Abstract
Haemophilus influenzae type b (Hib) is, in contrast to non-type b H. influenzae, associated with severe invasive disease, such as meningitis and epiglottitis, in small children. To date, accurate H. influenzae capsule typing requires PCR, a time-consuming and cumbersome method. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) provides rapid bacterial diagnostics and is increasingly used in clinical microbiology laboratories. Here, MALDI-TOF MS was evaluated as a novel approach to separate Hib from other H. influenzae. PCR-verified Hib and non-Hib reference isolates were selected based on genetic and spectral characteristics. Mass spectra of reference isolates were acquired and used to generate different classification algorithms for Hib/non-Hib differentiation using both ClinProTools and the MALDI Biotyper software. A test series of mass spectra from 33 Hib and 77 non-Hib isolates, all characterized by PCR, was used to evaluate the algorithms. Several algorithms yielded good results, but the two best were a ClinProTools model based on 22 separating peaks and subtyping main spectra (MSPs) using MALDI Biotyper. The ClinProTools model had a sensitivity of 100% and a specificity of 99%, and the results were 98% reproducible using a different MALDI-TOF MS instrument. The Biotyper subtyping MSPs had a sensitivity of 97%, a specificity of 100%, and 93% reproducibility. Our results suggest that it is possible to use MALDI-TOF MS to differentiate Hib from other H. influenzae. This is a promising method for rapidly identifying Hib in unvaccinated populations and for the screening and surveillance of Hib carriage in vaccinated populations.
INTRODUCTION
Haemophilus influenzae type b (Hib) has been, and in some regions still is, the dominating cause of severe invasive disease associated with the species H. influenzae. More specifically, it is (or used to be) a feared cause of meningitis and epiglottitis in small children (1, 2). The conjugate vaccines against Hib that were introduced in the early 1990s have resulted in a steep decline in invasive Hib disease (1, 3–5). However, Hib still causes 5 to 10% of invasive H. influenzae disease in Sweden, and occasional cases of fully vaccinated children with invasive Hib disease have been reported in several countries (6, 7). Hib is estimated to cause a substantial number of infections and deaths among young children each year on a global basis (8). PCR is at present required for accurate Hib capsule typing (9–11) but is a relatively time-consuming and laborious method. Agglutination with antisera is an alternative method used for capsular typing. Even though this is a faster method than PCR, studies have shown that its accuracy is comparatively poor (12).
Early studies have suggested that the Hib population consists of two genetically distinct clusters (13, 14). This population structure was later confirmed using multilocus sequence typing (MLST), since practically all Hib isolates that had been submitted to the MLST database from all over the world could be sorted into one of the two clusters (15). The dominant cluster comprises the main portion of Hib isolates and is centered on sequence type 6 (ST6) in the MLST database. The second Hib cluster is comparatively uncommon, and the predominant sequence type in this cluster is ST93. The fact that there are two distinct Hib clusters is an important factor to take into account when investigating methods of identifying Hib that do not focus on capsule identification per se.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has revolutionized clinical microbiological diagnostics and is used nowadays to identify fungal and bacterial species, including H. influenzae (16, 17). The technique has also been suggested to be a useful tool for various subtyping of bacteria, testing of antimicrobial susceptibility, and identification of virulent bacterial clones (18–21). However, MALDI-TOF MS is currently used only to identify H. influenzae at the species level and has not yet been established for use in subtyping.
The aim of this study was to investigate whether MALDI-TOF MS can be used to differentiate Hib from other H. influenzae capsular types and nontypeable (unencapsulated) H. influenzae (NTHi). The results demonstrate that MALDI-TOF MS can successfully be used to discriminate between Hib and non-b H. influenzae, but the choice of testing algorithm is essential to achieve optimal results. MALDI-TOF MS can thus be used as a highly valuable cost- and time-effective methodology for typing of Hib.
MATERIALS AND METHODS
Bacterial isolates.
The culture collection in this study comprised 127 isolates. The majority of the isolates (n = 114) were invasive H. influenzae isolates collected in four Swedish metropolitan areas (Gothenburg, Lund, Malmö, and Stockholm) between 1997 and 2010. The remaining isolates were collected from other countries, such as the United States, or were obtained from culture collections (Culture Collection, University of Gothenburg [CCUG], Sweden, and the National Collection of Type Cultures [NCTC], Public Health England, London, United Kingdom). Isolates were identified as H. influenzae using routine clinical methods (hemin and NAD required for growth) and standard MALDI-TOF MS analysis using the MALDI Biotyper software. All isolates were stored in glycerol at −80°C and cultured on chocolate agar plates at 37°C and 5% CO2 for 18 to 24 h before use.
Preparation of bacterial DNA.
To prepare DNA, a few colonies were suspended in 100 μl of distilled water and heated at 98°C for 10 min. After centrifugation, the supernatant was collected and used as the template for PCR. To obtain high-quality sequencing results, extraction of genomic DNA was performed with the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO).
Capsular typing by PCR.
All isolates in this study were capsule typed by PCR. To determine the presence of a capsule, PCR to detect the common capsule transport complex (bexB) was performed as previously described (9). On isolates positive for bexB, capsule type-specific PCRs were performed as described previously (10, 11).
MLST.
MLST was performed as described by Meats et al. (22) using primers available in the MLST database (www.mlst.net). The resulting PCR products were sequenced, analyzed using Geneious 7.1.3 (Biomatters, Auckland, New Zealand), and the STs of the isolates were determined using information in the MLST database.
MALDI-TOF MS data acquisition.
Isolates were prepared for acquisition of spectra using a standard ethanol-formic acid extraction protocol developed by the mass spectrometer manufacturer (Bruker Daltonics, Bremen, Germany). The bacterial pellets were allowed to dry completely (up to 1 h) following ethanol washing. The volume of formic acid and acetonitrile used (10 to 40 μl) was based on pellet size. Reference and test isolates were applied on 8 and 2 spots, respectively. Three spectra per spot were acquired, giving 24 spectra per reference isolate and 6 spectra per test isolate.
Mass spectra of the reference and test isolates were acquired using a microflex MALDI-TOF mass spectrometer with flexControl software (Bruker Daltonics, Bremen, Germany) using default settings (mass range of spectra, m/z 2,000 to 20,000 in linear positive-ionization mode). Each spectrum consisted of 240 shots, divided into 40-shot increments. For reproducibility testing of the generated classification methods, 19 Hib and 23 non-Hib test isolates were randomly selected for repeated extraction and measurement on a different MALDI-TOF MS instrument (ultrafleXtreme; Bruker Daltonics) in another laboratory using default settings (a total of 800 shots divided into 100-shot increments). Before each run, the instruments were calibrated with Bacterial Test Standard (BTS) (Bruker Daltonics).
Manual analysis of mass spectra and selection of reference isolates.
Manual analysis of mass spectra, using the ClinProTools 3.0 software (Bruker Daltonics), was performed to identify peak differences between Hib and non-Hib isolates. Reference Hib isolates were selected from a screen of 19 randomly chosen Hib isolates based on a manual analysis of their spectral profile and MLST. Reference isolates were chosen in order to cover the broadest possible range within the MLST Hib clusters. Isolates not chosen as reference isolates were included among the test isolates. The non-Hib reference isolates included all capsular types except Hib, as well as five NTHi.
Hib/non-Hib classification methods.
Four different methods were each tested in two separate ways, resulting in a total of 8 different classification algorithms, and these were evaluated for differentiation of Hib and non-Hib. ClinProTools was used for the generation of classification models using raw spectra of the reference isolates. The default settings were used for spectrum preparation (resolution, 800; baseline subtraction, top-hat, 10% minimal baseline width; recalibration, 1,000 ppm maximal peak shift and 30% match to calibrant peaks), peak calculation (peak picking on the total average spectrum; signal-to-noise ratio, 5), and peak selection (use all peaks). The models in ClinProTools were generated using three different algorithms (genetic, supervised serial network, and quick classifier). The number of peaks to be included in the genetic algorithm varied between 5 and 25. This was considered a suitable range, based on manual analysis for capsule-specific peak differences of the spectra of reference isolates. The other two algorithms detect the optimal number of peaks to be included automatically. The model with the best Hib/non-Hib differentiation (when analyzing 6 spectra per test isolate) was chosen for further testing with only two spectra per test isolate and reproducibility testing (see below).
The MALDI Biotyper 3.1 software was used to classify spectra using three different methods; regular main spectra (MSPs), subtyping MSPs, and specially designed MSPs. Before MSPs were created, the quality of the reference isolate spectra was checked using FlexAnalysis 3.4 (Bruker Daltonics). Smoothing and baseline subtraction were performed, and outlier (lacking a peak or having an extra peak) or low-quality (peaks outside a 500-ppm range) spectra were removed. The remaining spectra, ≥20 of each isolate, were used to create the different types of MSPs in Biotyper.
Regular MSPs of Hib and non-Hib reference isolates were created using default settings (maximum, 70 peaks; mass range, m/z 3,000 to 15,000). For the creation and testing of subtyping MSPs, the preprocessing, MSP creation, and subtyping MSP creation methods in Biotyper were modified to use a wider mass range (m/z 2,500 to 15,500) and to allow more peaks in the MSPs (maximum, 150 peaks). MSPs were created for all reference isolates. The subtyping MSPs of each Hib reference isolate were then individually generated against the MSPs of the non-Hib isolates (not together with other Hib MSPs) to avoid Hib subtyping MSPs to differentiate between different Hib clones.
Specially designed MSPs of the Hib reference isolates were created using default mass range settings (m/z 3,000 to 15,000) but with an increased number of peaks allowed (maximum, 150 peaks). The MSP peak lists were modified to include only two peaks with masses of approximately m/z 6,789 and m/z 8,348 (Fig. 1). These peaks were selected based on a manual evaluation of spectra for their capacity to differentiate between Hib isolates of the ST6 cluster and non-Hib isolates. H. influenzae strain KR194 is not a part of the ST6 cluster and does not express these peaks.
FIG 1.
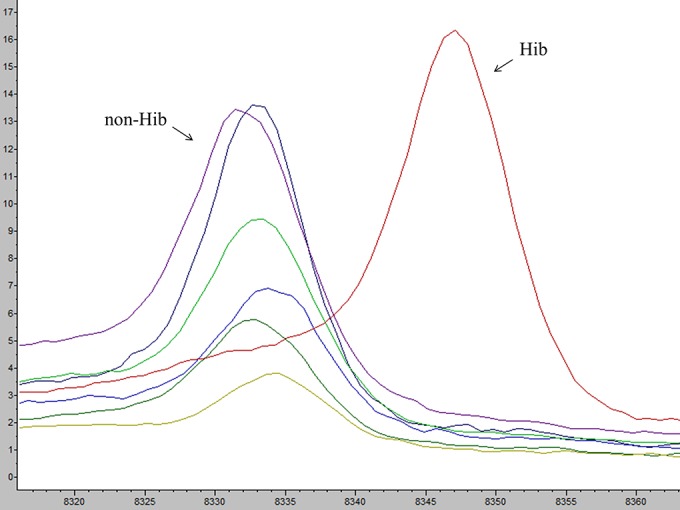
Illustration of peak m/z 8,332 and peak m/z 8,348 as potential differentiators between Hib and non-b H. influenzae. Non-Hib (Hia, Hic, Hid, Hie, Hif, and NTHi) isolates expressed a peak of approximately m/z 8,332. In contrast, Hib isolates expressed an offset peak of approximately m/z 8,348. Each capsular type is represented by a single isolate. x and y axes show m/z values and intensity (in arbitrary units), respectively.
Testing of different classification algorithms.
Each method was tested in two different ways using 33 Hib and 77 non-Hib isolates (Table 1). First, the methods were tested by classification of all six spectra acquired for each test isolate. Isolates with ≥5 of 6 spectra classified as Hib were identified as Hib, and isolates with ≤2 of 6 spectra classified as Hib were identified as non-Hib. Isolates with 3 to 4 of 6 spectra classified as Hib were identified as uncertain. Second, to mimic the everyday clinical standard procedure, methods were tested using only the first spectrum acquired per spot, giving two spectra per test isolate. In this setting, isolates with ≥1 spectrum classified as Hib were identified as Hib, isolates with no spectra classified as Hib were identified as non-Hib. The identification category uncertain was not used.
TABLE 1.
Clinical isolates and laboratory reference strains used to test Hib/non-Hib classification
| Isolate | Capsular type | Location | Yr | Site of isolation |
|---|---|---|---|---|
| Hib | ||||
| Clinical isolates | ||||
| M58 | b | Malmö | 1997 | Blood |
| M61a | b | Malmö | 1997 | Blood |
| KR181a | b | Stockholm | 1998 | CSFb |
| G4a | b | Gothenburg | 2002 | Blood |
| M41 | b | Malmö | 2004 | Blood |
| M35a | b | Malmö | 2004 | Blood |
| L40 | b | Lund | 2004 | Blood |
| L2a | b | Lund | 2004 | Blood |
| KR196 | b | Stockholm | 2004 | Blood |
| KR192a | b | Stockholm | 2004 | Blood |
| M31 | b | Malmö | 2005 | Blood |
| L35a | b | Lund | 2005 | Blood |
| M66 | b | Malmö | 2006 | Blood |
| KR211 | b | Stockholm | 2006 | Blood |
| KR209a | b | Stockholm | 2006 | Blood |
| L58 | b | Lund | 2007 | Blood |
| L63a | b | Lund | 2008 | Blood |
| G32a | b | Gothenburg | 2008 | CSF |
| M7a | b | Malmö | 2009 | Blood |
| G26 | b | Gothenburg | 2009 | Blood |
| S86a | b | Stockholm | 2010 | Blood |
| S79 | b | Stockholm | 2010 | Blood |
| M81a | b | Malmö | 2010 | CSF |
| M75 | b | Malmö | 2010 | Blood |
| G74a | b | Gothenburg | 2010 | CSF |
| G72 | b | Gothenburg | 2010 | Blood |
| G67a | b | Gothenburg | 2010 | Blood |
| Laboratory reference strains | ||||
| CCUG 18095 (Eagan)a | b | Boston, MA | 1968 | CSF |
| MinnAa | b | Minneapolis, MN | 1979 | CSF |
| DL-42 | b | Dallas, TX | Before 1984 | Unknown |
| 850530a | b | Netherlands | 1985 | Unknown |
| HK 691a | b | Netherlands | Before 1985 | Unknown |
| HK 729 | b | United States | Before 1988 | CSF |
| Non-Hib | ||||
| Clinical isolates | ||||
| L59a | e | Lund | 2007 | Blood |
| KR228 | e | Stockholm | 2008 | Blood |
| KR236 | e | Stockholm | 2008 | CSF |
| KR138a | e | Unknown | Unknown | Unknown |
| KR147 | e | Unknown | Unknown | Unknown |
| KR569 | e | Malmö | 2009 | Tonsil |
| M63 | f | Malmö | 1997 | Blood |
| KR179a | f | Stockholm | 1998 | CSF |
| M54a | f | Malmö | 1999 | Blood |
| KR187 | f | Stockholm | 2000 | CSF |
| L45 | f | Lund | 2005 | Blood |
| L44 | f | Lund | 2005 | Blood |
| KR200 | f | Stockholm | 2005 | Blood |
| M65 | f | Malmö | 2006 | Blood |
| M29 | f | Malmö | 2006 | CSF |
| L11a | f | Lund | 2006 | Blood |
| KR210 | f | Stockholm | 2006 | Blood |
| L50a | f | Lund | 2007 | Blood |
| L19 | f | Lund | 2007 | Blood |
| L18 | f | Lund | 2007 | Blood |
| L16 | f | Lund | 2007 | Blood |
| M14 | f | Malmö | 2008 | Blood |
| M12 | f | Malmö | 2008 | Blood |
| M10a | f | Malmö | 2008 | Blood |
| L64 | f | Lund | 2008 | punctate |
| L22 | f | Lund | 2008 | Blood |
| L21 | f | Lund | 2008 | Blood |
| KR233a | f | Stockholm | 2008 | Blood |
| G34 | f | Gothenburg | 2008 | Blood |
| G22 | f | Gothenburg | 2008 | Blood |
| G20 | f | Gothenburg | 2008 | Blood |
| G19 | f | Gothenburg | 2008 | Blood |
| G18a | f | Gothenburg | 2008 | Blood |
| L29 | f | Lund | 2009 | Blood |
| L25 | f | Lund | 2009 | Blood |
| L24 | f | Lund | 2009 | Blood |
| S78 | f | Stockholm | 2010 | Blood |
| M60 | NTc | Malmö | 1997 | Blood |
| M59 | NT | Malmö | 1997 | Blood |
| KR170a | NT | Stockholm | 1998 | Blood |
| G1a | NT | Gothenburg | 1999 | Blood |
| KR183 | NT | Stockholm | 2000 | Blood |
| M51a | NT | Malmö | 2001 | CSF |
| G2a | NT | Gothenburg | 2001 | Blood |
| M45 | NT | Malmö | 2002 | Blood |
| M36 | NT | Malmö | 2004 | Blood |
| L3a | NT | Lund | 2004 | Blood |
| L17 | NT | Lund | 2004 | Blood |
| KR189 | NT | Stockholm | 2004 | Blood |
| KR201 | NT | Stockholm | 2005 | Blood |
| L9 | NT | Lund | 2006 | Blood |
| L8 | NT | Lund | 2006 | Blood |
| L48 | NT | Lund | 2006 | Blood |
| M23 | NT | Malmö | 2007 | Blood |
| M21 | NT | Malmö | 2007 | Blood |
| L61 | NT | Lund | 2007 | Blood |
| L57a | NT | Lund | 2007 | Blood |
| L56 | NT | Lund | 2007 | Blood |
| L55a | NT | Lund | 2007 | Blood |
| L20 | NT | Lund | 2007 | Blood |
| G14 | NT | Gothenburg | 2007 | Blood |
| M9 | NT | Malmö | 2008 | Blood |
| M13 | NT | Malmö | 2008 | Blood |
| KR223a | NT | Stockholm | 2008 | Blood |
| G17 | NT | Gothenburg | 2008 | Blood |
| M5 | NT | Malmö | 2009 | Blood |
| M4a | NT | Malmö | 2009 | Blood |
| M38 | NT | Malmö | 2009 | Blood |
| L26 | NT | Lund | 2009 | Blood |
| G40a | NT | Gothenburg | 2009 | Blood |
| Laboratory reference strains | ||||
| CCUG 6881a | a | Unknown | Before 1973 | Unknown |
| CCUG 7315 | a | United States | 1941 | Respiratory tract |
| CCUG 4851a | c | United States | 1942 | Sputum |
| CCUG 4852 | c | New York, NY | Before 1950 | CSF |
| NCTC 8470a | d | Netherlands | 1937 | Throat |
| CCUG 15521a | e | Unknown | Before 1984 | Unknown |
| CCUG 6877 | NT | United States | 1941 | Unknown |
To test reproducibility, extraction and MALDI-TOF MS measurements were also performed on a different instrument in a second laboratory.
CSF, cerebrospinal fluid.
NT, nontypeable.
ClinProTools models classify all spectra as either of the classes used to generate the models (e.g., Hib or non-Hib). In MALDI Biotyper, scores are calculated for each spectrum. When using regular MSPs, a spectrum was classified as Hib if it had a Hib MSP as the best match (since all MSPs were of the same species, scores were expected to be high on all MSPs, regardless of the capsular type of the test isolates). When using subtyping MSPs, spectra were classified as Hib if the score was >2.0 for any Hib subtyping MSP. When using specially designed MSPs, spectra were classified as Hib if the score was >1.3 for any Hib MSP, as 1.301 was the score yielded by spectra expressing both peaks included in these MSPs.
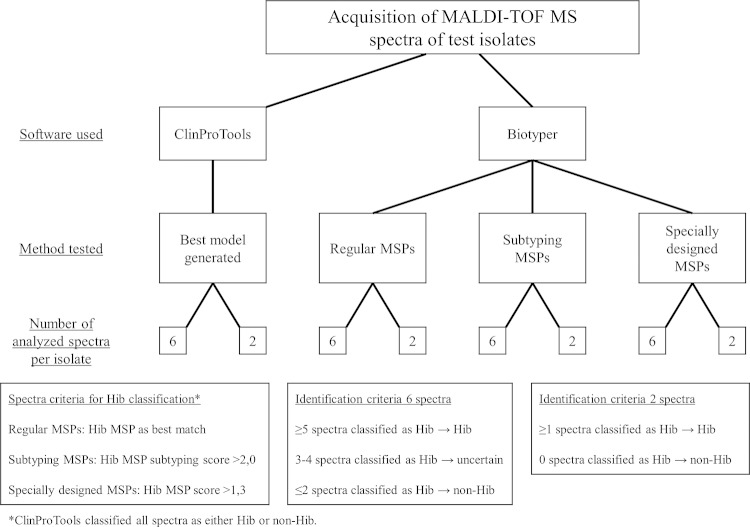
A flowchart of the testing procedure of the different classification methods is presented in Fig. 2.
FIG 2.
Flowchart presenting the testing procedures of different Hib/non-Hib classification methods. The ClinProTools and MALDI Biotyper softwares were used to separate Hib and non-Hib test isolates. In ClinProTools, the best model was chosen for further testing. In Biotyper, regular MSPs, subtyping MSPs, and specially designed MSPs were used to classify test isolate spectra. Each method was tested with both 6 and 2 spectra per test isolate. Extraction and MALDI-TOF MS spectra acquisition were repeated on 19 Hib and 23 non-Hib isolates on a different MALDI-TOF MS instrument to test the reproducibility of the classification methods.
Analysis of test results.
Sensitivity, specificity, and discriminatory accuracy (expressed as the area under the receiver operating characteristic [AU-ROC]) were calculated for each Hib/non-Hib classification algorithm based on the results generated on the microflex instrument. AU-ROC was calculated using SPSS Statistics 22.0 (IBM, Armonk, NY) based on correct or false identification compared to PCR (golden standard). All isolates identified as uncertain were considered misidentified in the analysis. The reproducibility of each classification method was calculated as the percentage of tested isolates that was sorted into the same category (Hib, uncertain, or non-Hib) in both measurements (microflex and ultrafleXtreme).
RESULTS
MLST analysis and selection of reference isolates.
MLST was performed on 19 Hib isolates. Seven different STs were represented in the group (Table 2). The isolates KR194, L2, KR211, and M17 belonged to new STs. All were single-allele variants from previously known STs. All isolates belonged to the worldwide dominant ST6 cluster, except KR194, which belonged to the established second cluster (single-allele variant from ST93). By choosing KR194 and 6 different variants from the ST6 cluster as reference isolates for MALDI-TOF MS classification, we aimed for optimal coverage of known Hib clusters.
TABLE 2.
MLST of Hib isolates and reference isolates used for creation of Hib/non-Hib classification methods
| Isolate | Capsular type | MLST results |
Location | Yr | Site of isolation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adk | atpG | frdB | fucK | mdh | pgi | recA | ST | |||||
| MLST of Hib isolatesa | ||||||||||||
| G4 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Gothenburg | 2002 | Blood |
| G6b | b | 31 | 14 | 4 | 5 | 4 | 7 | 8 | 95 | Gothenburg | 2004 | Blood |
| G26 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Gothenburg | 2009 | Blood |
| G32 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Gothenburg | 2008 | CSF |
| KR169b | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Stockholm | 1998 | Blood |
| KR181 | b | 31 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Stockholm | 1998 | CSF |
| KR191b | b | 10 | 14 | 5 | 7 | 57 | 7 | 8 | 120 | Stockholm | 2004 | Blood |
| KR194b | b | 6 | 30 | 23 | 9 | 33 | 29 | 7 | New STc | Stockholm | 2004 | Blood |
| KR209 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Stockholm | 2009 | Blood |
| KR211 | b | 10 | 14 | 4 | 5 | 4 | New | 8 | New STd | Stockholm | 2009 | Blood |
| L2 | b | 10 | New | 4 | 5 | 4 | 7 | 8 | New STe | Lund | 2004 | Blood |
| L35 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Lund | 2005 | Blood |
| L40 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Lund | 2004 | Blood |
| L58 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Lund | 2007 | Blood |
| L62b | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Lund | 2007 | Blood |
| L63 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Lund | 2008 | Blood |
| M16b | b | 10 | 14 | 4 | 1 | 4 | 7 | 8 | 206 | Malmö | 2007 | Blood |
| M17b | b | 10 | 14 | 5 | 7 | 4 | New | 8 | New STd | Malmö | 2007 | Blood |
| M41 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | Malmö | 2004 | Blood |
| Non-Hib reference isolates | ||||||||||||
| KR152f | a | 20 | 12 | 25 | 7 | 20 | 23 | 19 | 30 | Unknown | Unknown | Unknown |
| KR153f | c | 7 | 11 | 6 | 8 | 6 | 16 | 9 | 9 | Unknown | Unknown | Unknown |
| KR154f | d | 5 | 15 | 7 | 9 | 7 | 5 | 11 | 47 | Unknown | Unknown | Unknown |
| L13 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | Lund | 2006 | Blood |
| M1 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | Malmö | 2009 | Blood |
| G23 | NTg | 5 | 1 | 1 | 1 | 1 | 2 | 5 | 14 | Gothenburg | 2008 | Blood |
| G63 | NT | 14 | 7 | 13 | 7 | 17 | 13 | 17 | 57 | Gothenburg | 2010 | Blood |
| KR206 | NT | 11 | 2 | 15 | 8 | 28 | 26 | 3 | 34 | Stockholm | 2008 | Blood |
| L1 | NT | 1 | 1 | New | 14 | 9 | 14 | 3 | New STh | Lund | 2004 | Blood |
| M15 | NT | 1 | 1 | 1 | 1 | 7 | 2 | 5 | New STi | Malmö | 2008 | Blood |
MLST was performed on 19 Hib isolates. The selection of Hib reference isolates was based on MLST and spectral characteristics. Hib not selected as reference isolates were used as test isolates for Hib/non-Hib classification methods. Non-Hib reference isolates and their characteristics are also presented.
Selected as Hib reference isolate.
Most similar to ST93 among previously known STs.
Most similar to ST53 among previously known STs.
Most similar to ST6 among previously known STs.
Unfortunately, for the Hia, Hic, and Hid reference isolates, the location, year, and tissue from which they were isolated are unknown.
NT, nontypeable.
Most similar to ST103 among previously known STs.
Most similar to ST3 among previously known STs.
For the non-Hib reference group, one isolate of each non-Hib capsular type (a, c, d, e, and f) and five NTHi were selected as reference isolates. Five NTHi isolates with different spectral and MLST patterns were chosen as references. Even though the reference NTHi were genetically and spectrally heterogenous, we did not cover the full and very wide range of NTHi. The focus of the selection was based on their spectral variation in key Hib peaks. The selected reference isolates are presented in Table 2.
Identification of Hib using MALDI-TOF MS.
Four different classification methods were tested in two different ways (Fig. 2). Sensitivity, specificity, AU-ROC, and reproducibility were calculated for all classification algorithms. A full summary of the test results is presented in Table 3.
TABLE 3.
Evaluation of different MALDI-TOF MS Hib/non-Hib classification methods
| Results by no. of spectra for: | Hib (n = 33) | Non-Hib (n = 77) | Sensitivity (%)a | Specificity (%)a | AU-ROCb | Reproducibility (%)c |
|---|---|---|---|---|---|---|
| ClinProTools | ||||||
| 6 spectra | 100 | 98.7 | 0.994 | 97.6 | ||
| Hib | 33 | 1 | ||||
| Uncertain | 0 | 0 | ||||
| Non-Hib | 0 | 76 | ||||
| 2 spectra | 100 | 97.4 | 0.987 | 100 | ||
| Hib | 33 | 2 | ||||
| Non-Hib | 0 | 75 | ||||
| Biotyper MSPs: | ||||||
| Regular | ||||||
| 6 spectra | 100 | 98.7 | 0.994 | 90.5 | ||
| Hib | 33 | 0 | ||||
| Uncertain | 0 | 1 | ||||
| Non-Hib | 0 | 76 | ||||
| 2 spectra | 100 | 97.4 | 0.987 | 95.2 | ||
| Hib | 33 | 2 | ||||
| Non-Hib | 0 | 75 | ||||
| Subtyping | ||||||
| 6 spectra | 97.0 | 100 | 0.985 | 92.9d | ||
| Hib | 32 | 0 | ||||
| Uncertain | 1 | 0 | ||||
| Non-Hib | 0 | 77 | ||||
| 2 spectra | 100 | 100 | 1.000 | 100d | ||
| Hib | 33 | 0 | ||||
| Non-Hib | 0 | 77 | ||||
| Specially designed | ||||||
| 6 spectra | 81.8 | 100 | 0.909 | 76.2 | ||
| Hib | 27 | 0 | ||||
| Uncertain | 3 | 0 | ||||
| Non-Hib | 3 | 77 | ||||
| 2 spectra | 97.0 | 98.7 | 0.978 | 85.7 | ||
| Hib | 32 | 1 | ||||
| Non-Hib | 1 | 76 |
Sensitivity and specificity were calculated based on the results from the original laboratory (Malmö).
Area under the receiver operator characteristic curve calculated based on correct or false result from the measurements with the microflex instrument compared to those with PCR. All uncertain isolates were considered falsely identified. AU-ROC values were calculated using SPSS Statistics 22.0 (IBM, Armonk, NY).
Reproducibility was calculated as the percentage of isolates MALDI-TOF MS measured on both the microflex and ultrafleXtreme instrument, which were identified to the same category (Hib, uncertain, or non-Hib) on both instruments.
Reproducibility with adjusted cutoff.
Identification of Hib using ClinProTools.
The best model in ClinProTools was generated using the genetic algorithm and was based upon 22 peaks. Other models generated by the genetic algorithm achieved results close to those of the 22-peak model. The results from the 22-peak model (referred to as the ClinProTools model) are presented below.
When 6 spectra per isolate were used, all Hib (n = 33) and 98.7% of non-Hib (76 of 77) isolates were correctly identified. The non-Hib isolate KR183 was identified as Hib. All 6 spectra of all individual Hib isolates were correctly classified, and all KR183 spectra were incorrectly classified. Three non-Hib isolates each had one of six spectra that was incorrectly classified. The reproducibility was 97.6%, as all isolates except one Hib isolate (identified as uncertain by the ultrafleXtreme) were sorted into the same category by both instruments. When only two spectra per isolate were analyzed, the NTHi isolate M5 was incorrectly identified as Hib according to our definitions (since one of the two first spectra was incorrectly classified in this isolate) along with KR183. Here, all measured isolates were identified in the same category on the second instrument, giving a reproducibility of 100%.
Identification of Hib using regular Biotyper MSPs.
When using 6 spectra, all Hib isolates were correctly classified on all spectra. A large portion (98.7%) of the non-Hib isolates were correctly identified. Several of the non-Hib isolates had one or two spectra incorrectly classified but only one isolate was incorrectly identified as uncertain, with four spectra classified as Hib. When repeated on a different instrument, 16 of 19 Hib and 22 of 23 non-Hib isolates were identified to the same category, resulting in a reproducibility of 90.5%.
When two spectra per isolate were analyzed, all Hib isolates were correctly identified, with both spectra classified as Hib. The majority (97.4% [75 of 77]) of the non-Hib isolates were correctly identified. When measured using the other instrument, all Hib (n = 19) and 21 of 23 non-Hib isolates were identified to the same category, resulting in a reproducibility of 95.2%.
Identification of Hib using Biotyper subtyping MSPs.
When using subtyping MSPs, a spectrum was classified as Hib if it had any Hib subtyping MSP score of >2.0. When 6 spectra per isolate were analyzed, 97.0% (32 of 33) of the Hib isolates were correctly identified. One isolate was classified as uncertain. All non-Hib isolates were correctly identified, and all non-Hib spectra (6 per isolate) had Hib subtyping scores of ≤2.0. When the analysis was repeated with the ultrafleXtreme instrument, the absolute scores were lower in general. However, relative differences in scores between Hib and non-Hib isolates remained. The score cutoff on Hib subtyping MSPs for Hib classification was consequently adjusted for these spectra. The absolute reproducibility with this method was therefore low, but with the adjusted cutoff, the reproducibility was 92.9%, with 16 of 19 Hib and all non-Hib (n = 23) isolates were identified to the same category by the two instruments.
When using only two spectra per isolate, all Hib isolates and all non-Hib isolates were correctly identified. All isolates extracted and measured by the other instrument were also correctly identified, giving a reproducibility of 100% (implying adjusted cutoff).
Identification of Hib using specially designed Biotyper MSPs.
For all Hib reference isolates except KR194, which belongs to the less common of the two established genetic clusters of Hib, MSPs were generated. A score of >1.3 on any of the Hib MSPs was required for Hib classification.
When 6 spectra per isolate were used, 81.8% (27 of 33) of the Hib isolates were correctly identified. Three isolates were identified as uncertain, and three isolates were identified as non-Hib. All non-Hib isolates (n = 77) were identified as non-Hib. When analyzed using the ultrafleXtreme instrument, only 9 of 19 Hib isolates were assigned to the same category as by the microflex instrument. However, all non-Hib isolates were correctly identified, and the reproducibility was 76.2%.
When two spectra per isolate were used, 97.0% (32 of 33) of the Hib isolates and 98.7% (76 of 77) of the non-Hib isolates were correctly identified. When the measurements were repeated with the other instrument, 5 of 19 Hib isolates and 1 of 23 non-Hib isolates were assigned to a different category. Thus, 6 isolates changed categories, and the reproducibility was 85.7%.
DISCUSSION
In the present study, we show that MALDI-TOF MS can be used to differentiate Hib from non-b H. influenzae. All classification algorithms that were generated and tested identified Hib among the H. influenzae isolates, with variations in sensitivity and specificity. The ClinProTools model had the best and most robust results, performing similarly to or better than models based on this software in previous studies (23–25).
A major strength in our work is the generation, testing, and comparison of several different classification algorithms to test our hypothesis. The careful selection of genetically different reference isolates is also important, especially since there have been reports on increased genetic diversity within the ST6 cluster since vaccination against Hib was introduced (26). Another strength of this study is the large selection of well-characterized isolates used to validate the classification algorithms. The isolates were collected from several geographical areas during a wide time range to ensure that the algorithms are applicable not just in a Scandinavian setting. The robustness of the algorithms was validated by repeated extraction and spectral acquisition on another MALDI-TOF MS instrument in a different laboratory.
One limitation of our study is that it has not been possible to fully evaluate the capacity of the different algorithms to identify Hib from the less common genetic cluster (represented by KR194 among our reference isolates). None of our test isolates (neither Hib nor non-Hib) were fully similar to KR194 in its capsule type-defining peaks during manual analysis, indicating that this genetic cluster most likely was not represented among our test isolates. This is not surprising, since isolates from this cluster are uncommon both in Sweden and worldwide (22). A search in the MLST database (www.mlst.net) reveals that isolates from this cluster are found mostly in Russia but also in a few other countries. In these countries, they still represent only a very small portion of Hib isolates. KR194 was, however, separable from the non-Hib isolates in manual analysis, suggesting that identification of this cluster with MALDI-TOF MS should be possible. It is likely that our methods do not identify the type b capsule itself but rather identify proteins common to the Hib population due to the clonal relationship of the isolates. It is therefore important to keep this in mind and perhaps regularly evaluate the sensitivity of MALDI-TOF MS as a capsule typing method.
Two of the classification algorithms, the ClinProTools model and the Biotyper subtyping MSPs, showed better results. These algorithms reliably separated Hib from non-Hib isolates, and the separation could be fully or almost fully reproduced on another instrument in another laboratory. Reproducibility of a classification algorithm is important, since it increases the possibility of using MALDI-TOF MS in clinical routine diagnostics. The use of different instrument models (i.e., microflex and ultrafleXtreme) should not affect the test results. Each MALDI-TOF MS instrument should, however, be individually calibrated for optimal outcome. The Biotyper subtyping MSPs had lower sensitivity than the ClinProTools model when 6 spectra per isolates were used for testing. Our requirements for Hib identification (≥5 of 6 spectra classified as Hib) were high, considering that the Hib isolate that was not correctly identified in the original measurement had four spectra classified as Hib, while no isolate in the non-Hib group had any spectra classified as Hib using our method. It was therefore not surprising that the sensitivity for the Biotyper subtyping MSPs improved when using only two spectra per test isolate, while the specificity remained high.
Regular Biotyper MSPs performed somewhat less well in separating Hib and non-Hib isolates, even though it cannot be ruled out that the results could be improved with optimized instrument settings. The slightly poorer performance, however, was not unexpected, as regular MSPs take all peaks of an isolate into account when generating a score. Since this study investigated subtyping within one species, the spectral patterns as a whole were much alike, increasing the risk of misidentification using regular MSPs. The advantage of the ClinProTools model and the Biotyper subtyping MSPs is that spectral differences, rather than similarities, are highlighted. This might also explain the slightly lower reproducibility of the regular Biotyper MSPs. The creation of specially designed MSPs with only two peaks was performed to test if these peaks were sufficient for Hib/non-Hib differentiation. Regarding reproducibility, this classification method was not, however, as robust as the ClinProTools model and subtyping MSPs, indicating that it is more sensitive to differences in spectral quality and between different MALDI-TOF MS instruments.
One isolate (NTHi KR183) was incorrectly classified as Hib using ClinProTools. The known genetic variation within the NTHi population makes the design of a set model with full specificity for NTHi isolates challenging. However, if these novel methods are used in a clinical setting, it would likely be used as a screening for Hib among H. influenzae isolates. In this case, sensitivity, which was very good in both of our best methods, is of higher priority than specificity. Hib isolates identified in a MALDI-TOF MS screening should be confirmed by capsule PCR.
Several studies have illustrated the value of MALDI-TOF MS in difficult microbiological diagnostics and bacterial subtyping. Bruin et al. (27) showed that MALDI-TOF MS effectively differentiated H. influenzae from the commensal Haemophilus haemolyticus, which has been difficult with traditional microbiological techniques. A few recent studies have also been published that use ClinProTools software for bacterial classification, for example, for the differentiation of Shigella species from E. coli and Streptococcus pneumoniae from Streptococcus mitis (23, 24). These studies also used ethanol-formic acid extraction. The direct transfer method and extended direct transfer method were used on a sample of test isolates in this study to test our classification methods. However, the results were less accurate than when ethanol-formic acid extraction was used. It is likely that small spectral differences and low-intensity peaks, although constant, are not as clear when using the simpler preparation methods before mass spectrometry measurement. This is not surprising, since it is well known that the quality and reproducibility of weaker peaks are better when the extraction method is used, and the method results in improved scores with the MALDI Biotyper classification software (28).
In conclusion, the present study shows that rapid identification of Hib among H. influenzae is possible using a routine sample preparation and MALDI-TOF MS analysis of H. influenzae isolates. After methodological optimization, this novel finding may be implemented in clinical microbiology laboratories. H. influenzae isolates identified by standard MALDI-TOF MS analysis could then be directly reanalyzed using the ethanol-formic acid extraction procedure to identify Hib isolates. This would improve the efficiency of Hib identification and make rapid intervention possible in cases of vaccine failure or poor vaccination coverage. MALDI-TOF MS could also be used as a surveillance tool in vaccinated populations to monitor sustained vaccine efficacy and as an initial screening method of invasive H. influenzae isolates.
ACKNOWLEDGMENTS
We thank Åsa Johansson, Håkan Janson, and Gunnar Kahlmeter from the Department of Clinical Microbiology, Växjö, for fruitful discussions in the initial process, and Søren Lehmann, Bruker Daltonics, for technical support.
This work was supported by grants from the Alfred Österlund, Anna and Edwin Berger, and Greta and Johan Kock foundations, O. E. och Edla Johanssons, the Swedish Medical Research Council (grant K2015-57X-03163-43-4 [www.vr.se]), the Physiographical Society (Forssman's Foundation), and Skåne County Council's research and development foundation.
Markus Kostrzewa is an employee of Bruker Daltonics.
REFERENCES
- 1.Peltola H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev 13:302–317. doi: 10.1128/CMR.13.2.302-317.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jalalvand F, Riesbeck K. 2014. Haemophilus influenzae: recent advances in the understanding of molecular pathogenesis and polymicrobial infections. Curr Opin Infect Dis 27:268–274. doi: 10.1097/QCO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 3.Ishiwada N, Hishiki H, Nagasawa K, Naito S, Sato Y, Chang B, Sasaki Y, Kimura K, Ohnishi M, Shibayama K. 2014. The incidence of pediatric invasive Haemophilus influenzae and pneumococcal disease in Chiba prefecture, Japan before and after the introduction of conjugate vaccines. Vaccine 32:5425–5431. doi: 10.1016/j.vaccine.2014.07.100. [DOI] [PubMed] [Google Scholar]
- 4.Bajanca-Lavado MP, Simões AS, Betencourt CR, Sa-Leão R, Portuguese Group for Study for Haemophilus influenzae Invasive Infection. 2014. Characteristics of Haemophilus influenzae invasive isolates from Portugal following routine childhood vaccination against H. influenzae serotype b (2002–2010). Eur J Clin Microbiol Infect Dis 33:603–610. doi: 10.1007/s10096-013-1994-6. [DOI] [PubMed] [Google Scholar]
- 5.Resman F, Ristovski M, Ahl J, Forsgren A, Gilsdorf JR, Jasir A, Kaijser B, Kronvall G, Riesbeck K. 2011. Invasive disease caused by Haemophilus influenzae in Sweden 1997–2009; evidence of increasing incidence and clinical burden of non-type b strains. Clin Microbiol Infect 17:1638–1645. doi: 10.1111/j.1469-0691.2010.03417.x. [DOI] [PubMed] [Google Scholar]
- 6.Schouls L, van der Heide H, Witteveen S, Zomer B, van der Ende A, Burger M, Schot C. 2008. Two variants among Haemophilus influenzae serotype b strains with distinct bcs4, hcsA and hcsB genes display differences in expression of the polysaccharide capsule. BMC Microbiol 8:35. doi: 10.1186/1471-2180-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lepp T, Uhnoo I, Schölin A, Szirmai M, Lindstrand A, Tegnell A. 2014. The childhood immunization program in Sweden 2013, annual report. Public Health Agency of Sweden, Solna, Sweden: http://www.skane.se/Upload/Webbplatser/Smittskydd/Dokument/vaccinationst%C3%A4ckning/A%CC%8Arsrapport_barnvaccinationsprogrammet%202013%5B2%5D.pdf. [Google Scholar]
- 8.Watt JP, Wolfson LJ, O'Brien KL, Henkle E, Deloria-Knoll M, McCall N, Lee E, Levine OS, Hajjeh R, Mulholland K, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team. 2009. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet 374:903–911. doi: 10.1016/S0140-6736(09)61203-4. [DOI] [PubMed] [Google Scholar]
- 9.Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. 2011. Use of bexB to detect the capsule locus in Haemophilus influenzae. J Clin Microbiol 49:2594–2601. doi: 10.1128/JCM.02509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. 1994. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 32:2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam TT, Elias J, Frosch M, Vogel U, Claus H. 2011. New diagnostic PCR for Haemophilus influenzae serotype e based on the cap locus of strain ATCC 8142. Int J Med Microbiol 301:176–179. doi: 10.1016/j.ijmm.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Satola SW, Collins JT, Napier R, Farley MM. 2007. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol 45:3230–3238. doi: 10.1128/JCM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musser JM, Kroll JS, Moxon ER, Selander RK. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci U S A 85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musser JM, Kroll JS, Moxon ER, Selander RK. 1988. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun 56:1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erwin AL, Sandstedt SA, Bonthuis PJ, Geelhood JL, Nelson KL, Unrath WC, Diggle MA, Theodore MJ, Pleatman CR, Mothershed EA, Sacchi CT, Mayer LW, Gilsdorf JR, Smith AL. 2008. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol 190:1473–1483. doi: 10.1128/JB.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 17.Emonet S, Shah HN, Cherkaoui A, Schrenzel J. 2010. Application and use of various mass spectrometry methods in clinical microbiology. Clin Microbiol Infect 16:1604–1613. doi: 10.1111/j.1469-0691.2010.03368.x. [DOI] [PubMed] [Google Scholar]
- 18.Demirev PA, Hagan NS, Antoine MD, Lin JS, Feldman AB. 2013. Establishing drug resistance in microorganisms by mass spectrometry. J Am Soc Mass Spectrom 24:1194–1201. doi: 10.1007/s13361-013-0609-x. [DOI] [PubMed] [Google Scholar]
- 19.Lange C, Schubert S, Jung J, Kostrzewa M, Sparbier K. 2014. Quantitative matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid resistance detection. J Clin Microbiol 52:4155–4162. doi: 10.1128/jcm.01872-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christner M, Trusch M, Rohde H, Kwiatkowski M, Schluter H, Wolters M, Aepfelbacher M, Hentschke M. 2014. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS One 9:e101924. doi: 10.1371/journal.pone.0101924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogne C, Prod'hom G, Jaton K, Decosterd L, Greub G. 2014. A simple, robust and rapid approach to detect carbapenemases in Gram-negative isolates by MALDI-TOF mass spectrometry: validation with triple quadripole tandem mass spectrometry, microarray and PCR. Clin Microbiol Infect 20:O1106–O1112. doi: 10.1111/1469-0691.12715. [DOI] [PubMed] [Google Scholar]
- 22.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khot PD, Fisher MA. 2013. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:3711–3716. doi: 10.1128/JCM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikryannikova LN, Filimonova AV, Malakhova MV, Savinova T, Filimonova O, Ilina EN, Dubovickaya VA, Sidorenko SV, Govorun VM. 2013. Discrimination between Streptococcus pneumoniae and Streptococcus mitis based on sorting of their MALDI mass spectra. Clin Microbiol Infect 19:1066–1071. doi: 10.1111/1469-0691.12113. [DOI] [PubMed] [Google Scholar]
- 25.Xiao D, Zhao F, Zhang H, Meng F, Zhang J. 2014. Novel strategy for typing Mycoplasma pneumoniae isolates by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry coupled with ClinProTools. J Clin Microbiol 52:3038–3043. doi: 10.1128/JCM.01265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schouls LM, van der Ende A, van de Pol I, Schot C, Spanjaard L, VauteriVauterin P, Wilderbeek D, Witteveen S. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J Clin Microbiol 43:2741–2749. doi: 10.1128/JCM.43.6.2741-2749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruin JP, Kostrzewa M, van der Ende A, Badoux P, Jansen R, Boers SA, Diederen BM. 2014. Identification of Haemophilus influenzae and Haemophilus haemolyticus by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis 33:279–284. doi: 10.1007/s10096-013-1958-x. [DOI] [PubMed] [Google Scholar]
- 28.Khot PD, Couturier MR, Wilson A, Croft A, Fisher MA. 2012. Optimization of matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis for bacterial identification. J Clin Microbiol 50:3845–3852. doi: 10.1128/JCM.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]