Abstract
The quality of sample inoculation is critical for achieving an optimal yield of discrete colonies in both monomicrobial and polymicrobial samples to perform identification and antibiotic susceptibility testing. Consequently, we compared the performance between the InoqulA (BD Kiestra), the WASP (Copan), and manual inoculation methods. Defined mono- and polymicrobial samples of 4 bacterial species and cloudy urine specimens were inoculated on chromogenic agar by the InoqulA, the WASP, and manual methods. Images taken with ImagA (BD Kiestra) were analyzed with the VisionLab version 3.43 image analysis software to assess the quality of growth and to prevent subjective interpretation of the data. A 3- to 10-fold higher yield of discrete colonies was observed following automated inoculation with both the InoqulA and WASP systems than that with manual inoculation. The difference in performance between automated and manual inoculation was mainly observed at concentrations of >106 bacteria/ml. Inoculation with the InoqulA system allowed us to obtain significantly more discrete colonies than the WASP system at concentrations of >107 bacteria/ml. However, the level of difference observed was bacterial species dependent. Discrete colonies of bacteria present in 100- to 1,000-fold lower concentrations than the most concentrated populations in defined polymicrobial samples were not reproducibly recovered, even with the automated systems. The analysis of cloudy urine specimens showed that InoqulA inoculation provided a statistically significantly higher number of discrete colonies than that with WASP and manual inoculation. Consequently, the automated InoqulA inoculation greatly decreased the requirement for bacterial subculture and thus resulted in a significant reduction in the time to results, laboratory workload, and laboratory costs.
INTRODUCTION
The emergence of automation in bacteriology has opened a new era in clinical diagnostic laboratories. Automation is impacting laboratory management and workflow but also offers new perspectives for research and development in bacteriology by developing intelligent algorithms and driving innovation. Sample inoculation is a fastidious and repetitive process representing about 25% of a laboratory's workload (1). Thus, automated inoculation systems represent a need in diagnostic laboratories given the reductions in human, material, and financial resources and the increase in sample volumes (1). Moreover, the quality of inoculation is critical for achieving an optimal yield of discrete colonies in both monomicrobial and polymicrobial samples to facilitate rapid identification (ID) and antibiotic susceptibility testing (AST). Several inoculation and streaking instruments are currently available for routine diagnostic laboratories, including the Autoplak (NTE-SENER), the InoqulA (BD Kiestra), the Innova (BD), the PreLUD (i2a), the Previ Isola (bioMérieux), and the WASP (Copan). However, the true effectiveness of automated inoculation systems needs to be validated by independent routine clinical microbiology laboratories. Compared to manual streaking, a few studies have demonstrated that the InoqulA and Previ Isola automated systems produce more isolated colonies, show better reproducibility, have no cross-contamination, and exhibit a significant decrease in hands-on plating time (2–5). These studies concluded that such automated systems should improve laboratory workflow and shorten the time to results, but direct laboratory impact assessments remain to be performed to confirm these expectations. Moreover, the available few studies compared only automated to manual streaking performance, but direct comparative studies between the available automated systems remained to be performed.
Consequently, we compared the performances of manual inoculation, the automated inoculation InoqulA BT system (BD Kiestra, Netherlands), and the Walk-Away Specimen Processor (WASP) (Copan, Italy). Several parameters, including the yield of discrete colonies and colony distribution, were determined following inoculation of defined monomicrobial and polymicrobial samples. Moreover, the capacity of each inoculation system to reproducibly produce discrete colonies and the requirement to perform additional reisolation to obtain discrete colonies for subsequent ID and AST were prospectively evaluated on clinical cloudy urine samples. The need for reisolation, time to results, and laboratory analytical costs were determined to assess whether the performance of the different inoculation systems has an impact on laboratory financial and time-to-results outcomes.
MATERIALS AND METHODS
Strains, media, and bacterial suspension.
Escherichia coli strain ATCC 25922, Klebsiella pneumoniae strain ATCC BAA-1706, Staphylococcus aureus strain ATCC 29213, and Enterococcus faecalis strain ATCC 29212 were grown on Columbia agar with 5% sheep blood (Columbia III agar; BD, Franklin Lakes, NJ, USA) at 37°C in 5% CO2 atmosphere incubators. Colonies of each bacterial species were utilized to prepare a bacterial suspension in saline solution adjusted to a 0.5 McFarland turbidity measured with a DensiCheck densitometer instrument (bioMérieux, Marcy l'Etoile, France) and corresponding to a bacterial concentration of 108 CFU per ml. Different concentrations of monomicrobial suspensions in saline solution, ranging from 108 to 103 CFU/ml, were prepared by doing serial 10-fold dilutions in saline solution. All bacterial suspensions were plated on Columbia agar with 5% sheep blood to verify the number of CFU per milliliter. Polymicrobial suspensions containing 4 bacterial species at different ratios ranging from 1:1 to 1:1,000 (see Table S1 in the supplemental material) were obtained by mixing different concentrations of the diluted and nondiluted monomicrobial suspensions.
Sample collection.
Cloudy urine samples were collected during a 1-month period from ambulatory and hospitalized patients at the University Hospital of Lausanne (Switzerland). All urine specimens were deidentified prior to testing. A total of 75 cloudy urine specimens found positive for bacteria by Gram staining microscopy were selected to include in the study only urine samples containing ≥105 CFU/ml of bacteria. Selected urinary samples were transferred into sterile 5-ml Copan tubes (Copan, Brescia, Italy), vortexed, and inoculated with the WASP, InoqulA BT, or manual methods, as described below.
Inoculation and incubation.
According to specific guidelines for urine cultures (6–9), detection at the level of 102 CFU/ml is necessary for specific populations, such as women with acute cystitis, specimens obtained from patient catheters, and patients in the early stage of a urinary tract infection. Thus, the guidelines specifically state that ≥10 μl of urine should be plated using a back-and-forth streaking method to detect most of clinically relevant urinary tract infections. Therefore, inocula of 10 μl were streaked onto chromogenic agar (CHROMagar Orientation; BD, Franklin Lakes, NJ, USA) manually and with the automated inoculation systems InoqulA BT and WASP. Chromogenic agar is routinely used in many diagnostic laboratories for the analysis of urine samples and was used to facilitate the recognition and classification of bacterial colonies by the BD Vision Toolbox with embedded VisionLab version 3.43 imaging analysis software.
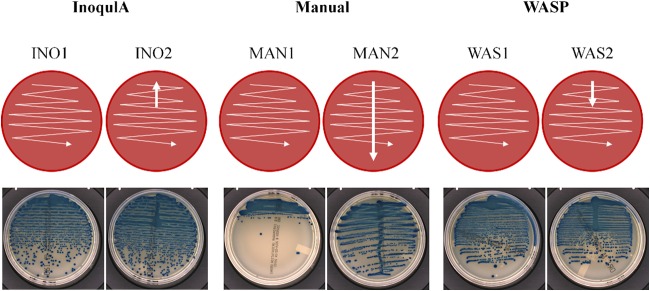
Manual and WASP streaking were performed with a 10-μl loop, whereas plate inoculation with the InoqulA was performed with a rolling magnetic bead. The volume of 10 μl was seeded onto chromogenic agar with a calibrated pipette for manual streaking and the InoqulA automated system and with a 10-μl loop for the WASP automated system. Two manual quantitative plate inoculation patterns were performed by an experienced microbiologist with 10-μl loops: (i) a zigzag streaking pattern (MAN1), and (ii) a central single streaking throughout the plate followed by a zigzag pattern (MAN2) (Fig. 1). Two similar automated quantitative plate inoculation patterns with the InoqulA BT and WASP were performed: (i) a zigzag streaking pattern (INO1 and WAS1, respectively), and (ii) a central single streaking of 20 mm followed by a zigzag pattern (INO2 and WAS2, respectively) (Fig. 1).
FIG 1.
Manual and automated semiquantitative streaking protocols. Two manual quantitative plate inoculation patterns were performed by an experienced microbiologist with 10-μl loops in a zigzag streaking pattern (MAN1), or a central single streaking throughout the plate followed by a zigzag pattern (MAN2). Two similar automated quantitative plate inoculation patterns with the InoqulA BT and the WASP were performed in a zigzag streaking pattern (INO1 and WAS1, respectively) or a central single streaking of 20 mm followed by a zigzag pattern (INO2 and WAS2, respectively). The InoqulA INO1 pattern and the WASP WAS2 pattern were used as optimized factory-designed semiquantitative inoculation protocols. The manual MAN2 streaking approach was chosen as the conventional semiquantitative manual inoculation used in most diagnostic laboratories. The INO2 is similar to the WAS2 streaking pattern, whereas the WAS1 and MAN1 are similar to the INO1 streaking pattern.
The manual MAN2 streaking pattern is a conventional semiquantitative approach used routinely by many diagnostic laboratories. The INO1 and WAS2 streaking approaches are semiquantitative patterns recommended by the manufacturers (BD and Copan, respectively) to obtain optimal quantitative and qualitative results. Thus, two different streaking approaches, the streaking 1 patterns (INO1, WAS1, and MAN1) and the streaking 2 patterns (INO2, WAS2, and MAN2), were used for a direct comparison of similar inoculation patterns between manual and automated systems.
The inoculated chromogenic agar plates were incubated in a normal ambient atmosphere for 20 h at 35°C, as recommended by the manufacturer, thus allowing us to obtain both an acceptable turnaround time (TAT) and enough microbiological material to perform ID and AST. Automated and manual inoculations of defined monomicrobial and polymicrobial samples were performed at least in three independent experimental runs, whereas inoculation of cloudy urine specimens was performed only once per sample.
Analysis of reporting times and laboratory costs.
A total of 75 cloudy urine specimens defined as positive by Gram stain were prospectively inoculated manually or with the InoqulA and the WASP automated systems. Among them, 41 urine specimens positive for E. coli recovered from both monomicrobial and polymicrobial cultures were analyzed with the VisionLab version 3.43 software to determine the yield of discrete colonies obtained by each inoculation system. The remaining 34 urine specimens considered to be contaminated urine specimens or including bacterial species not recognized by the VisionLab version 3.43 software were excluded from the analysis. The time to results and laboratory costs were calculated based on the ability of the different systems to produce a minimal number of isolated colonies to perform identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and AST. The minimal number of E. coli colonies grown on chromogenic BBL CHROMagar Orientation agar plates in a normal ambient atmosphere for 20 h at 35°C was determined, according to conventional laboratory procedures, with a minimum of 1 discrete colony required for ID by MALDI-TOF MS and a minimum of 5 colonies required to make a 2-ml bacterial suspension in saline solution with a turbidity of 0.5 McFarland standard for AST, as recommended in the EUCAST/CLSI guidelines. A delayed time to result of 1 working day (16 h to 24 h) and additional laboratory costs were applied when the minimal number of isolated colonies required to perform the ID and AST procedures was not obtained. The laboratory cost per reisolation was calculated in Swiss francs (CHF), European euros (EUR), and U.S. dollars (USD) based on consumable prices and labor costs, including social security charges applied at the University Hospital of Lausanne, Switzerland, as follows: agar plate, 1 CHF/0.96 EUR/1.06 USD; plastic loop, 0.1 CHF/0.096 EUR/0.106 USD; and 2 min working time to perform a reisolation, 2.3 CHF/2.2 EUR/2.4 USD, for a total of 3.4 CHF, 3.3 EUR/3.6 USD per reisolation. The conversion rates of 1 EUR to 1.04 CHF and 1 USD to 0.94 CHF were calculated in April 2015 and may be subjected to variations due to the volatility of the foreign exchange rate. The experimental working time of 2 min to perform reisolation includes the following tasks: (i) collecting the agar plate containing the sample for reisolation in the incubator, (ii) collecting a sterile plate for subculture in the cold room, (iii) collecting a plastic loop, (iv) plate labeling, (v) colony picking and 4-quadrant plate streaking, and (vi) storing the plates in the incubators. The working time of 2 min was measured and used for an experimental modelization of additional laboratory costs due to the requirement of subculture to perform ID and AST from discrete colonies in both automated and conventional laboratories. The measured working time strongly depends on the organization of the laboratory workflow and may vary greatly between laboratories.
Imaging and image analysis.
All images were taken using a specialized imaging device called the ImagA BT (BD Kiestra), which allowed us to obtain reproducible and consistent images with the different inoculation methods and sample preparations. The resolution of the camera allowed the recognition of objects ≥0.4 mm in diameter. Objects <0.4 mm were thus considered small noisy objects and were not considered discrete colonies. Image analysis was performed with the VisionLab version 3.43 software (Van de Loosdrecht Machine Vision BV, Buitenpost, The Netherlands). Image analysis was used to provide a reliable and objective measure for the properties of the colonies, minimizing the bias from manual observation. The parameters of the image analysis software were trained by an experienced lab technician by selecting objects and specifying their discreteness and bacterial species. The properties of the colonies were measured with the VisionLab version 3.43 software, enabling fast automated counting of discrete colonies and automatic recognition of specific bacterial species.
Classification of discreteness and bacterial species was done by a linear discriminant analysis (LDA)-based (10, 11) classifier. LDA is a linear model that uses statistics of the data to determine the optimal separation between the different classes. A data set of 3,379 images of discrete and nondiscrete colonies of E. coli, K. pneumoniae, S. aureus, and E. faecalis were defined by a technician, resulting in the defined data set. The LDA classifier was trained with samples from the defined data set, meaning that colony discreteness and species recognition were determined indirectly by the lab technician and not by the specific configuration of the image analysis software.
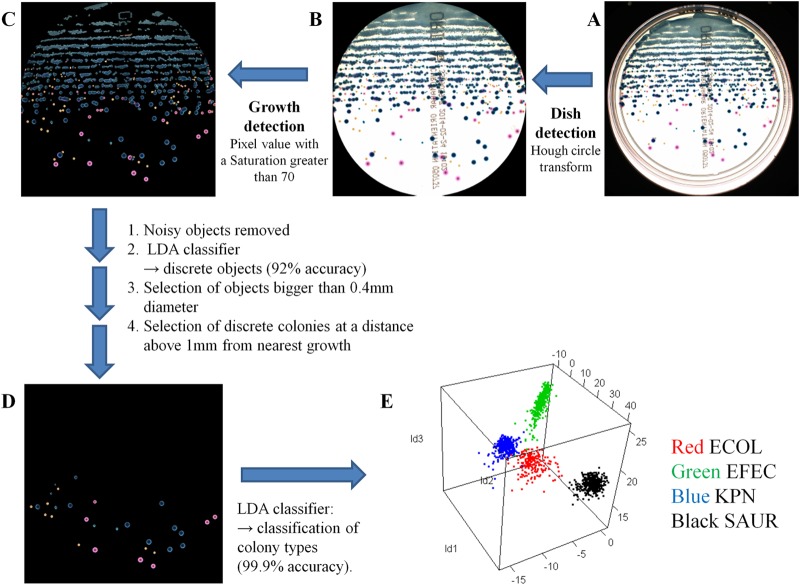
The image analysis was performed in 5 steps, as shown in Fig. 2. First, the petri dish surface was analyzed by using a Hough circle transform (12), which was specifically suited for detecting circles (Fig. 2B). The Hough circle transform (12) was used as a robust method, giving the correct position of the petri dish for every image in the set (Fig. 2A and B). Because of refractions and reflections at the dish border, a few millimeters of the outer border of the petri dish image were ignored by the image analysis software to increase measurement accuracy. The size of this border is equal among all images, preventing any bias toward any image (Fig. 2B).
FIG 2.
Image analysis procedure. Image analysis was performed in 5 steps (A to E). (A) Raw image of the petri dish. (B) Surface pixels of the petri dish. (C) Pixels considered to be growth. (D) Discrete colonies. (E) Four distinct clusters produced by linear discriminant analysis. Each color represents a different bacterial species. ECOL, E. coli; EFEC, E. faecalis; KPN, K. pneumoniae; SAUR, S. aureus.
Second, bacterial growth was determined by selecting pixels with high color saturation (i.e., if the color is different from white) (Fig. 2C) (13). Pixels with a high saturation value of 70 were considered nonwhite and were selected as object pixels (Fig. 2C). A lower value corresponded to more growth pixels around each colony, while a higher value corresponded to less growth. As a result, a white agar background was required for a reliable detection of bacterial growth.
Connected pixels were grouped into objects, and each object could be either one discrete colony or several connected colonies. Discrete objects were recognized by the discreteness LDA classifier. The LDA classifier for determining colony discreteness (discrete or nondiscrete) was trained and evaluated to classify objects into discrete and nondiscrete colonies based on their geometric features (Fig. 2D). A linear transformation of geometric features was automatically determined by LDA by using the defined data set. LDA minimizes the variance within a class and maximizes variance between classes, allowing the formation of clusters. Highly separated clusters yield high classification accuracy. Each object's features were transformed to the trained LDA space to form the discrete and nondiscrete clusters. The closest cluster was chosen as the proper class for each object. The real class for each object in the evaluation set was known (e.g., discrete or nondiscrete), so the results of the classification could be compared to the defined data set. Next, objects with a diameter of <0.4 mm and/or with a distance to the nearest growth of <1 mm were removed. All remaining objects were considered discrete colonies (Fig. 2D). These criteria were chosen to ensure that manual or automated colony picking could be easily performed without risk of contamination by nearby bacterial growth.
Finally, the bacterial species of each discrete colony was determined by a bacterial species LDA classifier trained for four bacterial species, E. coli, K. pneumoniae, E. faecalis, and S. aureus (Fig. 2E). Consequently, in this study, only these four bacterial species were automatically recognized on the agar plates. The LDA classifier for determining colony species was trained and evaluated to classify discrete colonies of E. coli, K. pneumoniae, E. faecalis, and S. aureus based on the color features of the discrete colony. A linear transformation of color features was automatically determined by the LDA using the defined data set favoring high cluster separation. The color features of each discrete colony in the defined data set were transformed to the trained LDA space, resulting in four clusters, one for each bacterial species (Fig. 2E). The closest cluster was chosen as the proper class for each discrete colony. The real class for each discrete colony was known, and the results of the classification could be compared to the defined data set.
An evaluation of the accuracy in classifying discrete and nondiscrete objects and identifying bacterial colony species was performed for each step involved in the image analysis process (Fig. 2C to E). The evaluation was performed using the defined data set containing 3,379 images of known objects belonging to discreteness (discrete or nondiscrete) and bacterial species classes (1,915 nondiscrete objects, 423 E. coli, 353 K. pneumoniae, 199 E. faecalis, and 489 S. aureus images). Objects from all the bacterial species classes were discrete. The defined data set was divided in a training set and an evaluation set to be used for a 2-fold cross-validation. The accuracy of LDA classifiers was defined as the percentage of colonies correctly classified compared to the defined data set compiled by a technician. A quantitative analysis of the evaluation results provided insight into the error that could be expected from the measurements (see Results). The error of classification was similar for each inoculation method and did not bias the results for any specific automated or manual inoculation method.
The median discrete colony distribution was determined as follows. The medium plate was delimited in 1,500 lines, starting from the border located close to the sample seeding zone (line 0) to the opposite plate border (line 1,500). For each line, the number of discrete colony pixels on that line was divided by the total number of growth pixels on that line, giving a normalized measure of the percentage of discrete colonies on each line. Finally, the concatenation of all lines was plotted for each inoculation method and each bacterial species.
Statistical analysis.
The statistical difference of the number of discrete colonies obtained following automated and manual inoculation of monomicrobial and polymicrobial samples was analyzed by multiple comparisons of means using contrasts in linear regression in R. The analysis was done using the lm() function in R, followed by the extraction of contrasts using the contrast() function from package contrast; multiple comparisons, including the confidence intervals around the differences between means, were computed by the glht() function from package multcomp.
A one-way analysis of variance (ANOVA) multiple comparison was performed using the GraphPad Prism 6.04 software to analyze the statistical difference of the number of discrete colonies obtained from cloudy urine samples with the automated and manual inoculations.
RESULTS
Image analysis.
The discreteness LDA classifier was trained and evaluated to classify objects into discrete and nondiscrete colonies based on their geometric features (Fig. 2D). Compared to the defined data set characterized by a trained technician, 92% of the objects were correctly classified as discrete or nondiscrete objects by the LDA classifier, thus giving a 92% probability of correct automated discreteness classification of undefined samples. Next, the bacterial species LDA classifier was trained and evaluated. Discrete colonies were classified as E. coli, K. pneumoniae, E. faecalis, or S. aureus based on the color features of the discrete colony (Fig. 2E). Compared to the defined data set characterized by a trained technician, 99.9% of the bacterial species colonies were correctly classified by the LDA classifier, thus giving a 99.9% probability of correct automated bacterial species colony classification of undefined samples.
Thus, the accuracy of both the discreteness classifier and the bacterial species classifier resulted in reliable measurement results of the properties of the colonies.
Quality of isolation of different bacterial concentrations of E. coli.
Quantitative streaking patterns used in this study are routinely performed with urine samples that require quantification of growing microorganisms for biomedical interpretation. E. coli is the most prevalent etiological agent of urinary tract infections (UTIs), responsible for 66% to 90% of cases in complicated and uncomplicated UTIs, respectively (9). The quality of isolation was thus assessed with different bacterial concentrations of E. coli ranging from 103 to 108 CFU/ml to measure the ability of the different systems to generate discrete colonies with a wide range of bacterial concentrations.
The different inoculation methods showed a gradual increase in the number of discrete colonies with rising bacterial concentrations but differed by reaching a peak or plateau of isolated colonies at different bacterial titers (Fig. 3; see also Fig. S1 in the supplemental material). A gradual increase in discrete colonies, reaching a plateau at 107 CFU/ml, was observed with the INO1 inoculation. The INO2 inoculation was able to generate more isolated colonies than the INO1 at lower bacterial concentrations, thus producing a high yield of discrete colonies at a wider range of bacterial concentrations. The MAN1 streaking showed a weak gradual increase in isolated colonies with rising bacterial concentrations, reaching a maximal median value at 108 CFU/ml. A high yield of discrete colonies was obtained with the MAN2 streaking at low to moderate bacterial concentrations, but significantly decreased performance was observed at high bacterial concentrations. Similarly, the WAS1 and WAS2 inoculations showed an increased yield of discrete colonies but exhibited weak performance at 108 CFU/ml. Thus, the INO1, INO2, and MAN1 inoculations showed a gradual increase in isolated colonies, reaching a plateau of discrete colonies at different bacterial concentrations, whereas the MAN2, WAS1, and WAS2 inoculation methods were characterized by an increased yield of discrete colonies, followed by a significantly reduced performance when reaching moderate (106 CFU/ml with the MAN2) to high bacterial concentrations (107 CFU/ml with the WAS1 and WAS2 inoculations), respectively. The automated inoculation systems InoqulA and WASP showed a statistically significantly higher yield of discrete colonies (P < 0.05, multiple comparisons of means) than that of manual inoculation at 107 CFU/ml, whereas the InoqulA produced statistically significantly more discrete colonies (P < 0.05, multiple comparisons of means) than that of the WASP and manual inoculation at 108 CFU/ml (see Table S2 in the supplemental material).
FIG 3.
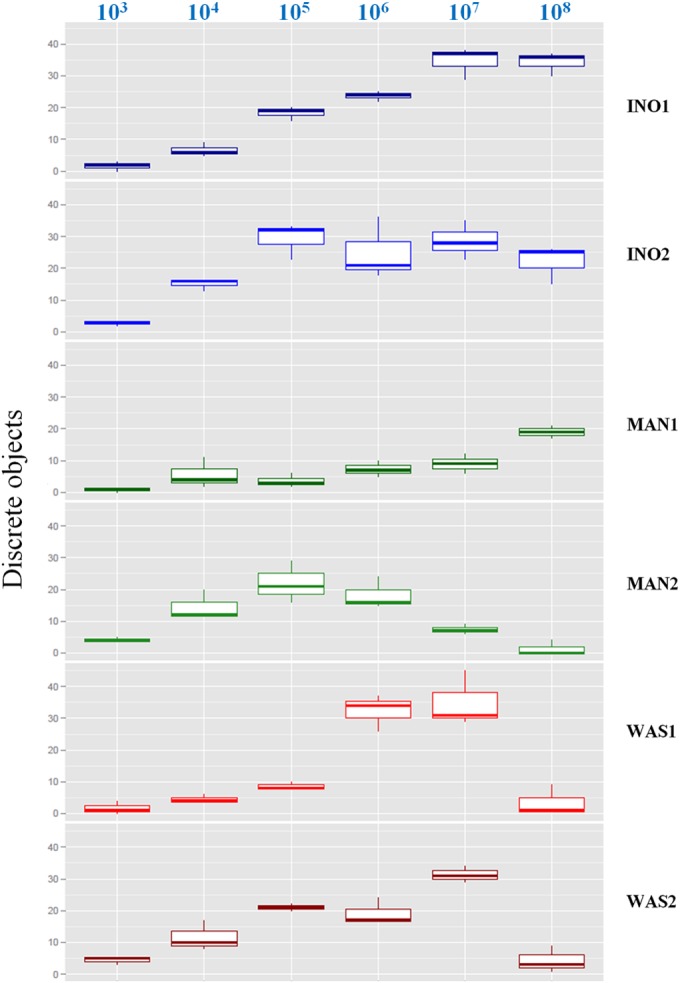
Performance of manual, InoqulA, and WASP plate inoculations at different bacterial concentrations of E. coli. Shown are box-and-whisker plots (representing the minimum, first quartile, median, third quartile, and maximum) of the number of discrete colonies following InoqulA (INO1 and INO2), manual (MAN1 and MAN2), and WASP (WAS1 and WAS2) plate inoculations of different bacterial concentrations of E. coli ranging from 103 to 108 CFU/ml (top).
Quality of isolation of defined monomicrobial samples.
As demonstrated with the inoculation of different bacterial concentrations of E. coli (Fig. 3), a significant difference between the inoculation systems was observed mainly at bacterial concentrations of ≥107 CFU/ml. The streaking quality of manual and automated inoculation was thus assessed by measuring the yield of discrete colonies following inoculation of four bacterial suspensions at a concentration of 108 CFU/ml. Two Gram-negative and two Gram-positive bacteria, E. coli, K. pneumoniae, S. aureus, and E. faecalis, were used to integrate morphological and physiological trait differences that may impact the streaking efficiency of the manual and automated systems. In addition, colonies of these 4 bacterial species growing on chromogenic agar exhibit different colors that facilitate the recognition and classification of discrete colonies by the VisionLab version 3.43 software. The yields of discrete colonies and the differences observed between the automated and manual inoculations were bacterial species dependent (Fig. 4). All the streaking methods except MAN2 were able to produce a high yield of discrete colonies of E. faecalis. However, the INO1 inoculation method produced a statistically significantly higher number of discrete colonies (P < 0.05, multiple comparisons of means) than that of manual and WAS1 inoculations (see Table S3 in the supplemental material). To the contrary, a lower yield of K. pneumoniae isolated colonies was obtained with the 6 streaking approaches than that with the other bacterial species, with no statistically significant difference between automated and manual inoculations (see Table S3). The yields of discrete colonies of E. coli and to a lesser extent of S. aureus were strongly dependent on the streaking method. A statistically significantly higher yield of E. coli discrete colonies (P < 0.05, multiple comparisons of means) was reproducibly obtained with the InoqulA instrument than that with manual or WASP plate streaking (Fig. 4; see also Table S3). A high yield of S. aureus discrete colonies was obtained with the InoqulA and WAS2 streaking methods, whereas a low number of isolated colonies was obtained manually or with the WAS1 streaking approaches. However, only the INO1 inoculation exhibited a statistically significantly higher yield of S. aureus discrete colonies (P < 0.05, multiple comparisons of means) than that with manual and WAS1 inoculations (see Table S3).
FIG 4.
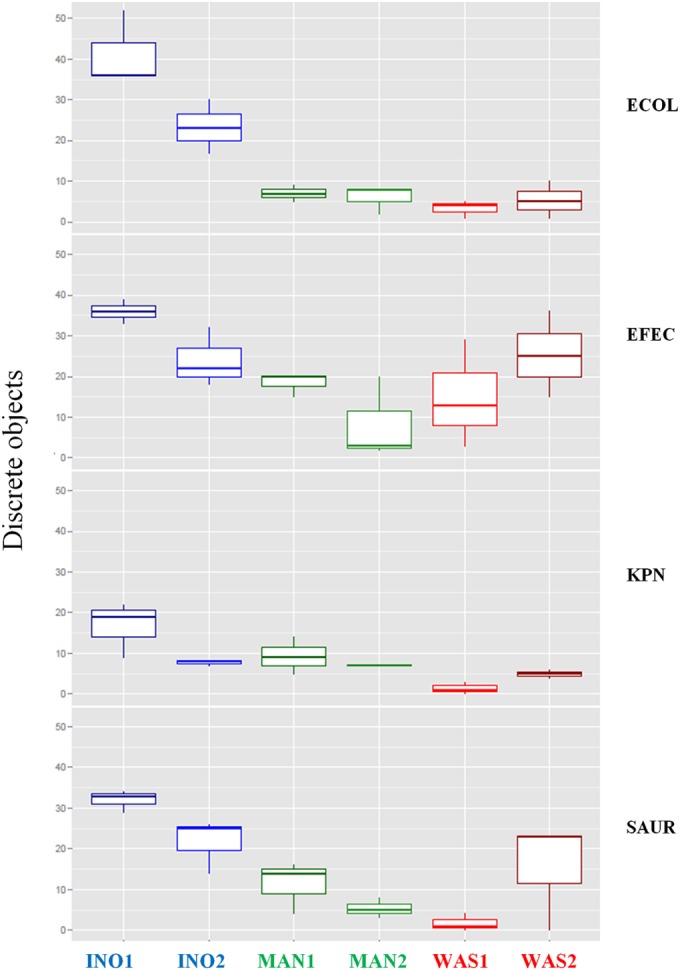
Performance of manual, InoqulA, and WASP inoculations following streaking of monomicrobial samples at a concentration of 108 CFU/ml. Shown are box-and-whisker plots (representing the minimum, first quartile, median, third quartile, and maximum) of the number of discrete colonies of E. coli (ECOL), E. faecalis (EFEC), K. pneumoniae (KPN), and S. aureus (SAUR) following InoqulA (INO1 and INO2), manual (MAN1 and MAN2), and WASP (WAS1 and WAS2) plate inoculations.
The automated and manual streaking approaches exhibited different discrete colony distribution patterns (see Fig. S2A and B in the supplemental material). A gradual increase in the number of discrete colonies following the inoculation path throughout the plate was observed with the InoqulA and WAS2 streaking approaches. Interestingly, the InoqulA magnetic bead inoculation method showed a larger zone of discrete colony distributions due to its capacity to cover the entire surface of the plate than that with manual or WASP loop streaking, which have limited access to the plate edges (Fig. 1; see also Fig. S1 and S2 in the supplemental material). Identical patterns of distribution were observed between the different tested bacterial species, except for K. pneumoniae. Unlike manual streaking, the distribution of K. pneumoniae with the WASP and InoqulA automated inoculations differed by showing later appearances of discrete colonies following the path of the streaking pattern compared to the other tested bacterial species (see Fig. S2B, and data not shown).
Quality of isolation of defined polymicrobial samples.
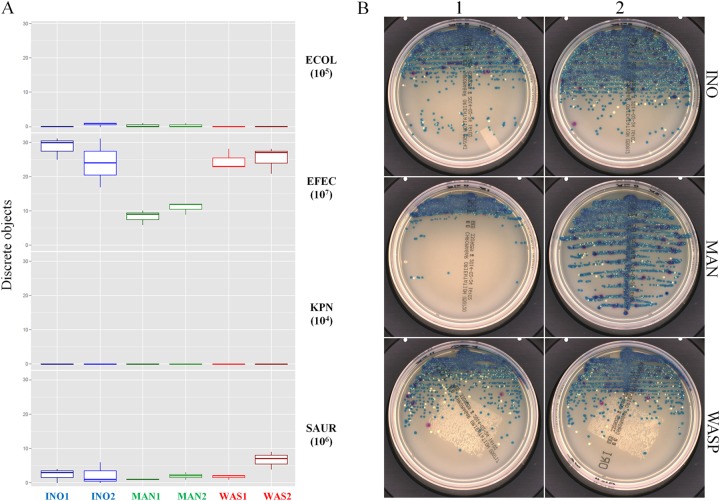
The ability of the different inoculation systems to obtain discrete colonies of each bacterial species contained in an artificial polymicrobial sample was assessed to determine their discriminative power. Eleven polymicrobial suspensions containing E. coli, K. pneumoniae, S. aureus, and E. faecalis were obtained by mixing the 4 bacterial species at different ratios ranging from 1:1 to 1,000:1 between the highest and lowest bacterial concentrations (see Table S1 in the supplemental material). The results obtained with mixes M01 to M10 (see Fig. S3 in the supplemental material) were similar to those observed in the polymicrobial suspension mix M11 (Fig. 5). Mix M11 was composed of E. faecalis at 107 CFU/ml, S. aureus at 106 CFU/ml, E. coli at 105 CFU/ml, and K. pneumoniae at 104 CFU/ml. The InoqulA and WASP inoculations produced a statistically significantly higher yield of E. faecalis discrete colonies (P < 0.05, multiple comparisons of means) than that with manual streaking (see Table S4 in the supplemental material). However, the 6 inoculation methods produced a low yield of colonies of S. aureus, which was present at a 10-fold lower concentration than E. faecalis. In addition, no significant statistical difference was observed between the automated and manual inoculation approaches (see Table S4). Discrete colonies of E. coli and K. pneumoniae present at 100- to 1,000-fold lower concentrations than the most concentrated E. faecalis population in the sample were not reproducibly recovered with the manual or automated inoculation methods used in this study. Thus, the results of the M01 to M11 mixes suggest that colonies of bacterial species present at concentrations ≤100-fold lower than the most concentrated bacterial population in a polymicrobial sample are likely not recovered following manual or automated inoculation with the streaking patterns used in this study.
FIG 5.
Recovery of discrete colonies of each bacterial species contained in polymicrobial samples following manual and automated inoculation. Shown are box-and-whisker plots (representing the minimum, first quartile, median, third quartile, and maximum) (A) and plate images (B) of the number of discrete colonies following InoqulA (INO1 and INO2), manual (MAN1 and MAN2), and WASP (WAS1 and WAS2) plate inoculations of a polymicrobial sample containing E. faecalis at 107 CFU/ml, S. aureus at 106 CFU/ml, E. coli at 105 CFU/ml, and K. pneumoniae at 104 CFU/ml representing 1:1, 10:1, 100:1, and 1,000:1 ratios between the highest and the lowest bacterial concentrations, respectively.
Performance of the manual and automated systems on clinical cloudy urine specimens.
The performance of the different systems and their impact on the time to results and on laboratory costs were assessed by determining (i) the yield of discrete colonies and (ii) the need for reisolation of colonies for identification (ID) by MALDI-TOF MS and antibiotic susceptibility testing (AST).
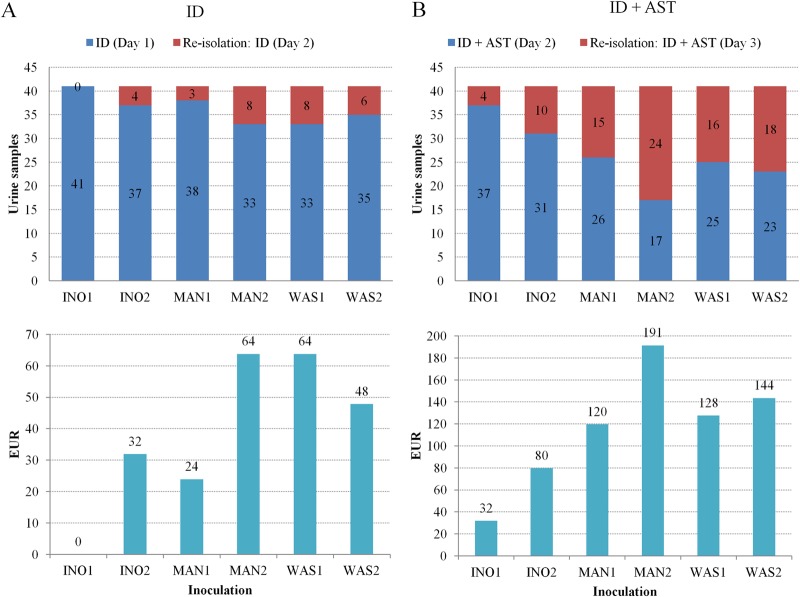
The INO1 inoculation showed a statistically significantly higher yield of discrete colonies (P < 0.05, one-way ANOVA multiple comparison) than that with the manual and WASP plate streaking (Fig. 6 and Table 1), whereas no significant difference was observed between the manual, INO2, and WASP inoculations. The number of discrete colonies grown on BBL chromogenic agar following manual and automated inoculation was measured to assess the need for reisolation resulting in delayed time to results of 1 working day (16 h to 24 h) and additional laboratory costs (Fig. 7A and B). The additional laboratory cost per reisolation, including consumables and technician time, was estimated to be 3.3 EUR (3.6 USD). All inoculation methods except the INO1 required reisolation for bacterial identification for 3 (7.3%) to 8 (19.5%) cloudy urine samples. Moreover, the additional laboratory costs due to reisolation for bacterial ID ranged between 10 and 26 EUR (11 and 29 USD), which represents an additional cost of 24 to 64 EUR (26 to 71 USD) when extrapolated to 100 samples for simplicity (Fig. 7A). The InoqulA INO1 method also showed the best performance by requiring reisolation of only 4 out of 41 (9.8%) cloudy urine samples to perform ID and AST (Fig. 7B). Reisolation with the other inoculation methods was required for 10 (24.4%) cloudy urine specimens with the INO2 and 24 (58.5%) cloudy urine specimens with the MAN2. A similar level of performance was observed between the MAN1, WAS1, and WAS2 inoculation methods, which showed a need for reisolation in 15 (36.6%) to 18 (43.9%) cloudy urine samples. The laboratory costs due to reisolation to perform ID and AST extrapolated to 100 samples showed a minimum laboratory cost of 32 EUR (35 USD) with the INO1 inoculation and a maximum laboratory cost of 191 EUR (212 USD) with the MAN2 streaking. Thus, a 2.5-fold (INO2) to 6-fold (MAN2) increase in laboratory costs was observed with the INO2, MAN1, MAN2, WAS1, and WAS2 inoculation methods compared to the INO1 inoculation method, which presented the best performance following semiquantitative inoculation of clinical urinary samples.
FIG 6.
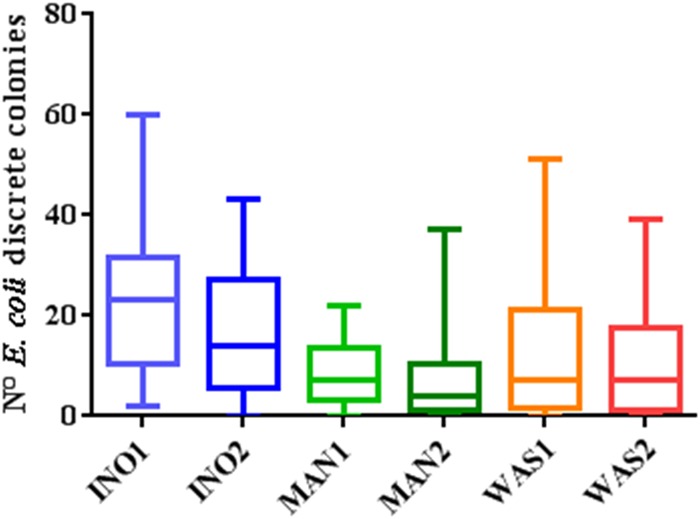
Performance of manual and automated inoculation on clinical urine samples. Shown are box-and-whisker plots (representing the minimum, first quartile, median, third quartile, and maximum) of the yield of discrete colonies from 41 cloudy urine specimen clinical samples positive for E. coli obtained following inoculation of 10 μl on chromogenic agar with the InoqulA (INO1 and INO2), manual (MAN1 and MAN2), and WASP (WAS1 and WAS2) methods. A statistically significantly higher number of discrete colonies (one-way ANOVA multiple comparison, P < 0.05) was observed between the INO1 and the MAN1, MAN2, WAS1, and WAS2 inoculations.
TABLE 1.
One-way ANOVA multiple comparisons of the number of discrete colonies from cloudy urine samples obtained with the InoqulA (INO1 and INO2), manually (MAN1 and MAN2), and the WASP (WAS1 and WAS2)
| Dunn's multiple-comparison test | Statistically significant? | P value |
|---|---|---|
| INO1 vs INO2 | No | 0.0993 |
| INO1 vs MAN1 | Yes | <0.0001 |
| INO1 vs MAN2 | Yes | <0.0001 |
| INO1 vs WAS1 | Yes | <0.0001 |
| INO1 vs WAS2 | Yes | <0.0001 |
| INO2 vs MAN1 | No | 0.0908 |
| INO2 vs MAN2 | Yes | 0.0010 |
| INO2 vs WAS1 | No | 0.9446 |
| INO2 vs WAS2 | No | 0.5419 |
| MAN1 vs MAN2 | No | >0.9999 |
| MAN1 vs WAS1 | No | >0.9999 |
| MAN1 vs WAS2 | No | >0.9999 |
| MAN2 vs WAS1 | No | 0.5038 |
| MAN2 vs WAS2 | No | 0.8836 |
| WAS1 vs WAS2 | No | >0.9999 |
FIG 7.
Impact of the performance of the different manual (MAN1 and MAN2) and automated inoculation InoqulA (INO1 and INO2) and WASP (WAS1 and WAS2) systems on the time to results and laboratory costs. (A) One discrete colony was required to perform identification by MALDI-TOF MS at day 1 postinoculation. Reisolation was performed when at least one colony was not obtained, leading to a delayed time to results of 1 working day (ID report at day 2). An additional laboratory cost of 3.3 EUR (3.6 USD) per reisolation was calculated for each subculture, and the results were extrapolated to 100 samples for clarity. (B) A minimum number of 6 discrete colonies grown on BBL chromogenic agar was required (i) to perform an ID by MALDI-TOF MS and (ii) to make a bacterial suspension in 2 ml of saline solution equivalent to a 0.5 McFarland standard turbidity to complete AST at day 1 and to report the results at day 2. Thus, each sample containing <6 colonies needed reisolation, leading to a delayed time to AST results of 1 working day (AST report at day 3). Similar to identification, an additional laboratory cost of 3.3 EUR (3.6 USD) per reisolation was calculated for each subculture, and the results were extrapolated to 100 samples for simplicity.
DISCUSSION
This is, to our knowledge, the first study comparing the performance of two automated systems, the WASP and the InoqulA, with manual inoculation on both defined and clinical samples. The quality of inoculation was assessed by measuring several parameters, including the yield of isolated colonies and their distribution on the agar plates. Quality of inoculation is a critical factor in clinical bacteriology, since a poor yield of discrete colonies significantly increases the time to results, hands-on time, and costs by adding steps of manual colony isolation and subculture, which often prolong the time to identification and antibiotic susceptibility testing results by 1 working day (16 to 24 h).
Images of the plates were taken with the ImagA BT digital imaging solution module (BD Kiestra) and were analyzed with VisionLab version 3.43 software to assess the quality of colony growth. Thus, the yield of discrete colonies and colony distribution were accurately measured by an image analysis software that removed the subjective interpretation of manual observation and allowed a precise quantification of the streaking quality of the different automated and manual approaches used in this study.
Only semiquantitative inoculation approaches were used in this study to determine the qualitative performance of the manual and automated quantitative streaking methods. The InoqulA INO1 pattern (zigzag) and the WASP WAS2 pattern (20 mm central streaking followed by zigzag streaking) were used as optimized factory-designed semiquantitative inoculation protocols. The manual MAN2 streaking approach (central streaking throughout the plate followed by zigzag streaking) was chosen as the conventional semiquantitative manual inoculation method used in our diagnostic laboratories. The INO2, MAN1, and WAS1 were chosen to use similar inoculation protocols, allowing a direct comparison between the different automated and manual systems. Thus, all the results obtained in this study should not be extrapolated to other inoculation methods that may exhibit a higher performance in colony isolation, such as the conventional nonquantitative 4-quadrant streaking methods, including a sterilization of the loop after streaking of the first quadrant that can be easily performed manually and by the WASP system. Thus, laboratories should carefully select and validate automated qualitative and quantitative patterns yielding the best performance for each sample type.
Similar to previous studies (2–5, 14), a higher number of discrete colonies was reproducibly obtained with the automated inoculation systems InoqulA and WASP than that with manual inoculation. Moreover, the difference in the recovery of microorganisms obtained between manual and automated inoculations increased with increasing bacterial concentrations in the sample. The manual and automated inoculation approaches, except for MAN1 and WAS1, showed similar performance, with a high recovery of discrete colonies at low to moderate bacterial concentrations. However, the automated systems allowed a significantly higher recovery of discrete colonies than that with manual inoculation at high bacterial concentrations of about 107 CFU/ml. Moreover, only the InoqulA INO1 and INO2 methods were able to reproducibly generate a high yield of discrete colonies at concentrations of >107 CFU/ml with all bacterial species tested in this study. The WASP inoculation system exhibited high performance up to 107 CFU/ml but was unable to allow efficient recovery of the isolated colonies of some bacterial species at high bacterial concentrations. Using pure bacterial cultures, the difference in performance observed between the InoqulA, WASP, and manual inoculation methods was bacterial species dependent. The InoqulA INO1 and INO2 methods showed significantly higher performance than that of manual and WASP streaking following inoculation of high concentrations of E. coli and, to a lesser extent, of K. pneumoniae, whereas no or little difference was observed between the InoqulA INO1/INO2 and the WASP WAS2 methods following inoculation of a high concentration of E. faecalis or S. aureus. Thus, the efficiency of each inoculation method to generate isolated colonies relies on multiple factors, including specific morphological and physiological traits of bacterial cells and colonies and the inoculation technology used. Bacterial cell membranes, shapes, and sizes likely exhibit different affinities for the inoculation support (magnetic beads, plastic loops, or metal loops) and for the agar surface that may impact the release of microorganisms during the streaking or the rolling process and thus the distribution gradient and yield of discrete colonies. For instance, we observed a slower release of the encapsulated K. pneumoniae strain by the InoqulA and WASP systems compared to that with other species (see Fig. S2B in the supplemental material, and data not shown), which resulted in a decreased yield of discrete colonies. This observation suggests that the capsular polysaccharide of K. pneumoniae may confer a stronger interaction of the bacteria with the inoculating device and thus decrease the rate of bacterial release during the streaking process. Moreover, bacterial colony growth kinetics and sizes likely also impact the recovery of discrete colonies. Finally, the higher performance of the InoqulA INO1 method with all bacterial species tested in this study is also likely based on its capacity to generate a gradual distribution of discrete colonies on a larger zone of the medium plate than other streaking approaches (as observed in Fig. 1; see also Fig. S1 and S2 in the supplemental material), thus optimizing the surface available for the recovery of isolated colonies.
Compared to automated inoculation, decreased reproducibility was observed following manual inoculation, with minor to moderate differences in the yields of discrete colonies obtained between different experiments (see Fig. 3 and 4). This observation is congruent with the findings in several studies showing that significantly higher reproducibility is observed with automated inoculation devices than that with manual inoculation (2–4). A greater variation in the yield of discrete colonies was observed with cloudy urine specimens due to significant bacterial load differences between samples. This was expected, since the yield of discrete colonies is largely determined by the bacterial concentration in a given sample, as shown in Fig. 3.
None of the manual or automated inoculation systems tested in this study allowed the recovery of discrete colonies of bacterial species present at concentrations 100- to 1,000-fold lower than those of the most concentrated species present in the sample. These results suggest that only a minor fraction of bacterial species present in polymicrobial samples are identified by routine laboratory culture procedures. Missing “minority species” has in most cases a small impact on the clinical outcome, since clinically relevant infectious agents are most often present at similar or higher concentrations than other microorganisms present in polymicrobial samples, such as urine samples. However, these results also indicate that the use of selective media in routine bacteriology is required to identify and recover true pathogens present in lower concentrations than the natural microflora in complex samples, such as respiratory samples.
The quality of inoculation is characterized by the ability of a system to obtain a high yield of discrete colonies for each bacterial species of a monomicrobial or polymicrobial sample. However, the real impact of an inoculation system on laboratory results and thus on clinical outcomes is not based on its ability to generate a maximal amount of isolated colonies but mainly on its ability to produce a critical minimal amount of discrete colonies required to perform downstream applications, including bacterial ID by MALDI-TOF MS, phenotypic and biochemical tests, and AST. According to EUCAST (www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/) and CLSI M02-A11 (15) guidelines, but also in predictions of automatic colony-picking technology, ID and AST should ideally be performed from isolated colonies and not from a bacterial lawn, even with pure culture (16). Thus, the impact of inoculation quality on the time to results and laboratory costs was assessed on clinical cloudy urine samples by determining the yield of discrete colonies and the need for reisolation to perform ID and AST. These results showed that the InoqulA INO1 system produces a statistically significantly higher yield of discrete colonies than that of manual and WASP inoculation but was also characterized by its greater ability to obtain the minimal amount of discrete colonies necessary to rapidly perform downstream applications. The INO1 system was the only tested inoculation approach that allowed direct identification by MALDI-TOF MS of the 41 cloudy urine specimens positive for E. coli. Moreover, subculture was required for only 4 out of 41 (9.7%) cloudy urine specimens following INO1 inoculation, indicating that AST could be performed directly for 37 (90.3%) of the E. coli strains recovered in the urine samples. Together, the conventional routine laboratory manual semiquantitative approach exhibited the lowest performance, clearly indicating that automation may efficiently improve laboratory productivity while reducing laboratory cost. This study showed that the ability of the InoqulA INO1 method to yield a high number of discrete colonies reduced the turnaround time (TAT) compared to that of the other inoculation approaches, allowing also significant reduced laboratory costs by reducing the need to make bacterial subculture for ID and AST procedures. Moreover, the reduced TAT observed with the InoqulA automated system should positively impact clinical management and thus clinical costs. However, the hypothetical benefits remain to be addressed in a specific study measuring the impact of partial and full laboratory automation on clinical outcomes and hospitalization costs.
In summary, this study showed that a higher number of discrete colonies was reproducibly obtained with the InoqulA and WASP automated systems than that with manual inoculation. The InoqulA exhibited a higher performance than that of the WASP system at bacterial concentrations of >107 CFU/ml. However, the difference observed was bacterial species dependent, since a significant difference was observed with E. coli and K. pneumoniae but not with S. aureus and E. faecalis. The prospective analysis of clinical cloudy urine specimens showed that the InoqulA (INO1) method provided a statistically significantly higher number of discrete colonies than that with the WASP and manual inoculations, resulting in a reduced time to ID and AST results and reduced laboratory costs due to a decreased need to perform colony reisolation. Finally, both the automated inoculation technology (magnetic bead versus loop) and the design of optimal streaking patterns had a significant impact on the performance of the inoculation methods observed in this study.
This work represents one of the first studies conducted by an independent clinical diagnostic laboratory that demonstrates the true effectiveness of automated inoculation systems to generate isolated colonies positively impacting both the TAT and costs. Unlike manual inoculation, automated streaking systems are highly reproducible and offer the possibility to investigate new technical inoculation approaches to improve the quality and the quantification of colony growth, thus further increasing the productivity of the diagnostic laboratory.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03076-14.
REFERENCES
- 1.Greub G, Prod'hom G. 2011. Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect 17:655–660. doi: 10.1111/j.1469-0691.2011.03513.x. [DOI] [PubMed] [Google Scholar]
- 2.Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. 2012. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J Clin Microbiol 50:2732–2736. doi: 10.1128/JCM.05501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Froment P, Marchandin H, Vande Perre P, Lamy B. 2014. Automated versus manual sample inoculations in routine clinical microbiology: a performance evaluation of the fully automated InoqulA instrument. J Clin Microbiol 52:796–802. doi: 10.1128/JCM.02341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbeau PP, Swartz BL. 2009. First evaluation of the WASP, a new automated microbiology plating instrument. J Clin Microbiol 47:1101–1106. doi: 10.1128/JCM.01963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones G, Matthews R, Cunningham R, Jenks P. 2011. Comparison of automated processing of flocked swabs with manual processing of fiber swabs for detection of nasal carriage of Staphylococcus aureus. J Clin Microbiol 49:2717–2718. doi: 10.1128/JCM.00504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103–e120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 7.Bennett JE, Dolin R, Blaser MJ. 2014. Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 8th ed, Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 8.Versalovic J, Carroll K, Funke G, Jorgensen J, Landry M, Warnock D. 2011. Manual of clinical microbiology, 10th ed, ASM Press, Washington, DC. [Google Scholar]
- 9.McCarter YS, Burd EM, Hall GS, Zervos M. 2009. Cumitech 2C: laboratory diagnosis of urinary tract infections. ASM Press, Washington, DC. [Google Scholar]
- 10.Fisher RA. 1936. The use of multiple measurements in taxonomic problems. Ann Eugen 7:179–188. doi: 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- 11.McLachlan GJ. 2004. Discriminant analysis and statistical pattern recognition. Wiley-Interscience, Hoboken, NJ. [Google Scholar]
- 12.Van de Loosdrecht J. 2014. Computer vision: distance and Hough transforms. NHL, Leeuwarden, The Netherlands: http://www.nhlcomputervision.nl/upload/course/13_transfor.pdf. [Google Scholar]
- 13.Joblove GH, Greenberg D. 1978. Color Spaces for Computer Graphics. SIGGRAPH Comput Graph 12:20–25. doi: 10.1145/965139.807362. [DOI] [Google Scholar]
- 14.Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. 2008. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J Clin Microbiol 46:1281–1284. doi: 10.1128/JCM.01687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard—11th ed. CLSI document M02-A11 Clinical and Laboratory Standards Institute, Wayne, PA: http://antimicrobianos.com.ar/ATB/wp-content/uploads/2012/11/01-CLSI-M02-A11-2012.pdf. [Google Scholar]
- 16.Matuschek E, Brown DF, Kahlmeter G. 2014. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect 20:O255–O266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.