis Trust (19). Samples were labeled with unique identifiers corresponding to the patient and clinic visit.
Sputum nucleic acid extraction.
Sputum specimens for culture-independent analysis were transported within 24 h to the regional virology laboratory in Manchester for processing as described previously (20). After liquefaction and dilution in extraction buffer within a containment level 3 laboratory (a standard precaution against unknown tuberculosis cases within respiratory specimens), sputum samples were processed in an extraction-dedicated containment level 2 laboratory. To avoid and monitor for contamination, a no-template control (nuclease-free deionized water) was included in all extraction batches, and environmental swabbing and testing of positive/negative extraction controls was carried out on a regular basis. Briefly, sputum was mixed in a 1:1 ratio with AL lysis buffer (Qiagen, Hilden, Germany) to a volume of 600 μl before being inactivated at 80°C for 20 min. Samples were stored at −80°C before undergoing DNA extraction. Total nucleic acids were extracted from sputum using the automated QIAamp virus Biorobot MDx instrument (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions and procedures to avoid sample cross-contamination. Extracted DNA samples were stored at −80°C until further analysis.
PCR diagnostics.
RISA was performed as described previously (18). Briefly, PCRs were set up with 1 μl of extracted sputum DNA (approximately to 20 to 40 ng) and 10 pmol of each RISA PCR primer (1406F, TGYACACACCGCCCGT, and 23SR, GGGTTBCCCCATTCRG) (16). RISA amplicons (2 μl of amplified DNA) were separated by microfluidics (Agilent 2100 Bioanalyzer; Agilent Technologies, California, USA), and their profiles were analyzed using Gelcompar II (Applied Maths, Sint-Martens Latem, Belgium) and clustered based on their similarity as described previously (21). DNA from pure bacterial cultures of reference CF pathogen species was used to generate control ITS amplicons for putative pathogen identification based on size correlation. Negative controls containing water were included with every batch of sputum DNA extracts analyzed and produced no amplification products. The in silico PCR tool (22) was used to predict ITS amplicon size from the genome sequences of selected CF pathogens and microbiota species.
16S rRNA gene pyrosequencing.
A subset of samples from 60 patients representative of the diversity observed by RISA profile cluster analysis (see Fig. S3 in the supplemental material) were selected for 16S rRNA gene pyrosequencing analysis (23). The first sputum sample (designated sample A) provided by each patient was used for 16S rRNA pyrosequencing, except for patients 39 and 58; for these individuals, the second sample (designated sample B) was used due to a lack of remaining extract in their first sample. Sufficient amplicon reads for statistical analysis were successfully obtained from 59 out of the 60 selected samples. Pyrosequencing was performed by Research and Testing Laboratory Inc. (Lubbock, TX, USA). Briefly, approximately 100 ng of sputum DNA was amplified using the universal 16S rRNA gene primers 530F (5′-GTG CCA GCM GCN GCG G) and 1100R (5′-GGG TTN CGN TCG TTG), targeting the V4 to V6 regions of the bacterial 16S rRNA gene. Resulting amplicons were sequenced using the Genome Sequencer FLX system (Roche, Nutley, NJ). No template controls were taken through the entire pyrosequencing pipeline to monitor for contamination. The 16S rRNA gene pyrosequencing reads were analyzed using MOTHUR version 1.33 (24) (www.mothur.org). After normalization, one sample (57A) was excluded from the final analysis due to the number of generated reads falling below the threshold level of 1,000. Sequences were assigned to genus level using the Ribosomal Database Project classifier (25). Diversity indices, species richness, and rarefaction analysis were carried out in MOTHUR (24). Additional microbial population analyses were performed using Statistical Analysis of Taxonomic and Functional Profiles (STAMP) version 2.07 (26).
Statistical analysis.
Statistical analyses were performed using R (27), and data are presented as means (± standard deviations) or medians (± interquartile ranges) as appropriate. One-way analysis of variance (ANOVA) was used to compare subgroups of patients with distinct RISA profiles. A pairwise t test was used to assess the diversity differences in samples dominated with selected pathogens, and the Bonferroni correction was applied to allow for multiple comparisons. Multiple linear regression was used to investigate the effect of demographic variables on indices of sputum bacterial diversity. The conventional P value of <0.05 was used to determine statistical significance, and multiple comparisons were corrected with the Bonferroni and Storey false discovery rate (FDR) algorithms, as indicated.
Nucleotide sequence accession number.
The 16S rRNA gene reads were submitted to the European Nucleotide Archive (accession number PRJEB7867).
RESULTS
Efficacy of sputum DNA and RISA for bacterial diversity profiling in CF.
A total of 200 sputum samples were analyzed from 93 adult CF patients between December 2010 and November 2011. Baseline demographics of the entire study cohort and those successfully subjected to 16S rRNA pyrosequencing analysis (n = 59) were similar, except for a greater number of B. cepacia complex-infected patients in the sequenced subset (Table 1). Patients contributed a median of 2 samples (range, 1 to 5) to the full data set, and 77 (38.5%) of 200 specimens were collected at the time of pulmonary exacerbation. RISA bacterial diversity profiles were generated successfully from all 200 samples, and paired microbial culture data were available for 179 of these. RISA profiles were reproducible for individual samples amplified on multiple occasions (see Fig. S1 in the supplemental material) but showed stability or variation across sequential samples typical of that seen in chronically infected CF patients (see Fig. S2 in the supplemental material) (13).
TABLE 1.
Baseline demographics of study participantsa
| Characteristic | Full RISA cohort | 16S rRNA gene pyrosequencing subset |
|---|---|---|
| No. of patients | 93 | 59 |
| Age (yr) | 28 (23–35) | 28 (23–34) |
| Gender (% female) | 53 | 47 |
| Body mass index (kg/m2) | 21.9 (2.9) | 22.1 (3.1) |
| Baseline FEV1 (% predicted) | 59.5 (21.7) | 57.4 (21.8) |
| % with: | ||
| Chronic P. aeruginosa infection | 74.2 | 69.5 |
| Chronic methicillin-sensitive S. aureus infection | 15.1 | 11.9 |
| Chronic methicillin-resistant S. aureus infection | 4.3 | 3.4 |
| Chronic B. cepacia complex infection | 14.0 | 18.6 |
RISA, ribosomal intergenic spacer analysis; FEV1, forced expiratory volume in 1s. Data are mean (SD), median (interquartile range), or percent, as appropriate.
Distinct bacterial diversity profiles correlated to pathogen culture data.
Cluster analysis identified multiple RISA profile similarity groups, with two designated the eNFGN and Pseudomonas groups, apparent when correlated to the paired sample CF pathogen culture data (Fig. 1; also, see Fig. S3 in the supplemental material). Sixty samples (Fig. 1) representative of the overall microbial diversity (see Fig. S3) were selected for 16S rRNA gene sequence analysis, and the same major RISA profile groups were observed for this subset. The eNFGN group profiles (50 samples representative of 26 patients) (see Fig. S3) were 50% culture positive for either Burkholderia, Achromobacter, Ralstonia, or Stenotrophomonas; only 20% of these samples grew P. aeruginosa. Their ITS profiles were dominated by amplicons correlating to those expected from each respective eNFGN species (Fig. 1; also, see Fig. S3). The Pseudomonas group RISA profiles (150 samples representative of 74 patients) (see Fig. S3) were mixed in terms of ITS diversity (Fig. 1) but were unified by a high proportion of P. aeruginosa culture positivity (74%) (see Fig. S3) for the paired sputum samples; only 10 of these 150 samples grew an eNFGN species (see Fig. S3). The samples selected for 16S rRNA gene bacterial diversity analysis demonstrated similar rates of pathogen culture positivity (13 of 24 [54%] positive for an eNFGN in that group; 29 of 36 [80%] Pseudomonas group samples positive for P. aeruginosa).
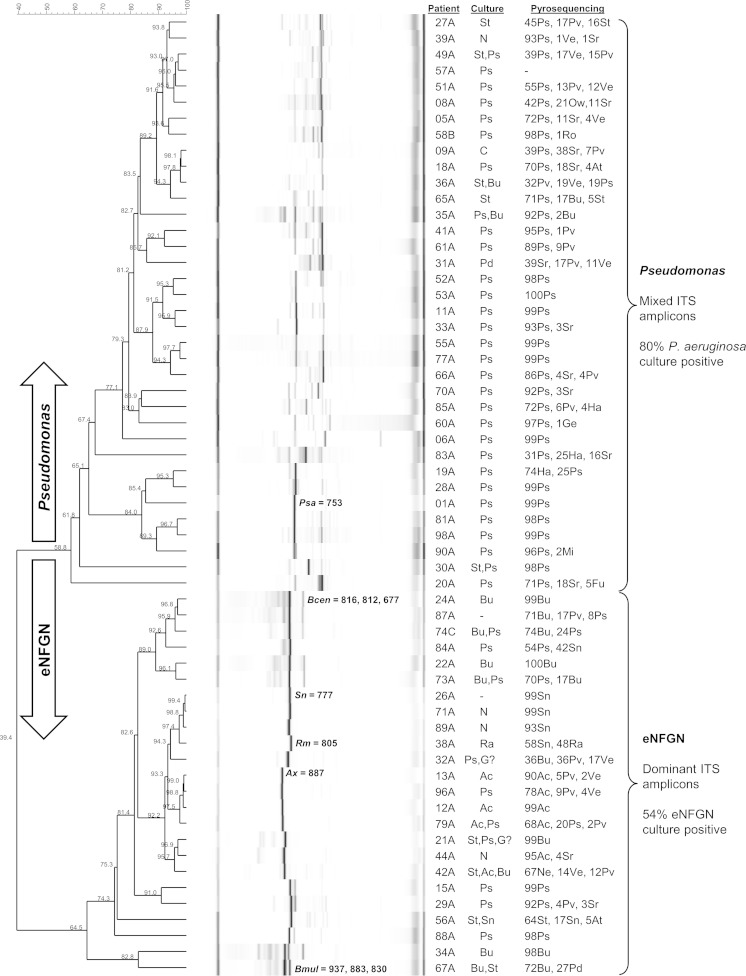
FIG 1.
RISA profiles for the 60 sputum DNA samples selected for 16S rRNA gene pyrosequencing. Cluster analysis of the 60 samples selected for bacterial diversity sequencing analysis is shown; sample 57A was subsequently excluded from further analysis because it produced fewer than 1,000 16S rRNA gene pyrosequencing reads. The two similarity groups correlating to cultivated pathogens—eNFGN species and Pseudomonas—are labeled on the right. The size and correlation of specific ITS amplicons are indicated for samples from which the same species had been cultured. Patient number, microbiology data, and the top 3 bacterial genera (percent relative abundance) in the 16S rRNA sequencing analysis are listed (right columns). The percentage profile similarity is shown on the scale bar (top left) and dendrogram branches. Microbial identifications are abbreviated as follows: Ac, Achromobacter; At, Actinomyces; Bu, Burkholderia; C, Candida; Fu, Fusobacterium; Ha, Haemophilus; G?, unidentified Gram-negative rod; Ge, Gemella; N, normal flora; Mi, Micrococcus; Ne, Neisseria; Pd, Pandoraea; Pv, Prevotella; Ps, Pseudomonas; Ra, Ralstonia; Ro, Rothia, Sn, Stenotrophomonas; St, Staphylococcus; Sr, Streptococcus; Ve, Veillonella; and Ow, Owenweeksia.
Dominant ITS amplicons correlating to Burkholderia, Achromobacter, Ralstonia, and Stenotrophomonas were clearly visible in the eNFGN group profiles, with few other ITS fragments being amplified from these samples (Fig. 1). 16S rRNA gene pyrosequencing analysis substantiated the presumptive bacterial identity based on ITS amplicon size, with sequence reads for each predicted eNFGN genus detected in 20 of the 24 samples clustered in this group (Fig. 1). For example, samples 24A, 26A, 38A, and 13A were positive by ITS amplicon, 16S rRNA gene sequence, and culture analysis for Burkholderia, Stenotrophomonas, Ralstonia, and Achromobacter, respectively (Fig. 1). A dominant 753-bp amplicon correlating to the ITS size of P. aeruginosa was clearly visible in 16 of the Pseudomonas group samples (e.g., sample 01A) (Fig. 1); 32 of 36 (88%) of these samples had Pseudomonas 16S rRNA gene sequence reads as the most abundant genus (Fig. 1). Since culture data and 16S rRNA gene sequencing had validated the identity of the dominant pathogens present in the eNFGN and Pseudomonas RISA groups, respectively, this nomenclature and group designation was adopted for the subsequent microbial diversity analysis.
RISA identified CF pathogens missed by standard culture.
In 11 out of 93 patients (11.8% of cases) where a CF pathogen was not isolated on sputum culture, the RISA profile rapidly identified a specific CF pathogen based on the presence of a dominant, distinctive ITS amplicon, which was confirmed by subsequent 16S rRNA gene-based identification (Table 2). Of these 11 cases, 8 were linked to infection with a difficult-to-identify eNFGN species; 3 samples were associated with dominant Stenotrophomonas species, 3 with Burkholderia species, and 2 with Achromobacter species. For example, samples 71A and 26A were both noted as containing “normal flora” by culture but had a RISA profile containing a single dominant ITS and 16S rRNA gene reads correlating to Stenotrophomonas (Fig. 1). Sample 44A was also identified as containing “normal flora” by culture but was dominated by an ITS and sequence reads correlating to Achromobacter (Fig. 1). The remaining 3 cases were dominated by Pseudomonas; however, one of these (84A) clustered within the eNFGN group, with Stenotrophomonas (42%) being second in abundance to Pseudomonas (54%), yet only P. aeruginosa was reported on culture.
TABLE 2.
CF pathogens missed by culture but captured by the microbiota profiling
| Patient sample | Pathogen or organism isolated by culturea | Result of molecular analysis-based diagnostics |
|
|---|---|---|---|
| RISA cluster | 16S rRNA pyrosequencing | ||
| 9A | Candida | Pseudomonas | 39% Pseudomonas |
| 21A | S. aureus, P. aeruginosa, GNR? failed ID, and yeasts | eNFGN | 99% Burkholderia |
| 26A | None | eNFGN | 99% Stenotrophomonas |
| 32A | P. aeruginosa, GNR? failed ID | eNFGN | 36% Burkholderia |
| 39A | Normal flora | Pseudomonas | 93% Pseudomonas |
| 44A | Normal flora | eNFGN | 95% Achromobacter |
| 71A | Yeasts | eNFGN | 99% Stenotrophomonas |
| 84A | None | eNFGN | 54% Pseudomonas |
| 42% Stenotrophomonas | |||
| 87A | None | eNFGN | 72% Burkholderia |
| 89A | Normal flora | eNFGN | 93% Stenotrophomonas |
| 96A | P. aeruginosa | eNFGN | 78% Achromobacter |
GNR? failed ID, Gram-negative rod with failed initial identification.
RISA profiling rapidly detects dominant CF pathogens and low bacterial diversity.
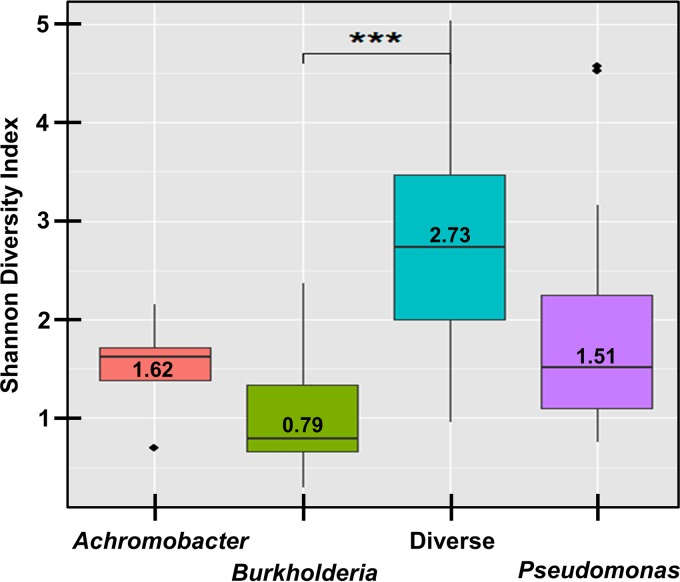
The microbiota dysbiosis between the Pseudomonas group and eNFGN group (Fig. 1) was borne out with respect to a greater proportional abundance of 16S rRNA gene reads for Pseudomonas (P < 0.001), and Burkholderia (P = 0.008), respectively (see Fig. S4 in the supplemental material). Prior to statistical correction, Achromobacter and Stenotrophomonas 16S rRNA gene reads were also associated with the eNFGN group, correlating with presence of dominant ITS amplicons for these respective pathogens in the RISA profiles (Fig. 1). The presence of dominant pathogens was also observed to reduce the overall bacterial diversity seen in 16S RNA gene reads for the samples (Fig. 2). A significantly lower Shannon diversity index was seen in samples dominated by Burkholderia (P < 0.001) compared to samples not dominated by this genus, Stenotrophomonas, or Pseudomonas (Fig. 2). The presence of dominant Pseudomonas and Stenotrophomonas 16S rRNA gene reads also considerably reduced the overall bacterial diversity but this trend did not reach statistical significance (Fig. 2).
FIG 2.
Effect of dominant pathogens on total sputum microbial diversity. The nonparametric Shannon diversity index range (the median is indicated by the line and value) for samples that were dominated by 16S rRNA gene reads for Achromobacter (n = 5), Burkholderia (n = 8), and Pseudomonas (n = 37), respectively, are shown. The diversity index range for samples not dominated by these CF pathogen operational taxonomic units was plotted as a single group designated “Diverse” (n = 9). The difference between the Burkholderia and “Diverse” group samples was significant (P < 0.001; t test with Bonferroni correction).
Pathogen dominance and reduced bacterial diversity correlate to poor lung function.
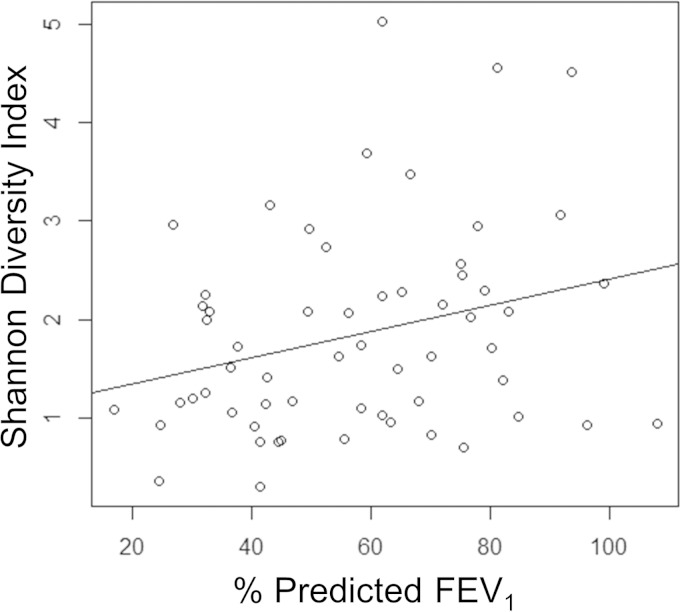
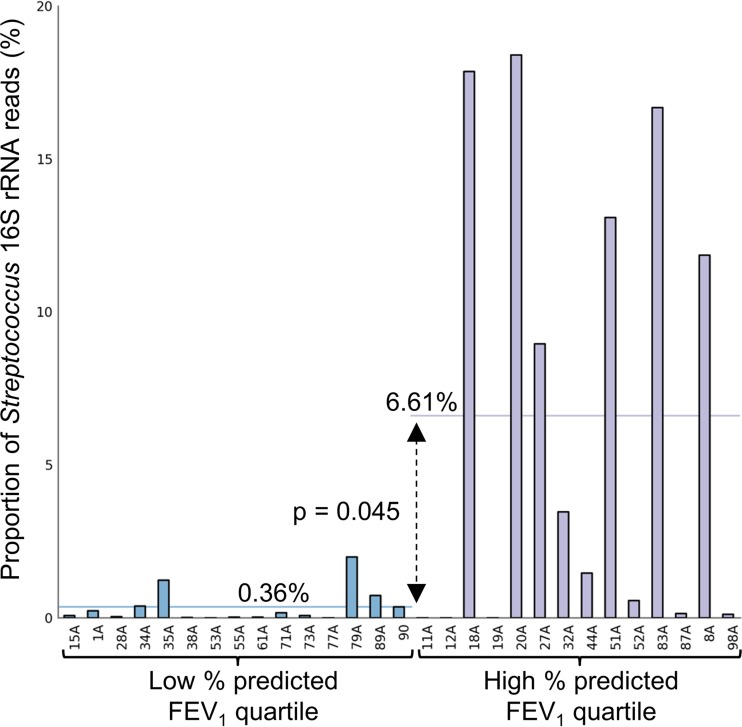
The bacterial diversity data were also compared to clinical parameters for the 59 patients where sufficient depth of 16S rRNA gene reads were obtained. Correlation of lung function (percent predicted force expiratory volume in 1 s [FEV1]) to bacterial diversity revealed a significant (P = 0.033) trend between low diversity and poor lung function (Fig. 3). Correlation of the microbiota sequencing data with patient age and body mass index (BMI) also demonstrated that low bacterial diversity was associated with increased age and low BMI, although these trends did not reach significance (see Fig. S5 in the supplemental material). To identify the bacterial genera that were reduced in number as lung disease severity increased, patients were divided into those with upper-quartile and lower-quartile percent predicted FEV1. The proportional abundance of Streptococcus was significantly (P = 0.045) reduced in patients with lung function in the lowest quartile (Fig. 4). A trend in reduction for 16S rRNA gene reads for Gemella, Granulicatella, Prevotella, and Veillonella was also observed in the patients with low-quartile lung function but did not reach statistical significance (see Fig. S6 in the supplemental material).
FIG 3.
Loss of lung function correlates with reduced microbial diversity. The nonparametric Shannon diversity index for the 59 patients with complete 16S rRNA pyrosequencing data was plotted against their percent predicted forced expiratory volume in 1 s (FEV1). The trend shown by the regression line was significant (P = 0.033; adjusted R2, 0.06078; F statistic, 4.754 on 1 and 57 degrees of freedom).
FIG 4.
The relative abundance 16S rRNA gene reads for Streptococcus is significantly reduced in patients with low lung function. Bacterial diversity for patients with lung function in the lowest quartile (n = 15) was compared by ANOVA to that for individuals with top quartile percent predicted FEV1 (n = 14 for each group [indicated by brackets]). The proportion of reads for Streptococcus has been plotted for each patient. The mean abundances for the low- and high-quartile groups (horizontal lines) were significantly different (P = 0.045; Storey FDR correction).
DISCUSSION
Multiple studies have shown that considerable microbial diversity is present within the infected lungs of people with CF (7, 8). The recent demonstration that high pathogen abundance (12) and low microbial diversity are clinically linked to severe lung disease (11, 13, 14) provides compelling arguments for the introduction of diagnostic approaches that account for CF microbiome types into routine clinical practice. Applying microbiome-based surveillance on a large scale for CF infection has been limited by multiple factors, including the technical complexity of extracting template DNA and the specialist sequencing or bioinformatics expertise required for 16S rRNA gene analysis. We have shown that these issues can be overcome by linking CF bacteriology with sputum DNA samples extracted for clinically accredited sputum virology and applying basic PCR profiling (16) ahead of state-of-the-art microbiota sequencing approaches (11, 13, 14). This simple, PCR-led molecular diagnostic scheme should be widely applicable in current CF microbiology laboratories and can accurately detect dominant infections with problematic eNFGN bacteria and the loss of microbial diversity in patients with severe lung disease.
Recent studies have shown that low bacterial diversity in CF is associated with severe lung disease (11–14) and this observation was corroborated in the present study, one of the largest surveys of the CF microbiome to date (Fig. 3). P. aeruginosa has been shown to be one of the pathogens that may dominate when microbial diversity is lost (11, 13, 14). Evidence for Achromobacter and Burkholderia dominance in the explanted lungs of individual patients with end-stage lung disease has been established (14). Dominant Achromobacter (1 patient) and Stenotrophomonas (5 patients) was also observed in bronchoalveolar lavage samples from children with CF (28). Our data expands the range of eNFGN pathogens observed to dominate during CF infection to include the genus Ralstonia (Fig. 1). We have also shown for the first time that the presence of Burkholderia is statistically correlated to reduced microbial diversity in CF (Fig. 2), correlating to their known ability to secrete antagonistic antibiotics capable of killing other CF bacteria (29). Extrapolation of the systematic identification data from the 59 16S rRNA gene-sequenced samples (Fig. 1) to the 200 RISA profiles (see Fig. S3 in the supplemental material) suggests that over 40% of CF adults have infections dominated by a single Gram-negative antimicrobial-resistant pathogen.
While identification of P. aeruginosa is straightforward by conventional culture, the eNFGN species identified by our CF microbiota analysis pose challenges to correct isolation and identification (3, 30). Lack of identification of eNFGN species despite their domination of the 16S rRNA gene sequencing data or RISA analysis was seen in 8 cases, with two of these (32A and 21A) (Fig. 1) being reported as culture positive for P. aeruginosa and an unknown Gram-negative organism (Table 2). The relative abundance of Pseudomonas in the two latter samples was 3.4% and 0.001%, respectively, suggesting that the status of P. aeruginosa as the major CF pathogen leads to its being reported by CF microbiology laboratories, even if its numbers are highly diminished in a sample. Given that one quarter of the 200 CF sputum samples examined were dominated by either Burkholderia, Stenotrophomonas, Ralstonia, or Achromobacter (see Fig. S3 in the supplemental material), P. aeruginosa may be overreported and eNFGN species underreported by routine CF microbiology. Since these eNFGN species are innately antibiotic resistant and are associated with both low diversity and severe lung infection, measures to improve their quantitative identification and subsequent therapeutic targeting should be considered.
It has been recognized that in addition to routine culture-based methodologies, CF microbiology needs to embrace a range of molecular techniques capable of dealing with the complexity of the lung microbiome, in particular using direct sampling methods to identify unconventional or emerging CF pathogens (30). Clinical microbiology laboratories considering introducing a simple microbiota profiling scheme into their CF diagnostic practices should consider the following. In terms of high-throughput robotic DNA extraction systems, multiple platforms on the market perform at a level equivalent to that of the system used in this study. To evaluate their suitability, the ability to lyse certain target organisms, such as mycobacteria and fungi, which may be difficult to break open, should be assessed. After RISA PCR, care should be taken with the interpretation of the RISA ITS amplicon profiles, as these are a measure of bacterial diversity rather than an absolute means to identify specific pathogens. ITS standards for P. aeruginosa and the common eNFGN species should be generated as controls, but correlation of ITS amplicon size should not be taken as unequivocal proof of identity; follow-up using species-specific PCRs or 16S rRNA gene sequencing should be employed for this purpose. Overall, by using a basic RISA microbiota profiling scheme, clinical laboratories should be able to (i) rapidly differentiate sputum samples dominated by P. aeruginosa or an eNFGN species; (ii) follow up with further identification of eNFGN pathogens, such as Burkholderia, Achromobacter, Stenotrophomonas, and Ralstonia, that were missed by culture and are difficult to identify; and (iii) report a status of either “diverse” or “pathogen dominated” for sputum samples, to help with future understanding of how to select antibiotics and manage polymicrobial CF infections.
Applying cultivation-independent molecular approaches based on DNA has been integral in advancing our current understanding of the CF microbiome (31) and clinically effective in surveillance and control of highly transmissible pathogens, such as Burkholderia (32). Obtaining template nucleic acid representative of the infectious organisms in sputum is a fundamental consideration for the translation of molecular diagnostics into routine clinical use. Because of the difficulties associated with the culture of viruses, clinical virology has already validated multiple molecular techniques such as sputum PCR and DNA sequencing for routine use (33). We have shown that CF sputum DNA isolated using a clinically accredited, automated protocol for respiratory virus diagnosis (20) serves as a clinically valid template for bacterial diversity analysis in CF. Linking of virology diagnostic resources with CF bacteriology should be exploited in future to allow the evaluation of the respiratory microbiome in day-to-day clinical practice. Such strategies could also prove highly relevant to other respiratory diseases, such as non-CF bronchiectasis or chronic obstructive pulmonary disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Manchester Adult Cystic Fibrosis Centre charity and research development funding from the Microbiology and Infection Translational Research Group, Cardiff, Wales, United Kingdom. A.S. and J.R.M. also acknowledge the Biotechnology and Biological Sciences Research Council (grant BB/J019143/1) for funding the establishment of the 16S rRNA gene microbial diversity analysis pipelines.
We have no conflicts of interest to declare in relation to this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00432-15.
REFERENCES
- 1.O'Sullivan B, Freedman S. 2009. Cystic fibrosis. Lancet 373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Lipuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahenthiralingam E. 2014. Emerging cystic fibrosis pathogens and the microbiome. Paediatr Respir Rev 15(Suppl 1):S13–S15. doi: 10.1016/j.prrv.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 5.Kerem E, Corey M, Gold R, Levison H. 1990. Pulmonary function and clinical course in patients with cystic fibrosis after pulmonary colonization with Pseudomonas aeruginosa. J Pediatr 116:714–719. doi: 10.1016/S0022-3476(05)82653-8. [DOI] [PubMed] [Google Scholar]
- 6.Jones AM, Dodd ME, Govan JR, Barcus V, Doherty CJ, Morris J, Webb AK. 2004. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59:948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers GB, Daniels TW, Tuck A, Carroll MP, Connett GJ, David GJ, Bruce KD. 2009. Studying bacteria in respiratory specimens by using conventional and molecular microbiological approaches. BMC Pulm Med 9:14. doi: 10.1186/1471-2466-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tunney MM, Field TR, Moriarty TF, Patrick S, Doering G, Muhlebach MS, Wolfgang MC, Boucher R, Gilpin DF, McDowell A, Elborn JS. 2008. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 9.Worlitzsch D, Rintelen C, Böhm K, Wollschläger B, Merkel N, Borneff-Lipp M, Döring G. 2009. Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clin Microbiol Infect 15:454–460. doi: 10.1111/j.1469-0691.2008.02659.x. [DOI] [PubMed] [Google Scholar]
- 10.Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, Lory S, Brodie EL, Lynch SV, Bohannan BJ, Green JL, Maurer BA, Kolter R. 2010. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 12:1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 11.Blainey PC, Milla CE, Cornfield DN, Quake SR. 2012. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med 4:153ra130. doi: 10.1126/scitranslmed.3004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL, Lynch SV. 2010. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. 2012. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers GB, Hoffman LR, Whiteley M, Daniels TW, Carroll MP, Bruce KD. 2010. Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends Microbiol 18:357–364. doi: 10.1016/j.tim.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borneman J, Triplett EW. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63:2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Martínez J, Acinas SG, Antón AI, Rodríguez-Valera F. 1999. Use of the 16S–23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods 36:55–64. doi: 10.1016/S0167-7012(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 18.Baxter CG, Rautemaa R, Jones AM, Webb AK, Bull M, Mahenthiralingam E, Denning DW. 2013. Intravenous antibiotics reduce the presence of Aspergillus in adult cystic fibrosis sputum. Thorax 68:652–657. doi: 10.1136/thoraxjnl-2012-202412. [DOI] [PubMed] [Google Scholar]
- 19.Anonymous. 2010. Laboratory standards for processing microbiological samples from people with cystic fibrosis. UK Cystic Fibrosis Trust, Bromley, United Kingdom. [Google Scholar]
- 20.Flight WG, Bright-Thomas RJ, Tilston P, Mutton KJ, Guiver M, Morris J, Webb AK, Jones AM. 2014. Incidence and clinical impact of respiratory viruses in adults with cystic fibrosis. Thorax 69:247–253. doi: 10.1136/thoraxjnl-2013-204000. [DOI] [PubMed] [Google Scholar]
- 21.White J, Gilbert J, Hill G, Hill E, Huse S, Weightman A, Mahenthiralingam E. 2011. Culture-independent analysis of bacterial fuel contamination provides insights into the level of concordance with the standard industry practice of aerobic cultivation. Appl Environ Microbiol 77:4527–4538. doi: 10.1128/AEM.02317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bikandi J, San Millan R, Rementeria A, Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics 20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 23.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 28.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. 2007. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci U S A 104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, Boaisha O, Paine J, Knight D, Challis GL. 2011. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem Biol 18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Burns JL, Rolain JM. 2014. Culture-based diagnostic microbiology in cystic fibrosis: can we simplify the complexity? J Cyst Fibros 13:1–9. doi: 10.1016/j.jcf.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Rogers GB, Carroll MP, Bruce KD. 2009. Studying bacterial infections through culture-independent approaches. J Med Microbiol 58:1401–1418. doi: 10.1099/jmm.0.013334-0. [DOI] [PubMed] [Google Scholar]
- 32.Drevinek P, Vosahlikova S, Dedeckova K, Cinek O, Mahenthiralingam E. 2010. Direct culture independent strain typing of Burkholderia cepacia complex from cystic fibrosis sputum. J Clin Microbiol 48:1888–1891. doi: 10.1128/JCM.02359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratcliff RM, Chang G, Kok T, Sloots TP. 2007. Molecular diagnosis of medical viruses. Curr Issues Mol Biol 9:87–102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.