Abstract
Unbiased nontargeted metagenomic RNA sequencing (UMERS) has the advantage to detect known as well as unknown pathogens and, thus, can significantly improve the detection of viral, bacterial, parasitic, and fungal sequences in public health settings. In particular, conventional diagnostic methods successfully identify the putative pathogenic agent in only 30% to 40% of respiratory specimens from patients with acute respiratory illness. Here, we applied UMERS to 24 diagnostic respiratory specimens (bronchoalveolar lavage [BAL] fluid, sputum samples, and a swab) from patients with seasonal influenza infection and 5 BAL fluid samples from patients with pneumonia that tested negative for influenza to validate RNA sequencing as an unbiased diagnostic tool in comparison to conventional diagnostic methods. In addition to our comparison to PCR, we evaluated the potential to retrieve comprehensive influenza virus genomic information and the capability to detect known superinfecting pathogens. Compared to quantitative real-time PCR for influenza viral sequences, UMERS detected influenza viral sequences in 18 of 24 samples. Complete influenza virus genomes could be assembled from 8 samples. Furthermore, in 3 of 24 influenza-positive samples, additional viral pathogens could be detected, and 2 of 24 samples showed a significantly increased abundance of individual bacterial species known to cause superinfections during an influenza virus infection. Thus, analysis of respiratory samples from known or suspected influenza patients by UMERS provides valuable information that is relevant for clinical investigation.
INTRODUCTION
Influenza has a severe impact on our health system, not only owing to its potential to cause worldwide pandemics but also due to the high number of seasonal infections. Bacterial and/or viral coinfections and subsequent pneumonia can lead to enhanced illness in elderly and immunosuppressed patients. Approximately 0.5% of all influenza A infections in healthy younger adults and 2.5% of influenza A infections in the elderly and younger children are accompanied by severe bacterial-induced pneumonia (1, 2). These numbers are significantly higher during pandemic episodes (3–5). The most common causes of coinfections observed in both pandemic and seasonal episodes of influenza A infections are Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, and Streptococcus pyogenes (6, 7). This relationship between influenza virus and bacterial pathogenicity is underlined by several studies using animal models. For example, mice infected with influenza virus or Streptococcus pneumoniae alone showed mortality rates of 35% and 15%, respectively, whereas mice coinfected with influenza virus and Streptococcus pneumoniae displayed a 100% mortality rate (8). Furthermore, several studies in humans indicate that colonization with Streptococcus pneumoniae increases the risk of severe complications associated with influenza A viral infection (9, 10), thus highlighting the importance of rapid diagnosis of bacterial coinfections or superinfections.
Next-generation sequencing (NGS) approaches hold a unique potential to overcome challenges in diagnostics and detection and likely will significantly improve our ability to detect and diagnose pathogenic infections. Recent advances in genome sequencing and bioinformatics, with declining costs of NGS methods, enable the application of this technique in routine diagnostic settings, where gold-standard techniques fail to detect a putative pathogen. NGS techniques provide us with an unprecedented opportunity to directly identify pathogens in clinical samples of hitherto idiopathic diseases.
The reliable, unbiased, and comprehensive metagenomic analysis of clinical samples requires the establishment of streamlined protocols with regard to sample preparation (e.g., key variables, such as processing and handling of different diagnostic specimens, and preanalytical reduction of sample complexity by removal of host nucleic acids), usage of different sequencing platforms (e.g., preferential use of short-read/high-coverage versus long-read/medium-coverage techniques, depending on the nature of the clinical sample and diagnostic question), as well as subsequent bioinformatic processing (e.g., de novo assembly or phylogenetic analysis of viral or bacterial genomes).
Given the above, the goals of this study were to (i) compare the sensitivity of influenza virus quantitative PCR (qPCR) and metagenomic sequencing of routine diagnostic material, (ii) evaluate the potential to extract full genome information of influenza viruses from the latter, (iii) analyze the detection of pathogens known to cause superinfections in patients with influenza virus infection, and (iv) estimate the feasibility of RNA sequencing from respiratory specimens as a putative diagnostic application in specific public health settings.
MATERIALS AND METHODS
Diagnostic samples.
Samples were received from the University Medical Center Hamburg-Eppendorf, Institute for Medical Microbiology. Respiratory samples included bronchoalveolar lavage (BAL) fluid, sputum samples, or swabs from patients with respiratory illness and suspected influenza infection. Swab samples were received from ambulatory patients with signs of influenza virus infection, whereas sputum samples, BAL fluid, and tracheal secretions were received from hospitalized patients with underlying diseases (mostly immunosuppressed patients) and suspected influenza virus infection. All samples were screened by standard diagnostic quantitative real-time PCR (RT-PCR) for influenza A and B virus.
Five BAL fluid samples collected from immunosuppressed patients (patients with hematopoietic stem cell transplantation) hospitalized because of severe pneumonia were included. These samples tested negative for influenza A and B by routine diagnostic RT-PCR. The samples were analyzed in a blinded fashion; diagnostic findings received by conventional diagnostics were not known to the scientists performing library preparation, sequencing, or primary data analysis.
The study was approved in compliance with relevant laws and institutional guidelines by the local ethics committee, Freie Hansestadt Hamburg, WF-025/12. The study was conducted retrospectively on anonymously stored clinical samples. Information which would allow the identification of the patient (name, address, birth date, hospitalization number) was removed. The samples were collected between November 2012 and March 2013.
Nucleic acid extraction.
Nucleic acid from 0.2 ml BAL fluid or sputum sample was extracted using an automated extraction system, NucliSens easyMag, from bioMérieux following the manufacturer's instructions. Swabs were incubated with 1 ml D solution containing guanidine thiocyanate, 1 M Tris HCl (pH 6.4), and 1% beta-mercaptoethanol, followed by nucleic acid extraction as described above using 0.2 ml of the solution.
Library preparation and high-throughput sequencing.
Illumina libraries from RNA were generated using a modified protocol of the Script Seq v2 RNA-seq kit (Epicentre Biotechnologies) (11). A total of 15 ng total RNA quantified by Qubit (Invitrogen) after DNase treatment was subjected to size fragmentation, followed by cDNA synthesis and the addition of a terminal tagged oligonucleotide. Di-tagged cDNA was purified with Agencourt AMPure XP beads followed by amplification (15 cycles). Fragment length distribution of all libraries was analyzed on a BioAnalyzer high-sensitivity LabChip. Diluted libraries (2 nM) were multiplex sequenced on the Illumina MiSeq (2- by 250-bp paired-end run, 2 to 3 million reads/sample) or HiSeq 2500 instrument (2- by 100-bp paired-end run, 30 to 40 million reads/sample).
De novo contig assembly and taxonomic classification.
Analysis of total RNA sequencing data were performed as recently described (11, 12), with modifications to detect taxonomic ambiguity among the detected sequences. To subtract reads originating from the host, reads were first aligned to the human reference assembly (NCBI 37.2) using Bowtie2 (v2.1.0). Trinity (r2013-02-25) was used to assemble contigs from reads not producing significant host alignments. Contigs assembled from 2- by 100-bp paired-end HiSeq reads were subsequently filtered for sequences of a minimal length of 300 bp. For MiSeq reads (2- by 250-bp paired-end reads), an increased length cutoff of 400 bp was used. To estimate contig abundance, all reads not aligning to human sequences were remapped to the filtered contigs using Bowtie2. Putative PCR duplicates were excluded from the abundance estimation.
For taxonomic classification, filtered contigs were aligned to the NCBI nucleotide database using the BLAST+ package (v2.2.30). A first round of alignments was performed with megaBLAST. All sequences not producing significant megaBLAST hits (E value cutoff, 0.01) were subsequently included in a second alignment round employing BLASTn. Contigs failing to produce alignments with an E value of ≤0.01 in either round were classified as unknown sequences. For each of the remaining contigs, all BLAST hits with a maximum bit score difference of 7 (corresponding to a maximum difference of P values of <0.01) relative to the hit with the highest observed bit score were retained. In order to establish the level of taxonomic ambiguity for each contig sequence, the lowest shared taxonomic ancestor was subsequently determined by moving up the taxonomic tree until an unambiguous assignment could be made for all retained BLAST hits. For downstream analysis on a given taxonomy level, only contigs with an unambiguous assignment at or below the chosen taxonomic level were used.
To avoid taxonomic assignments which are of doubtful significance (for example, due to sequences which exhibit nucleotide homology only across a minor fraction of the entire length of the contig), only contigs with at least one BLAST hit that extended over at least 80% of the entire contig length and exhibited at least 80% nucleotide identity were considered principally classifiable and retained for downstream analysis.
Identification of rRNA contigs.
All contigs were screened for 16S and 23S rRNA signatures using HMMER (v3.1b1) (13). Hidden Markov models (HMMs) were derived from 732 and 1,409 sequences obtained from RefSeq. Using Usearch (v7.0.1090), (14), the sequences were clustered at 90% sequence similarity, and one representative centroid was selected for each resulting cluster. Multiple sequence alignments, of which the HMMs were eventually built, were generated with Muscle (v3.8.31) (15). The whole set of originally obtained sequences was used to verify the sensitivity of the models built. To increase power at the expense of speed, all heuristic filters were turned off whenever hmmsearch was invoked. All contigs with an observed E value of maximally 0.01 were considered to be of ribosomal origin.
In silico modeling.
The detection sensitivity of the analysis pipeline was assessed using simulated influenza reads and, as a background, randomly selected reads from the pooled reads of the influenza virus-negative control samples (samples 1 to 5). The total number of paired-end reads to be included in each analysis was fixed as 50,000,000, whereas the absolute abundance of simulated influenza virus reads was increased in four steps from 125 up to 1,000 reads, corresponding to a relative abundance of 0.00025% up to 0.002%. In addition, different mutations rates, ranging from 0.05% to 25%, were introduced in the simulated influenza virus reads. The resulting 20 distinct combinations of relative abundance and mutation rates among the influenza virus reads were independently processed three times, each time with a newly generated set of simulated influenza virus reads and randomly selected background reads. To allow assessment of the background's influence on viral read recovery, all simulations were additionally carried out solely with simulated influenza reads.
For the simulation of 100-bp paired-end reads from the influenza reference sequences (GenBank accession numbers FJ966079.1, FJ966080.1, FJ966081.1, FJ966082.1, FJ966083.1, FJ966084.1, FJ966085.1, and FJ966086.1), the program wgsim of the SAMtools package (16) was employed in haplotype mode. Aside from parameters explicitly mentioned above, the program was invoked with default parameters.
All simulated data sets were analyzed by the same pipeline used for the analysis of clinical samples.
Variant calling.
Reads were aligned to the corresponding reference assemblies using Bowtie2 (v2.2.3). SAMtools (v0.1.18) was employed to remove putative PCR duplicates. Alignments of samples belonging to the same reference assemblies were merged. For each of the resulting two pools, variants were called with FreeBayes (v0.9.18-1-g4233a23) (17). Putative variants were filtered for quality (threshold 20), and positions at which at least one sample supported both the reference and an alternative sequence with at least five reads were visually assessed using the integrative genomics viewer (v2.3.40) (18).
RT-PCR.
The PCR primers and specific probes for influenza virus quantitative PCR used have been described previously (19–21). The following primers and probes were used: InflA_F (GACAAGACCAATCCTGTCACYTCTG), InflA_R (AAGCGTCTACGCTGCAGTCC and HEX-TTCACGCTCACCGTGCCCAGTGAGC-BHQ2 [HEX indicates 5′-hexachlorofluorescein; BHQ2, black hole quencher 2]), InflB_F (TCGCTGTTTGCAGACACAAT), InflB_R (TTCTTTCCCACCGAACCA and Cyan500-AGAAGATGGAGAAGGCAAAGCAGAACT-dabcyl-dT [DB]) (19), M_InflA_F (AAGACCAATCCTGTCACCTCTGA), M_InflA_R (CAAAGCGTCTACGCTGCAGTCC and FAM-TTTGTGTTCACGCTCACCGT-BHQ1 [FAM indicates 6-carboxyfluorescein], for the detection of the FLU A matrix protein) (21), HA_H1SWAS (ATGCTGCCGTTACACCTTTGT), and HA_H1SWS (CATTTGAAAGGTTTGAGATATTCC and FAM-ACAAGTTCATGGCCCAATCATGACTCG-BHQ1, for H1N1 subtyping) (20).
PCRs were performed using the Quantifast pathogen RT-PCR kit +IC (Qiagen). A total of 5 μl of eluted nucleic acid was amplified in a total volume of 25 μl on Roche Lightcycler 480 instruments using the following conditions: 20 min at 50°C, 5 min at 95°C, 45× 15 s at 95°C, and 30 s at 60°C. PCRs were carried out in a routine diagnostic environment which underlies internal and external quality controls. With regard to diagnostic accreditation, all diagnostic PCR tests are validated with clinical specimens and multicenter tests.
Sequence alignments and phylogenetic trees.
Maximum likelihood phylogeny analysis was performed using CLC main work bench 6.6.1.
Sequence data for all 29 samples have been submitted to the European Nucleotide Archive (ENA) and will be publicly available at http://www.ebi.ac.uk/ena/data/view.
RESULTS
Influenza RT-qPCR.
The influenza-positive specimens analyzed in our study represented 24 respiratory samples (from BAL fluid, n = 7; sputum sample, n = 5; swabs, n = 11; and a tracheal secretion, n = 1) from patients with seasonal influenza virus infection collected during the winter season of 2012-2013. All samples had previously tested positive for influenza A or B virus by routine diagnostic RT-qPCR using influenza-specific TaqMan probes and were stored at −80°C. To exclude false-negative findings resulting from degradation of stored samples, RT-qPCR for influenza A or B virus was repeated using three primer pairs and TaqMan probes: (i) influenza A primers (FluA) able to detect both H1N1 and H3N2 genotypes, (ii) a primer set specific for H1N1, and (iii) primers specific for influenza B virus (FluB). As shown in Table 1, a single sample was influenza B positive, while 23 samples were positive for influenza A viral sequences; 11 samples produced significant threshold cycle (CT) values for H1N1 specific primers, while 12 samples were negative in the H1N1 PCR. These sequences were not further genotyped, assuming that the subtypes of influenza A currently circulating in the human population are H1N1 and H3N2. In general, CT values ranged between 23 and 40, with 5 samples (samples 677, 2,535, 1,689, 768, and 2,544) exhibiting CT values ≥35. In addition to the influenza-positive samples, 5 respiratory samples (BAL fluid) from patients with respiratory infections which had tested negative for influenza A or B sequences were included in our study.
TABLE 1.
Summary of clinical samples and routine diagnostic results
| Sample no. | Diagnostic entity | PCR influenza CT valuesa |
Influenza genotype | ||
|---|---|---|---|---|---|
| FluA | Matrix | H1 | |||
| 104 | BAL fluid | 26 | 28 | Neg | fluAb |
| 755 | BAL fluid | 33 | 32 | 30 | H1N1 |
| 1,116 | BAL fluid | 30 | 28 | 29 | H1N1 |
| 1,721 | BAL fluid | 26 | 28 | Neg | fluAb |
| 2,535 | BAL fluid | 40 | NT | Neg | fluAb |
| 2,292 | BAL fluid | 24 | NT | 23 | H1N1 |
| 3,157 | BAL fluid | 30 | NT | Neg | fluAb |
| 1,773 | Sputum | 25 | 27 | Neg | fluAb |
| 1,168 | Sputum | 23 | 24 | Neg | fluAb |
| 853 | Sputum | 27 | 25 | 25 | H1N1 |
| 677 | Sputum | Neg | 35 | 34 | H1N1 |
| 208 | Sputum | 30 | Neg | 27 | H1N1 |
| 2,373 | Swab | 31 | NT | Neg | fluAb |
| 2,098 | Swab | 33 | NT | Neg | fluAb |
| 1,689 | Swab | 36 | 40 | Neg | fluAb |
| 1,647 | Swab | 31 | 34 | Neg | fluAb |
| 1,539 | Swab | 30 | 30 | 28 | H1N1 |
| 1,538 | Swab | 30 | 29 | 29 | H1N1 |
| 768 | Swab | 36 | 35 | 34 | H1N1 |
| 83 | Swab | 29 | 29 | 25 | H1N1 |
| 848 | Swab | 28 | 26 | 26 | H1N1 |
| 2,295 | Swab | 32 | 28 | Neg | fluAb |
| 2,544 | Swab | 35 | NT | Neg | fluAb |
| 14,087 | Secretion | Neg | NT | NT | InflB (CT of 29) |
Neg, negative; NT, not tested.
These sequences were negative in H1N1 genotyping; influenza A subtypes currently circulating in the human population are H1N1 and H3N2. InflB, influenza B.
RNA sequencing to detect influenza virus sequences in diagnostic samples.
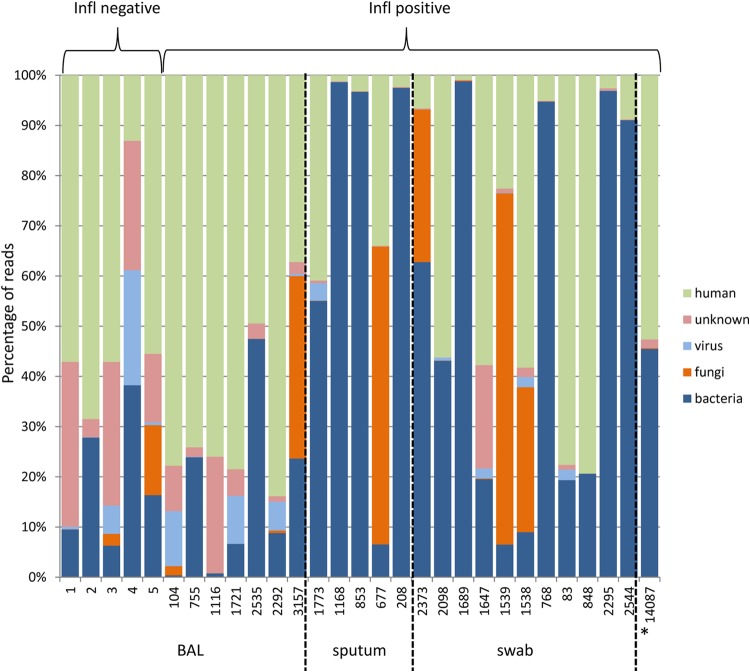
From each sample, strand-specific RNA-seq libraries were constructed and multiplex sequenced with 1.5 to 3.5 million or 25 to 45 million reads per sample on an Illumina MiSeq or HiSeq2500 instrument, respectively (see Table S1 in the supplemental material). Reads passing quality filters were aligned to the human reference genome to deplete sequences of host origin. The complete workflow is outlined in Fig. S1 in the supplemental material. As expected, the samples exhibited marked variation with regard to the frequency of human sequences (Fig. 1; see also Table S2 in the supplemental material). The BAL fluid samples contained an average 60.77% of human reads (minimum, 13.06%; maximum, 83.86%), sputum sample, an average of 16.3% (minimum, 1.2%; maximum, 40.91%), and swab samples, an average of 33.61% (minimum, 0.93%; maximum, 79.34%).
FIG 1.
Diagnostic sample composition at the phylum level for digitally subtracted host reads as well as reads mapped to taxonomically classified sequence contigs. Phylum profiles (proportion of all reads) for bacteria, fungi, and viruses are shown for the individual samples. Sample 14,087 (labeled with *) originated from a tracheal secretion.
Filtered reads were subjected to de novo contig assembly, producing between 510,402 and 17,953,404 contigs from HiSeq data sets and between 129,072 and 1,175,340 contigs from MiSeq data sets (see Table S1 in the supplemental material). After filtering for contigs of at least 300 or 400 nucleotides (HiSeq and MiSeq data sets, respectively), between 369 and 29,688 contigs of nonhuman origin remained for samples processed on the HiSeq, whereas between 16 and 2,799 contigs were retained for samples processed on the MiSeq instrument. The complete set of host-depleted reads was then aligned to the contigs to allow estimation of the abundance of individual sequences. For each contig, we calculated a normalized value representing the number of reads per million mapped reads (RPM). For homology-based taxonomic classification, all contigs were compared to the NCBI nucleotide database. Contigs with more than one BLAST hit were classified according to the lowest common taxonomic ancestor of individual hits (see the Material and Methods section for details). When visualizing the taxonomic assignment of the contigs to cellular organisms, viruses, or unclassified sequences (Fig. 2 and 3; Table 2; see also Table S2 in the supplemental material), it was obvious that 18 of the 24 samples contained a significant number of reads matching ssRNA viruses, family Orthomyxoviridae, genus Influenzavirus, encompassing 0.0003% to 29.6% of reads in matched contigs (RPM values between 4 and 365,373; Table 2). In accordance with the RT-qPCR results, 17 libraries contained contigs matching influenza A, whereas the library from sample 14,087 contained influenza B sequences. Among the 6 libraries which had been positive by PCR but negative by NGS, all but one (sample 1,116) had CT values above 35, indicating that failure to detect influenza virus infection was due to a very low abundance of viral sequences.
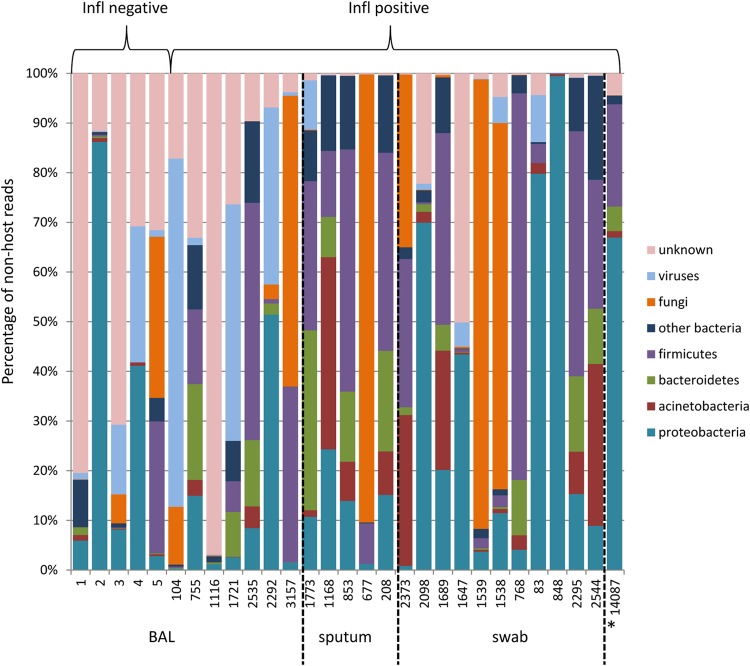
FIG 2.
Diagnostic sample composition at the phylum level without host sequences. Phylum profiles (percentage of reads mapping to nonhost contigs) for bacteria, fungi, and viruses are shown for the individual samples. Sample 14,087 (labeled with *) originated from a tracheal secretion.
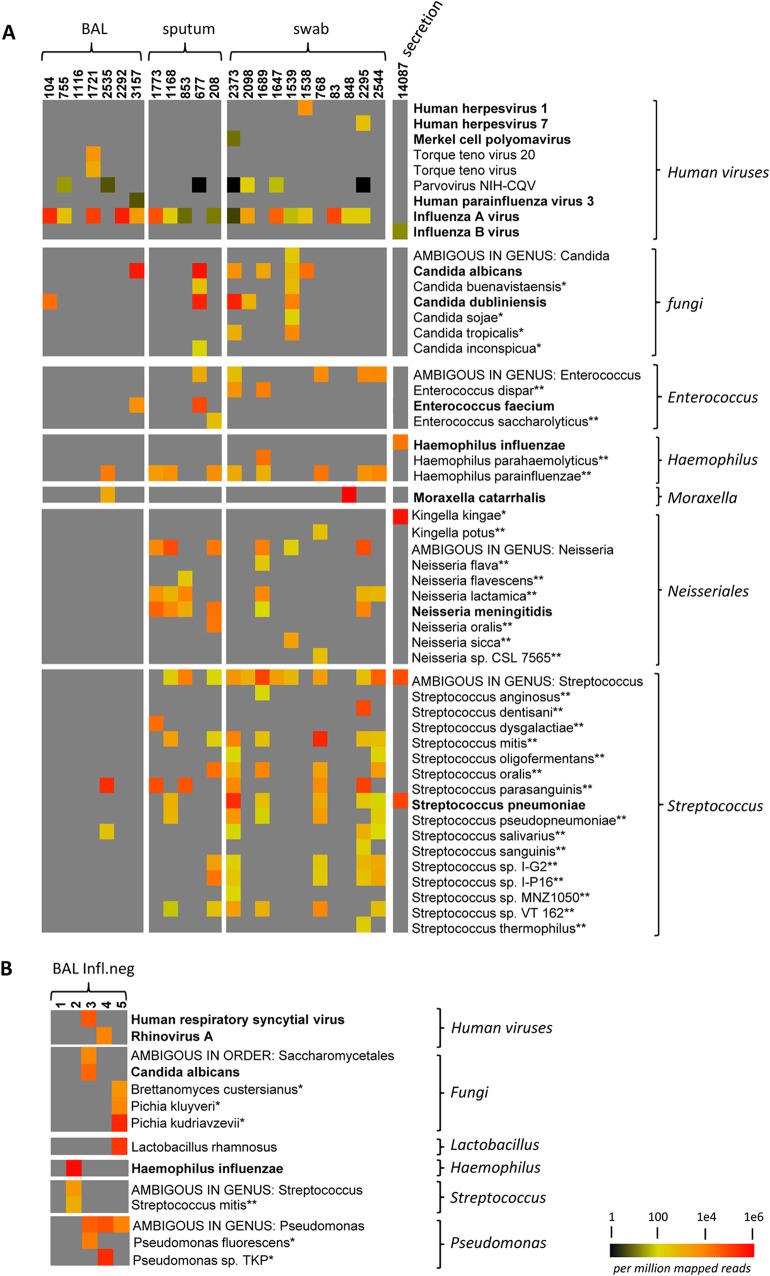
FIG 3.
Next-generation sequencing of RNA (RNA-seq) of clinical respiratory samples. (A) RNA-seq results obtained from BAL fluid, swab, or sputum samples with influenza A (InflA) or influenza B (InflB) (sample 14,097) infection. (B) RNA-seq results of RNA isolated from influenza-negative BAL fluid samples. The relative and normalized abundance of reads mapping to bacterial or viral species (in reads per million mapped reads [RPM]) is represented according to the heat map legend shown in the lower right corner. A gray rectangle indicates no reads were detected. For bacteria and fungi, only contigs with >2,000 RPM and BLAST hits covering at least 80% of the contig sequence with at least 80% sequence identity, were included. In addition, only contigs with an unambiguous classification on the selected taxonomy level were considered. Bacteria, fungi, or viruses associated with respiratory diseases are indicated in bold letters. *, common environmental microorganism; **, nonpathogenic commensal bacteria.
TABLE 2.
NGS data (on influenza-positive samples) showing pathogens known to cause respiratory infections and results of diagnostic culture
| Sample source and no. | RPMa of: |
Culture and PCR for virusesb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| InflA/InflB | S. pneumoniae | N. meningitidis | H. influenzae | M. catarrhalis | S. aureus | C. dubliniensis | C. albicans | ||
| BAL fluid | |||||||||
| 104 | 211,170 | 20,044 | Candida, alpha-hemolytic streptococci | ||||||
| 755 | 380 | Commensal bacteria | |||||||
| 1,116 | Negative | ||||||||
| 1,721 | 109,031 | Commensal bacteria | |||||||
| 2,535 | 1,281 | Commensal bacteria | |||||||
| 2,292 | 365,373 | na | |||||||
| 3,157c | 2,517 | 412,404 | HSV, yeast; commensal bacteria | ||||||
| Sputum samples | |||||||||
| 1,773 | 57,493 | 28,049 | Commensal bacteria | ||||||
| 1,168 | 200 | 863 | 6,520 | Commensal bacteria | |||||
| 853 | 10 | 1,075 | na | ||||||
| 677 | 310,610 | 566,218 | Commensal bacteria | ||||||
| 208 | 14 | 17,323 | Commensal bacteria | ||||||
| Swabs | |||||||||
| 2,373 | 4 | 202,406 | 310,950 | 4,224 | na | ||||
| 2,098 | 1,955 | na | |||||||
| 1,689 | 382 | 100 | 1,472 | na | |||||
| 1,647 | 32,113 | na | |||||||
| 1,539 | 55 | 9,448 | 712 | na | |||||
| 1,538d | 319 | 18,848 | na | ||||||
| 768 | 1,258 | na | |||||||
| 83 | 82,211 | na | |||||||
| 848 | 208 | 989,231 | na | ||||||
| 2,295e | 192 | 266 | 8,084 | na | |||||
| 2,544 | 89 | na | |||||||
| Secretions | |||||||||
| 14,087f | 14 | 90,138 | 14,648 | na | |||||
Values shown represent normalized number of reads (reads per million mapped reads [RPM]).
na, not applicable. Diagnostic culturing was not performed from swab samples.
Viral pathogen detected: hPIV (6 RPM).
Viral pathogen detected: HSV-1 (4,040 RPM).
Viral pathogen detected: HHV-7 (365 RPM).
Influenza virus b; Kingella kingae (536,079 RPM).
High correlation between the numbers of influenza virus NGS reads and the CT values of real-time influenza PCR in BAL fluid samples.
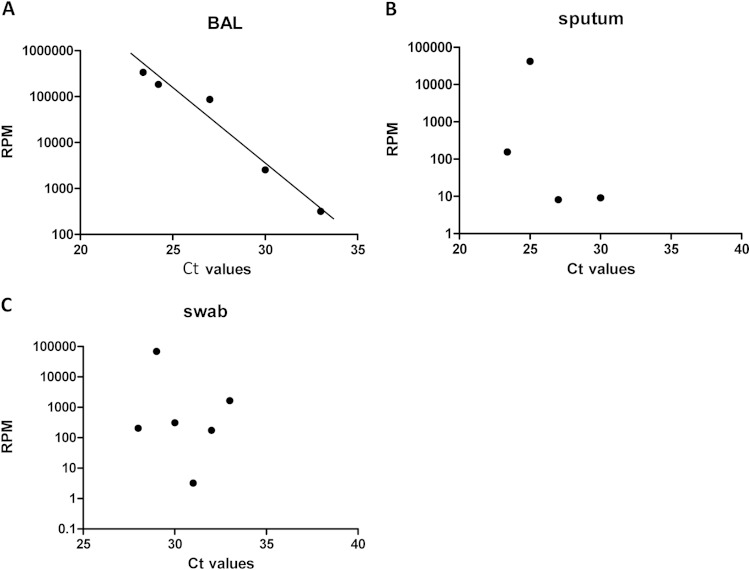
We calculated Pearson correlation coefficients between CT values observed by RT-qPCR and the relative abundance of influenza-specific reads. While we found a strong relationship between unbiased nontargeted metagenomic RNA sequencing (UMERS) and RT-qPCR for BAL fluid samples (Pearson correlation coefficient, −0.8112) (Fig. 4A), we found only a weak association for sputum samples and swabs, with Pearson correlation coefficients of −0.433 and −0.456, respectively (Fig. 4B and C).
FIG 4.
Correlation between reads per million mapped reads (RPM) matching influenza A virus sequences and CT values of influenza A virus-specific RT-qPCR in BAL fluid samples (A), sputum samples (B), and swab samples (C).
Recovery of influenza virus genomic segments.
Contig assembly allowed the recovery of influenza genomic fragments in all influenza virus-positive samples. In 8 of 17 cases, we were able to assemble full-length genome sequences, while, in 3 additional samples (samples 1,538, 1,539, and 83), 7 of 8 influenza A segments were successfully assembled. We applied the Web tool FluGenome (http://flugenome.org/) to assign the lineage and genotype of influenza A sequences and to perform a comprehensive genetic analysis of the entire viral genome. As shown in Table 3, the analysis confirmed the presence of seasonal circulating genotypes H3N2 (A [PB2], D [PB1], B [PA], 3A [HA], A [NP], 2A [NA], B [MP], and 1A [NS]) and H1N1 (C, D, E, 1A, A, 1F, F, and 1A). Different from the circulating strain, in sample 1,538, we observed segment 7 (MP) assigned to the B lineage.
TABLE 3.
Summary of contigs aligning to influenza A or B
| Sample no. | No. of influenza-aligning contigs | No. of influenza-aligning reads | Contig size, bp | Influenza A genotypea |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | MP | NS | ||||
| 104 | 8 | 25,643 | 907–2,350 | C | D | E | 1A | A | 1F | F | 1A |
| 755 | 7 | 111 | 333–958 | na | na | E | 1A | na | na | F | 1A |
| 1,116 | |||||||||||
| 1,721 | 10 | 7,630 | 858–2,349 | A | D | B | 3A | A | 2A | B | 1A |
| 2,535 | |||||||||||
| 2,292 | 8 | 2,016 | 877–2,425 | C | D | E | 1A | A | 1F | ||
| 3,157 | 9 | 1,365 | 442–2,344 | A | D | B | 3A | A | 2A | B | 1A |
| 1,773 | 8 | 173,941 | 909–2,381 | A | D | B | 3A | A | 2A | B | 1A |
| 1,168 | 8 | 6,408 | 874–2,345 | A | D | B | 3A | A | 2A | B | 1A |
| 853 | 18 | 295 | 309–1,414 | na | D | na | 1A | na | 1F | F | 1A |
| 677 | |||||||||||
| 208 | 12 | 273 | 300–691 | na | na | na | 1A | A | 1F | F | 1A |
| 2,373 | 1 | 118 | 362 | na | na | na | na | na | na | na | na |
| 2,098 | 17 | 4,814 | 347–1,033 | na | na | na | 3A | A | 2A | B | 1A |
| 1,689 | |||||||||||
| 1,647 | 10 | 40,170 | 347–2,328 | A | D | B | 3A | A | 2A | B | 1A |
| 1,539 | 17 | 890 | 308–2,241 | C | D | E | 1A | A | na | F | 1A |
| 1,538 | 16 | 615 | 310–1,153 | D | na | E | 1A | A | 1F | B | 1A |
| 768 | |||||||||||
| 83 | 9 | 232 | 718–1,203 | C | D | E | 1A | A | 1F | F | na |
| 848 | 2 | 21 | 715–726 | na | na | na | na | na | na | na | 1A |
| 2,295 | 9 | 6,231 | 624–2,309 | A | D | B | 3A | A | 2A | B | 1A |
| 2,544 | |||||||||||
| 14,087 | 5 | 33 | 410–860 | Influenza B | |||||||
na, not applicable; not sufficient sequence information available. The genotype data were determined using the online Web tool FluGenome (http://www.flugenome.org/genotyping.php) with the following settings: BLAST, 75% identity and 75% coverage.
Sensitivity of UMERS detection algorithm calculated by in silico analysis.
Given that our bioinformatic detection pipeline employs the analysis of sequence contigs, failure to assemble contigs of appropriate length (for example, due to the presence of interfering background sequences) may potentially lead to false-negative results. We therefore applied a benchmarking analysis to measure the performance of our bioinformatics analysis. As shown in Fig. S2A and Table S3 in the supplemental material, the analysis pipeline reliably detects influenza at abundances of only 250 simulated paired-end reads of a 100-bp size.
Employing ≥500 viral reads exhibited marked robustness even at the highest mutation rate. While 500 reads correspond to a relative abundance of only 0.001%, it is worth pointing out that relative abundance has no immediate impact on the detection capability of the pipeline. The integrated assembly step requires a minimal absolute number of influenza reads which allows efficient generation of sequence contigs, whereas the presence of background sequences is not expected to impair the assembly of viral contigs. In support of this notion, similar results were obtained when we repeated the analysis without the background of transcriptome reads (see Fig. S2B and Table S4 in the supplemental material).
Phylogenetic analysis of the hemagglutinin- and neuraminidase-encoding genes.
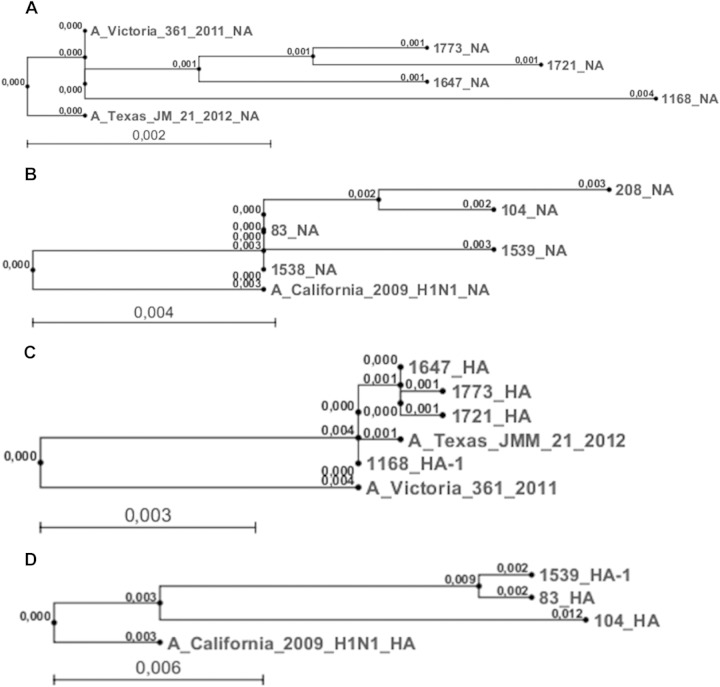
Phylogenetic analysis was performed to elucidate the sequence variation of the antigenic epitopes hemagglutinin (HA) and neuraminidase (NA) during the 2012-2013 season. Overall, 1,400 nucleotides of the NA-encoding gene (Fig. 5A and B) and 1,701 nucleotides of the HA-encoding gene (Fig. 5C and D) were aligned to the HA- and NA-encoding genes of the corresponding vaccine strains of that season (A_Victoria_361_2011_H3N2 and A_California_2009_H1N1). A 99.3% to 99.8% sequence identity was observed between NA gene fragments of the NA2 lineage (Fig. 5A), while a 98.9% to 99.5% sequence identity was observed for the fragments of the NA1 lineage (Fig. 5B). For the HA-encoding gene alignment, we observe a 99.8% sequence identity between the HA sequences isolated from the different patients and a 99% sequence identity to the corresponding HA sequence of the vaccine strain A_Victoria_361_2011 (Fig. 5C).
FIG 5.
Phylogenetic tree summarizing NA sequence alignment to vaccine strain NA A/Victoria/361/2011 (GenBank accession number KC342647.1), H3N2, and vaccine strain A/Texas/21/2012 (KC891013.1) (A) and vaccine strain A/California/4/2009 (FJ966084), H1N1 (B). (A) A total of 1,400 nucleotides were aligned by applying Clustal W alignment. The phylogenetic tree was created using neighbor-joining tree alignment and CLC workbench. Phylogenetic tree summarizing HA sequence alignment to vaccine strain HA A/Victoria/361/2011 (KC306165.1), H3N2, and vaccine strain A/Texas/21/2012 (KC891060.1) (C) and vaccine strain A/California/4/2009 (FJ966082), H1N1 (D). (C) A total of 1,701 nucleotides were aligned using Clustal W alignment. The phylogenetic tree was created using neighbor-joining tree alignment and CLC workbench. Scale bar represents substitutions per site; numbers at node points indicate the branch length.
Detection of viral sequence variants and therapy resistance markers.
We analyzed influenza sequences from 6 patients (only samples with influenza sequence reads >20,000 RPM were included) to search for genomic subpopulations identified by single nucleotide polymorphisms at specific nucleotide positions. For H3N2 and H1N1 sequences, samples from 4 (samples 104, 1,721, 1,773, and 1,647) and 2 (samples 2,292 and 83) patients, respectively, were included. Using a frequency cutoff of 10%, we did not find any evidence for quasispecies among the H1N1 sequences analyzed. In the case of H3N2 sequences, we identified in sample 1,773 one variant in the HA segment at nucleotide position 441 (T to C) resulting in a silent mutation in the triplet encoding amino acid (aa) 147. Likewise, in sample 1,647, a silent mutation at nucleotide position 1,050 (A to G) in the region encoding the HA segment was identified.
We analyzed all H1N1 and H3N2 NA segments for mutations previously described to contribute to NA inhibitor resistance (see Table S5 in the supplemental material). We found all H3N2 strains to likely be sensitive to neuraminidase inhibitors (oseltamivir, zanamivir, peramivir) (22). The H1N1 strains for which sufficient sequence information was available were found to likely be sensitive to oseltamivir and peramivir, since an H275Y mutation was not present. Furthermore, all strains were predicted to exhibit medium resistance against zanamivir due to the presence of the N70S mutation (22). Additionally, for all sequences with sufficient sequence information, we detected the presence of the S31 mutation in the M2 segment, which is responsible for adamantane resistance (data not shown).
Recovery of viral sequences other than Orthomyxoviridae from RNA libraries.
We recovered sequences of human viruses other than influenza virus in 5 of 24 samples. In 1 of the samples, we identified viral sequences matching to the human herpesvirus 1 (HSV-1) (4,040 RPM). One sample contained sequences corresponding to human parainfluenzavirus 3 (hPIV-3) (6 RPM), and we identified human herpesvirus 7 (365 RPM) in another sample (Fig. 3A). One sample showed a significant number of reads (902 RPM) belonging to the highly abundant, nonpathogenic anellovirus family TT virus. Furthermore, one sample exhibited 53 RPM for the human Merkel cell polyomavirus (MCPyV). This virus has previously been detected in respiratory samples (23); however, whether it may potentially play a role in respiratory disease remains unknown.
Furthermore, sequences of human-pathogenic viruses other than influenza were recovered in 2 of the 5 analyzed influenza negative samples. Sample 3 contained viral reads corresponding to human respiratory syncytial virus, and sample 4 contained sequences with significant homology to human rhinovirus A (Fig. 3B; see also Table S6 in the supplemental material).
In the majority of samples, viral sequences corresponding to retroviral sequences can be detected (data not shown). These sequences showed high identity to alpha-retroviruses, murine leukemia virus, or avian leukemia virus-related sequences, which can be explained by the fact that reverse transcriptase enzyme preparations (as used during the preparation of sequencing libraries) are frequently contaminated with retroviral sequences. All samples displayed sequences with significant homologies to Circoviridae, which was recently shown to be a common contaminant in commercial silica gel-based nucleic acid extraction spin columns (24, 25).
Bacterial sequences in influenza-positive respiratory specimens.
NGS reads corresponding to bacterial sequences, mostly representing commensal flora, were recovered in each of the RNA samples from respiratory material (Table 2; see also Table S2 in the supplemental material) in a range of 4,829 to 999,446 RPM (normalized numbers of reads per million mapped reads on nonhost origin). In accordance with previous studies, the phylogenetic composition of the upper respiratory samples (from sputum and swabs) at the phylum level is dominated by proteobacteria, acinetobacteria, and firmicutes (Fig. 1 and 2). By comparison, BAL fluid samples representing the lower respiratory tract were generally lower on bacterial sequences (Fig. 1 and 3). While some of the detected sequences may result from contamination with saliva, we also detected bacteria with the potential to cause severe coinfections or superinfections (Fig. 3, Table 2). Streptococcus pneumoniae, Neisseria meningitidis, Haemophilus influenzae, Staphylococcus aureus, and Moraxella catarrhalis were identified in most sputum and swab samples, while BAL fluid samples were generally low on bacterial sequences. Since all of these bacteria are part of the commensal flora of the respiratory tract, discrimination between colonization and infection cannot be achieved by laboratory detection alone but ultimately requires further evaluation of the clinical data. However, two samples displayed significant numbers of reads of a single individual bacterial species: in sample 2,373, 20.2% of all nonhuman reads (202,406 RPM) could be contributed to Streptococcus pneumoniae, which is part of the normal upper respiratory tract flora but can contribute to respiratory infections under certain conditions. In sample 848, 98.9% of all nonhuman reads (989,231 RPM) were mapped to Moraxella catarrhalis, which is a common respiratory tract pathogen in children and adults. Interestingly, in sample 14,087, 536,079 RPM corresponded to Kingella kingae, which in rare cases can contribute to respiratory infection in immunosuppressed patients.
Table 2 summarizes the results of diagnostic bacterial and fungal culture assays which were part of the routine diagnostics of the sputum and BAL fluid samples of patients with influenza infection. In all samples included in diagnostic culturing, commensal flora of the respiratory tract was detected. In general, swab samples were not subjected to routine culturing processes.
Analysis of bacterial ribosomal sequences.
Given that a large fraction of bacterial reads is likely to stem from highly conserved ribosomal sequences, we analyzed our data sets to identify sequence contigs of ribosomal origin. These contigs were evaluated to determine to what extent the corresponding sequences allow classification on different taxonomic levels. As shown in Table S7 in the supplemental material, although ribosomal contigs represented only a minor fraction of the total number of assembled sequences (on average, 3.3%), they contributed disproportionally large to the number of reads that were of nonhuman origin (average, 27%). However, determination of the lowest common ancestor among all BLAST alignments also shows that the majority of assembled sequences allow taxonomic classification on the species level. Hence, similar to targeted amplicon sequencing of bacterial rRNAs, UMERS-derived contigs originating from highly conserved sequences can hold sufficient discriminatory power to allow accurate taxonomic classification.
Recovery of fungal sequences.
We found fungal sequences with significant homology to Candida albicans and Candida dubliniensis in 7 of 24 samples. Three of these samples (BAL fluid sample 3,157, sputum sample 677, and swab sample 2,373) exhibited RPM values between 310,610 and 566,218, suggesting high abundance of these Candida sequences (Table 2). Candida dubliniensis was also detected by culture in 3 samples (BAL fluid samples 104 and 3,157 and sputum sample 677).
DISCUSSION
Unbiased DNA and/or RNA sequencing by NGS from diagnostic samples could be superior to current diagnostic technologies, since it has the potential to detect known as well as unknown pathogens (viruses, bacteria, fungi, and parasites) in a single application. NGS techniques to detect infectious agents have been used in the past; however, a systematic analysis of this technique in the detection of pathogens from different diagnostic entities is lacking. Furthermore, most studies searching for novel viruses applied specific strategies to enrich for viral particles or viral sequences (e.g., ultracentrifugation, filtration, or amplification strategies) (26) or applied amplicon sequencing to describe the microbiome associated with specific diseases (27–29). However, with regard to diagnostic application in the clinic, standard operating procedures for nucleic extraction, library preparation, and bioinformatics analysis together with validation of NGS are urgently needed. Since large-scale validation of NGS with regard to costs and complexity is rather challenging and hardly feasible for individual groups, small-scale studies applying this technique to different diagnostic entities are of significant value (30–32).
Assuming that a particular pathogen should be of relatively high abundance in an acute disease-associated metabiome, we applied unbiased metagenomic analysis to routine diagnostic respiratory samples (n = 29) to test this method as a putative diagnostic strategy in respiratory specimens in specific public health or clinical settings where unbiased diagnostic methods can efficiently complement classical diagnostic methods.
The objective of this study was to evaluate the potential of metagenomic analysis performed directly from RNA material isolated from diagnostic samples of patients suffering from seasonal influenza virus infection. For BAL fluid samples, we were able to show that there is high concordance between the sensitivity of influenza virus qPCR and metagenomic sequencing, which is in line with previous observations applying NGS to nasopharyngeal aspirates subtracted for host sequences (32). Our results clearly demonstrate that the application of NGS methods is strongly dependent on the type of diagnostic entity which is analyzed. BAL fluid samples, a generally reliable source of material for the diagnosis of pneumonia and other pulmonary infections, show a strong correlation between the percentage of NGS influenza sequence reads and influenza qPCR CT values. In contrast, there was no (or only a very poor) correlation between qPCR and relative read abundance values in sputum and swab samples, even though influenza virus sequence contigs were readily recovered from these specimens. The absence of correlation is most likely the result of greater heterogeneity among microbial sequences (see Fig. 1 to 3): Since the currently available NGS platforms can only provide relative abundance values, it is to be expected that correlation with absolute quantitation values as determined by qPCR will decrease as the variability of the nonhuman background increases.
The greater heterogeneity among nonhuman sequences in swab and sputum samples is likely to reflect differences of the interindividual microbiome composition as well as relatively poor sample uniformity (e.g., due to frequent but variable contamination of induced sputum samples with bacteria or viruses from the pharynx). Until single-molecule high-throughput sequencing technologies which do not require library amplification (and thus will allow a more direct determination of absolute sequence abundance) are available, our observations thus underline the fact that, especially for highly variable diagnostic samples, classical methods such as qPCR are still required to further evaluate the results of diagnostic metagenomic or metatranscriptomic analyses.
In addition, our results demonstrate that swab samples significantly differ in the amount of human sequences. While, according to our in silico analysis, the human background does not generally hinder the detection of pathogen reads, it nevertheless reduces the total number of nonhuman reads at a given read depth such that either the sequencing depth has to be increased or the human sequences have to be physically subtracted prior to sequencing.
Notably, we deliberately did not employ strategies such as enrichment of viral particles by ultracentrifugation, filtration, and nuclease digestion of nucleic acid outside virion particles (33). Our goal, rather, was to use the automatic nucleic acid extraction protocols which are commonly used in routine diagnostics and to subject these diagnostic specimens to unbiased sequencing to detect not only viruses but also other putative pathogens. A similar approach has recently been used by Kuroda and colleagues, who analyzed a lung biopsy specimen of a patient who died because of H1N1 pneumonia (34). Unlike in our study, Kuroda et al. analyzed a single sample which presumably showed a rather high viral load compared to the samples used in our study.
We successfully performed whole-genome genotyping from 8 of 17 influenza-positive samples, thereby confirming the seasonal circulating genotypes H3N2 and H1N1. Since we succeeded in recovering influenza virus whole genomes in 47% of the samples, we were able to immediately recognize new influenza virus types as a consequence of reassortment.
Furthermore, phylogenetic analysis of influenza HA and NA sequences was performed in all influenza A-positive cases to elucidate the sequence variation of the antigenic epitopes HA and NA during the 2012-2013 season. To our knowledge, this is the first comprehensive study to sequence complete genomes of seasonal influenza virus infections directly from diagnostic nucleic acid samples. Previous studies applying NGS with regard to influenza genome analysis were performed from smaller cohorts of pandemic influenza cases (30), from lung tissue (34), or after subculturing of the virus (35–37), which is laborious; in addition, dependent on the viral load, not all subculturing is successful. Other approaches applied amplicon sequencing approaches to selectively sequence all influenza genome segments (38).
Variant calling performed on samples with higher influenza virus sequence reads identified one variant in two samples. Considering the high error rates of RNA polymerase during influenza virus replication, one may expect more intrahost variants to be generated within a single infection. However, our results are in concordance with previously published studies (with most of them preselecting for influenza virus sequences) reporting a relatively low genetic diversity of influenza virus in patients (34, 39, 40).
With regard to sensitivity, we find our results to be in concordance with observations made in previously published studies (30, 32, 41, 42). Similar to these reports, we find a strong correlation between the percentages of NGS reads mapping to influenza A sequences and CT values from qPCR. With the exception of one sample (sample 1,116), the influenza virus sequences from samples with CT values <35 were detected by UMERS, which is comparable to the results obtained by Greninger and colleagues. They included 17 samples (nasopharyngeal swabs) in the NGS analysis, with 15 of 17 samples containing between 105 and 109 viral particles/ml (30). Furthermore, in a study focusing on the analysis of the human virome in febrile children, the sensitivity of unbiased NGS compared to qPCR was estimated for human adenovirus and human bocavirus sequences. A strong correlation between NGS reads and CT values was observed, and adenoviral sequences were detected up to a CT of 35, while human bocavirus was detected up to only a CT value of 30 (42). Similar results were obtained by a very recent study, in which enterovirus and rhinovirus sequences were detectable by NGS up to a CT value of 30 (32).
In BAL fluid samples, we observed a generally high correlation between the percentages of NGS reads matching to influenza genomes and the CT values of RT-qPCR. We were able to detect influenza virus sequences up to a CT of 35 and assembled full-length viral genomes from samples with CT values as high as 31. Furthermore, our benchmarking analysis employing different numbers of influenza virus reads and mutation rates clearly demonstrates that our bioinformatics analysis is highly capable of detecting influenza viral reads at a very low abundance.
Clearly, a major advantage of sequencing total RNA moieties is the ability to analyze the presence of putative superinfecting bacterial, viral, and fungal pathogens in a single approach. We recovered sequences of human-pathogenic viruses other than influenza virus in 7 samples. Human herpesvirus 1 (HSV-1), human herpesvirus 7 (HHV-7), human parainfluenzavirus 3 (hPIV-3), human rhinovirus (hRV), and respiratory syncytial virus (RSV) were identified. These viruses are well known to cause respiratory infections in immunocompetent patients, and HSV-1 and HHV-7 are well-known causes of respiratory infections in immunodeficient patients. In cases where a high viral load was observed, whole-genome information for viruses was recovered, which allows epidemiological evaluation of the viral infection as well as statements about putative treatments in cases where treatment is available. Interestingly, we were able to recover in one case (sample 1,538) >56% of the HSV-1 genome, which allowed us to analyze the potential resistance of this virus against acyclovir. Moreover, the UL23 open reading frame, encoding thymidine kinase (TK), showed 100% sequence identity (data not shown) to an HSV-1 strain recently recovered from a bone marrow transplant patient which was clinically resistant against acyclovir treatment (43).
Furthermore, we detected significant numbers of reads belonging to the nonpathogenic anellovirus family TT virus in one patient and, in another sample, the human Merkel cell polyomavirus (MCPyV). TT viruses and MCPyV have been described in respiratory samples before (23, 32); however, to date, there is no indication that these viruses play a role in respiratory diseases.
Although one previous study by Greninger et al. included microbiome analyses of pandemic influenza H1N1-positive samples (30), to our knowledge this is the first study using an unbiased NGS approach to investigate the microbiome of seasonal influenza A-positive diagnostic specimens. We did not observe differences in the microbiome at the phylum level between the influenza-positive specimens and respiratory samples from healthy patients, as previously published (44).
BAL fluid and sputum samples were routinely analyzed by culture to detect bacteria and fungi known to be involved in pneumonia or pulmonary infections. There is a generally high concordance between the results obtained by culture and NGS for bacteria and fungi showing a high abundance of reads. In addition, we identified 2 swab samples (which are not routinely analyzed by culture) that exhibited a high abundance of reads (with 20.2% to 98.9% of all nonhuman reads belonging to a single bacterial species) of the putative bacterial pathogens S. pneumoniae or M. catarrhalis.
Given that a large number of bacteria can colonize the nasopharynx, a caveat of NGS-based analyses of respiratory tract samples is that it is challenging to discriminate between colonization and coinfection events which may be of putative clinical relevance. While the detection of only a few unambiguously mapped reads may be sufficient to conclude that a sample is positive for a given agent, owing to the limited amount of presently available data, it is difficult to define abundance thresholds that may indicate a pathogenic infection, even if the clinical context is supportive of such a conclusion. While established conventional diagnostics, such as qPCR, suffer from the same principal limitations, extensive optimization and validation over several decades has led to the empirical determination of universally agreed-upon conventions as to when a given PCR result may be sufficient to identify a potential pathogenic infection. Hence, to implement NGS technology in the clinical laboratory, there is an urgent need for studies that systematically address the standardization of NGS methods and the definition of parameters for analytical and clinical validation. In addition, comparative studies are needed to determine the relative abundance of viral, bacterial, and fungal sequences not only in diagnostic specimens from patients suffering from infectious diseases but also in cohorts of healthy individuals.
Nevertheless, the results reported in this pilot study demonstrate that unbiased RNA sequencing is a valuable tool for complementing routine diagnostics, in particular in clinical or public health settings where routine diagnostics remain repeatedly negative and comprehensive surveillance for emerging viruses is needed.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a project grant of the German Center for Infection Research (DZIF) given to Nicole Fischer and Adam Grundhoff.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02495-14.
REFERENCES
- 1.Metersky ML, Masterton RG, Lode H, File TM Jr, Babinchak T. 2012. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis 16:e321–331. doi: 10.1016/j.ijid.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Seki M, Kosai K, Yanagihara K, Higashiyama Y, Kurihara S, Izumikawa K, Miyazaki Y, Hirakata Y, Tashiro T, Kohno S. 2007. Disease severity in patients with simultaneous influenza and bacterial pneumonia. Intern Med 46:953–958. doi: 10.2169/internalmedicine.46.6364. [DOI] [PubMed] [Google Scholar]
- 3.Chertow DS, Memoli MJ. 2013. Bacterial coinfection in influenza: a grand rounds review. JAMA 309:275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 4.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Molbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 5.Joseph C, Togawa Y, Shindo N. 2013. Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses 7(Suppl 2):S105–S113. doi: 10.1111/irv.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett JG, Mundy LM. 1995. Community-acquired pneumonia. N Engl J Med 333:1618–1624. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 7.Morens DM, Taubenberger JK, Fauci AS. 2008. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCullers JA, Rehg JE. 2002. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 9.McCullers JA. 2006. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palacios G, Hornig M, Cisterna D, Savji N, Bussetti AV, Kapoor V, Hui J, Tokarz R, Briese T, Baumeister E, Lipkin WI. 2009. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One 4:e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer N, Rohde H, Indenbirken D, Gunther T, Reumann K, Lutgehetmann M, Meyer T, Kluge S, Aepfelbacher M, Alawi M, Grundhoff A. 2014. Rapid metagenomic diagnostics for suspected outbreak of severe pneumonia. Emerg Infect Dis 20:1072–1075. doi: 10.3201/eid2006.131526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becher P, Fischer N, Grundhoff A, Stalder H, Schweizer M, Postel A. 2014. Complete genome sequence of bovine pestivirus strain PG-2, a second member of the tentative pestivirus species giraffe. Genome Announc 2:e00376–e00314. doi: 10.1128/genomeA.00376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 15.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN]. http://arxiv.org/abs/1207.3907
- 18.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen RR, Schinkel J, Koekkoek S, Pajkrt D, Beld M, de Jong MD, Molenkamp R. 2011. Development and evaluation of a four-tube real-time multiplex PCR assay covering fourteen respiratory viruses, and comparison to its corresponding single-target counterparts. J Clin Virol 51:179–185. doi: 10.1016/j.jcv.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panning M, Eickmann M, Landt O, Monazahian M, Olschlager S, Baumgarte S, Reischl U, Wenzel JJ, Niller HH, Gunther S, Hollmann B, Huzly D, Drexler JF, Helmer A, Becker S, Matz B, Eis-Hubinger A, Drosten C. 2009. Detection of influenza A(H1N1)v virus by real-time RT-PCR. Euro Surveill 14:pii=19329 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19329. [PubMed] [Google Scholar]
- 21.Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, Snowden BW, Tisdale M. 2004. Design and performance testing of quantitative real-time PCR assays for influenza A and B viral load measurement. J Clin Virol 29:179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamali A, Holodniy M. 2013. Influenza treatment and prophylaxis with neuraminidase inhibitors: a review. Infect Drug Resist 6:187–198. doi: 10.2147/IDR.S36601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babakir-Mina M, Ciccozzi M, Lo Presti A, Greco F, Perno CF, Ciotti M. 2010. Identification of Merkel cell polyomavirus in the lower respiratory tract of Italian patients. J Med Virol 82:505–509. doi: 10.1002/jmv.21711. [DOI] [PubMed] [Google Scholar]
- 24.Naccache SN, Greninger AL, Lee D, Coffey LL, Phan T, Rein-Weston A, Aronsohn A, Hackett J Jr, Delwart EL, Chiu CY. 2013. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol 87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smuts H, Kew M, Khan A, Korsman S. 2014. Novel hybrid parvovirus-like virus, NIH-CQV/PHV, contaminants in silica column-based nucleic acid extraction kits. J Virol 88:1398. doi: 10.1128/JVI.03206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokili JL, Rohwer F, Dutilh BE. 2012. Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox AJ, West NP, Cripps AW. 2015. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol 3:207–215. doi: 10.1016/S2213-8587(14)70134-2. [DOI] [PubMed] [Google Scholar]
- 28.Hill JM, Bhattacharjee S, Pogue AI, Lukiw WJ. 2014. The gastrointestinal tract microbiome and potential link to Alzheimer's disease. Front Neurol 5:43. doi: 10.3389/fneur.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. 2014. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 54:309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 30.Greninger AL, Chen EC, Sittler T, Scheinerman A, Roubinian N, Yu G, Kim E, Pillai DR, Guyard C, Mazzulli T, Isa P, Arias CF, Hackett J, Schochetman G, Miller S, Tang P, Chiu CY. 2010. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS One 5:e13381. doi: 10.1371/journal.pone.0013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, Weir JC, Quince C, Smith GP, Betley JR, Aepfelbacher M, Pallen MJ. 2013. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA 309:1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 32.Prachayangprecha S, Schapendonk CM, Koopmans MP, Osterhaus AD, Schurch AC, Pas SD, van der Eijk AA, Povoorawan Y, Haagmans BL, Smits SL. 2014. Exploring the potential of next-generation sequencing in diagnosis of respiratory viruses. J Clin Microbiol 52:3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren X, Yang F, Hu Y, Zhang T, Liu L, Dong J, Sun L, Zhu Y, Xiao Y, Li L, Yang J, Wang J, Jin Q. 2013. Full genome of influenza A (H7N9) virus derived by direct sequencing without culture. Emerg Infect Dis 19:1881–1884. doi: 10.3201/eid1911.130664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroda M, Katano H, Nakajima N, Tobiume M, Ainai A, Sekizuka T, Hasegawa H, Tashiro M, Sasaki Y, Arakawa Y, Hata S, Watanabe M, Sata T. 2010. Characterization of quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009) by de novo sequencing using a next-generation DNA sequencer. PLoS One 5:e10256. doi: 10.1371/journal.pone.0010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kampmann ML, Fordyce SL, Avila-Arcos MC, Rasmussen M, Willerslev E, Nielsen LP, Gilbert MT. 2011. A simple method for the parallel deep sequencing of full influenza A genomes. J Virol Methods 178:243–248. doi: 10.1016/j.jviromet.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Lee HK, Tang JW, Kong DH, Loh TP, Chiang DK, Lam TT, Koay ES. 2013. Comparison of mutation patterns in full-genome A/H3N2 influenza sequences obtained directly from clinical samples and the same samples after a single MDCK passage. PLoS One 8:e79252. doi: 10.1371/journal.pone.0079252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutvisuttinunt W, Chinnawirotpisan P, Simasathien S, Shrestha SK, Yoon IK, Klungthong C, Fernandez S. 2013. Simultaneous and complete genome sequencing of influenza A and B with high coverage by Illumina MiSeq platform. J Virol Methods 193:394–404. doi: 10.1016/j.jviromet.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Hoper D, Hoffmann B, Beer M. 2011. A comprehensive deep-sequencing strategy for full-length genomes of influenza A. PLoS One 6:e19075. doi: 10.1371/journal.pone.0019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fordyce SL, Bragstad K, Pedersen SS, Jensen TG, Gahrn-Hansen B, Daniels R, Hay A, Kampmann ML, Bruhn CA, Moreno-Mayar JV, Avila-Arcos MC, Gilbert MT, Nielsen LP. 2013. Genetic diversity among pandemic 2009 influenza viruses isolated from a transmission chain. Virol J 10:116. doi: 10.1186/1743-422X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakrishnan MA, Tu ZJ, Singh S, Chockalingam AK, Gramer MR, Wang P, Goyal SM, Yang M, Halvorson DA, Sreevatsan S. 2009. The feasibility of using high-resolution genome sequencing of influenza A viruses to detect mixed infections and quasispecies. PLoS One 4:e7105. doi: 10.1371/journal.pone.0007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheval J, Sauvage V, Frangeul L, Dacheux L, Guigon G, Dumey N, Pariente K, Rousseaux C, Dorange F, Berthet N, Brisse S, Moszer I, Bourhy H, Manuguerra CJ, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J Clin Microbiol 49:3268–3275. doi: 10.1128/JCM.00850-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauerbrei A, Bohn K, Heim A, Hofmann J, Weissbrich B, Schnitzler P, Hoffmann D, Zell R, Jahn G, Wutzler P, Hamprecht K. 2011. Novel resistance-associated mutations of thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 and type 2. Antivir Ther 16:1297–1308. doi: 10.3851/IMP1870. [DOI] [PubMed] [Google Scholar]
- 44.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1:pii=e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.