Abstract
Legionella, a large group of environmental Gram-negative bacteria, represents an occasional cause of pneumonia. We analyzed the microbiological and clinical features of 33 consecutive cases of Legionella infections that occurred at the University of Texas MD Anderson Cancer Center, Houston, TX, from 2002 to 2014. The Legionella strains were isolated from bronchoscopy specimens (32 strains) and a blood culture (1 strain) and were identified by sequencing analysis of the full-length 16S rRNA gene. The 33 strains involved 12 Legionella species or subspecies: 15 strains of L. pneumophila subsp. pneumophila, 3 strains of L. pneumophila subsp. fraseri or L. pneumophila subsp. pascullei, 4 strains of “L. donaldsonii,” 3 strains of L. micdadei, and one each of L. bozemanae, L. feeleii, L. gormanii, L. longbeachae, L. maceachernii, L. parisiensis, L. sainthelensi, and Legionella sp. strain D5382. All patients except one asymptomatic carrier showed pneumonia, including one with concurrent bacteremia. Nine patients died, with this infection being the immediate cause of death in six. Twenty-seven patients had underlying hematologic malignancies. Twenty-three patients were leukopenic. Six patients were recipients of allogeneic hematopoietic stem cell transplant, with their infections caused by five Legionella species. Together, these results suggest that diverse Legionella species infect patients with cancer in the Houston area and its vicinity. The five cases of pneumonia due to L. donaldsonii and Legionella sp. D5382 are likely the first reports of human infection with these organisms.
INTRODUCTION
The genus Legionella was proposed in 1979 as the causative agent of Legionnaires' disease, a form of severe pneumonia (1, 2). Currently, this genus contains 58 species and three subspecies (www.bacterio.net). Legionella organisms are environmental Gram-negative bacteria that can be found in bodies of water and soil (3, 4). They are intracellular parasites of freshwater protozoa. They are aerobic, nonfermentative, asaccharolytic, and fastidious and require l-cysteine and iron salt for growth and isolation on solid culture medium. Of the 61 species and subspecies comprising >70 serogroups, ≥20 have been implicated as causative agents of pneumonia. L. pneumophila is the most common species of Legionella pneumonia; it includes ≥16 serogroups, of which serogroups 1, 4, and 6 are most common (5). Known human pathogens also include Legionella longbeachae, Legionella micdadei, Legionella bozemanae, Legionella dumoffii, and others.
The clinical spectrum of Legionella infection ranges from mild Pontiac fever to severe pneumonia. Rare extrapulmonary manifestations of infection have been reported, such as necrotizing cellulitis, abscesses, and endocarditis (6, 7). The principal known risk factor for Legionella pneumonia is the suppression of cellular immunity (5, 8). Thus, the patients at greatest risk include those being treated with corticosteroids, immunosuppressive medications after organ transplantation, and antineoplastic chemotherapy. The nonspecific presentation of Legionella pneumonia necessitates empirical therapy for patients diagnosed with community-acquired pneumonia, particularly those presenting with respiratory failure or shock (9).
The laboratory tests available to diagnose Legionella infection include culture of lower respiratory secretions, tissues, or body fluids, direct fluorescent staining of specimens with polyvalent antibodies and microscopy, immunoassays for Legionella pneumophila serogroup 1 antigen in the urine, and molecular amplification. The direct fluorescent antibody (DFA) staining of clinical specimens lacks sensitivity (10), and its sensitivity and specificity are not precisely known for species aside from L. pneumophila (7). Culture may be the only valid option for non-pneumophila Legionella species. Thus, most clinical investigations on Legionella have focused on the role of L. pneumophila. The fastidious growth and limited yet overlapping biochemical reactions also make the accurate identification of various Legionella species difficult in most hospital settings, where the majority of infection occurs.
Analysis of several genes, singly or in combination, has been shown to accurately differentiate Legionella species (11, 12). For instance, sequencing of the 16S rRNA gene, in particular the full-length gene, resolves most Legionella species with considerable confidence. The universal nature of the 16S gene also means there is a broad application for bacterial identification. In this study, we used the 16S gene sequencing method to identify 33 strains of Legionella species that were isolated consecutively from patients with cancer. We also paid attention to the clinical and pathogenic features of those non-pneumophila Legionella species.
MATERIALS AND METHODS
Study setting and location.
The Legionella strains were consecutive (sporadic) isolates collected from 2002 to December 2014 at the University of Texas MD Anderson Cancer Center, Houston, TX. The institution is a comprehensive cancer center that had 500 beds in 2002, which increased to 620 beds in 2014. The majority of patients were from the greater Houston area and its vicinity (other parts of Texas and nearby Louisiana). In view of the environmental origin of Legionella, the climate in Houston includes abundant rainfall (around 140 cm each year) and warm temperatures (monthly means, 12.4°C in the coldest January to 29.1°C in the warmest July and August). The greater Houston area is known for abundant bodies of water (lakes, rivers, bayous, and the Gulf of Mexico coast).
Microbiological tests and cultures.
Lower respiratory specimens, mostly bronchoalveolar lavage (BAL) fluid and bronchial washings that were obtained through bronchoscopy, were stained by a direct fluorescent antibody assay for Legionella species (Remel, Lenexa, KS). The assay is designed to detect 22 Legionella species with 33 serogroups, including serogroups 1 to 10 of L. pneumophila, L. bozemanae, L. dumoffii, L. feeleii, L. gormanii, L. longbeachae, L. maceachernii, L. micdadei, L. parisiensis, L. sainthelensi, and others.
The respiratory specimens were also set up to culture Legionella (3). They were diluted approximately 5 times with low-tone acidic KCl solution (pH 2.2) and then plated. Four buffered charcoal yeast extract (BCYE) agar plates, two selective and two nonselective (BBL; BD Diagnostic Systems, Sparks, MD), were inoculated and incubated at 35°C for 7 days. Colonies of Gram-negative slender bacilli were subcultured onto sheep blood agar plates, and those with little or no growth on this agar represented potential Legionella species. Starting in 2011, BCYE plates with or without cysteine were also used for subcultures, which indicated Legionella growth with cysteine and no growth without cysteine. Potential Legionella isolates, particularly those with light blue- or pink-tinted colonies on BCYE agar, were further identified.
Blood cultures were performed using the Bactec 9240 automated culturing system (BD Diagnostic Systems) and manual Isolator tubes (Wampole Laboratories, Princeton, NJ). A blood culture set typically consisted of a 20-ml blood draw from a patient, which was divided equally and inoculated into a Bactec Plus Aerobic/F bottle and an Isolator tube. The Bactec bottles were incubated for 7 days at 35°C, with aeration. The Isolator tubes were processed by centrifugation, and the resulting blood sediments were spread on two chocolate agar plates and two sheep blood agar plates for incubation at 35°C with 5% carbon dioxide for 4 days. The Isolator plates allowed quantitation of bacterial colonies from the 10-ml blood draw. Approximately 30,000 sets of blood cultures were performed annually in the institution.
Definitive identification of Legionella species.
Definite species identification was achieved by sequencing the 16S rRNA gene, as previously described (13). Briefly, for DNA extraction, the isolated colonies were suspended in 100 μl of extraction buffer (PrepMan Ultra; Applied Biosystems, Inc., Carlsbad, CA), boiled for 10 min at 105°C, and centrifuged (8,000 × g for 10 min). One microliter of the supernatant was used as a template in a 25-μl PCR, including activation of the enzyme (at 92°C for 2 min), 35 cycles of denaturation (at 95°C for 20 s), annealing (at 62°C for 20 s), and extension (at 72°C for 40 s), and a final extension for 5 min. At the time of isolation of each strain, a 594-bp PCR amplicon in the mid-region of the 16S gene using two universal bacterial primers (5′-TGCCAGCAGCCGCGGTAATAC [forward, UBFO] and 5′-CGCTCGTTGCGGGACTTAACC [reverse, UBRE]) was sequenced. During this study from 2009 to 2014, each sequence was extended to near full length (∼1,460 bp) of the gene for more accurate species assignment. Two additional sets of primers were used for sequence extension, including 5′-GCGTGCTTAACACATGCAAGTC (forward, AFBFO) and 5′-GCACAAGCGGTGGAGCATGTG (reverse, 16R3), and 5′-GGTGCAAGCGTTAATCGGA (forward, 16F4) and 5′-AGGAGGTGATCCAACCGCA (reverse, 16R). Sequence matches were made through the GenBank Basic Local Alignment Search Tool (BLAST), and the best match, usually at 99.4% to 100% with a type or reference strain, yielded species identification.
Clinical correlation and GenBank deposition.
Clinical data were extracted from the medical records during isolation of a strain and/or later review and included demographics, underlying disease, presentation of symptoms and signs, physical and laboratory examination findings, radiographs, treatments, clinical course, and outcome. At least two physician authors assessed the clinical data for consistency. In view of the severity of these infections, nearly all patients also had chest computed tomography (CT) scans in addition to plain chest X rays. The review of the records was approved by the University of Texas MD Anderson Cancer Center institutional review board.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of an L. donaldsonii strain and an L. donaldsonii-like strain in this study were deposited in the GenBank under accession numbers KM504126 and KM504127.
RESULTS
General clinical features.
The clinical characteristics of the 33 patients are detailed in Table 1. These patients included 19 men and 14 women, with a mean age of 60 years (range, 28 to 74 years). Twenty-seven patients had underlying hematologic malignancies, while six patients had solid tumors. Nine patients were also hematopoietic stem cell transplant (HSCT) recipients; six HSCTs were allogeneic, and three were autologous. Twenty-three of the 33 patients were leukopenic, with total white blood cell (nonleukemic) counts ranging from 0 to 3.8 × 109 cells/liter; the absolute neutrophil count ranged from 0 to 1.66 ×109 cells/liter. Thirty-one patients acquired infection in the greater Houston area and its vicinity, and two patients likely acquired infection farther away, with case 4 acquired in Puerto Rico and case 16 acquired in Florida, due to their short arrival time on presentation.
TABLE 1.
Microbiological and clinical features of 33 cancer patients with legionellosis
| Case no., age (yr)/sex; culture sourcea | Cancer type, HSCT status; baseline WBC, baseline ANC (× 109 cells/liter)b | Presentation | Best match % (no./total no.) to species/strain, GenBank accession no.; serogroup | Treatment; outcome, notec |
|---|---|---|---|---|
| 15 cases with L. pneumophila subsp. pneumophila | ||||
| 24, 51/F; BAL fluid | Lung; 3.8, 2,4 | Pneumonia, resp. failured | 100 (1,475/1,475) to strains Paris, Lorraine, and Phily-1; 1e | FEP, AZM, VAN; death @ 5 days |
| 6, 64/F; BAL fluid | AML; 3.1, 0.26 | Pneumonia, neutropenic fever | 99.6 (1,401/1,406) to strains Paris, Lorraine, and Phily-1; 1 | LVX, MXF, AZM; recovered |
| 28, 75/M; BAL fluid | AML, allo-HSCT; 7.0, 6.5 | Pneumonia, resp. failure | 100 (1,478/1,478) to strains Paris, Lorraine, and Phily-1; 1 | MEM, MXF, VAN; recovered, urine antigen (+) |
| 32, 74/F; BAL fluid | Myeloma, auto-HSCT; 5.0, 3.0 | Pneumonia | 100 (1,443/1,443) to strains Paris, Lorraine, and Phily-1; 1 | CIP, CRO, AZM; recovered, urine antigen (−) |
| 15, 55/F; BAL fluid | AML; 0.1, 0 | Pneumonia, neutropenic fever | 100 (1,451/1,451) to strains Paris, Lorraine, and Phily-1; 6 | TZB, LZD, CIP; recovered |
| 16, 66/M; BAL fluid | Thyroid; 7.3, 4.42 | Pneumonia, resp. failure | 99.8 (1,449/1,451) to strains Paris, Lorraine, and Phily-1; 6 | AZM, CIP; recovered, infected in Florida |
| 4, 60/F; BAL fluid | Breast; 0.8, 0.4 | Pneumonia, resp. failure, neutropenic fever | 100 (1,444/1,444) to strains Paris, Lorraine, and Phily-1; not serogrouped | VAN, MEM, AZM, SXT; death @ 18 days, infected in Puerto Rico |
| 21, 61/M; BAL fluid | Large-cell lymphoma; 0.1, 0 | Pneumonia, resp. failure, shock, neutropenic fever | 100 (1,450/1,450) to strains Paris, Lorraine, and Phily-1; 3 | MEM, VAN, LVX; recovered, direct DFA of BAL fluid (+) |
| 14, 56/M; BAL fluid | Acute promyelocytic leukemia; 34.7-leukemic, 0 | Pneumonia, resp. failure, neutropenic fever | 99.9 (1,452/1,454) to strain Corby, CP000675; 1 | CIP, MEM, VAN; death @ 16 days |
| 29, 58/M, BAL fluid | Multiple myeloma; 4.1, 2.4 | Pneumonia | 100 (1,477/1,477) to strain Corby, CP000675; 1 | FEP, LZD, MXF; recovered |
| 22, 60/M; BAL fluid | CLL; 2.7, 1.56 | Pneumonia | 100 (1,434/1,434) to strain Corby, CP000675; 5 | VAN, FEP, LVX; recovered |
| 17, 56/F; BAL fluid | Myeloma, auto-HSCT; 3.1, 1.66 | Pneumonia, neutropenic fever | 100 (1,451/1,451) to strain Lens, CR628337; 1 | TZB, AZM, LVX; recovered |
| 19, 69/M; BAL fluid | AML; 51.4-leukemic, 0 | Pneumonia | 100 (1,443/1,443) to strain Lens, CR628337; 1 | FEP, TGC, MXF; recovered |
| 30, 68/M; BAL fluid | AML; 0, 0 | Pneumonia, resp. failure, neutropenic fever | 99.9 (1,458/1,459) to strain Lens, CR628337; 1 | FEP, LZD, IPM, CIP; death @ 30 days |
| 10, 55/F; BAL fluid | AML, allo-HSCT; 0.4, 0.28 | Pneumonia, neutropenic fever | 99.6 (1,403/1,408) to strain Alcoy, CP001828; 1 | FEP, LZD; recovered |
| 3 cases with L. pneumophila subsp. fraseri or subsp. pascullei | ||||
| 18, 56/M; bronchial washings | Esophageal; 3.6, 2.63 | No symptoms | 99.9 (1,443/1,445) to HQ287902; 6 | LVX after culture; no infection |
| 5, 74/M; blood | Renal cell; 8.3, 4.92 | Pneumonia-bacteremia, resp. failure, coagulopathy | 100 (1,441/1,441) to HQ287902; not serogrouped | FEP, CIP, VAN; recovered |
| 7, 54/F; BAL fluid | AML; 1.8, 1.10 | Pneumonia, neutropenic fever | 99.9 (1,436/1,438) to HQ287902; not serogrouped | LVX; recovered |
| 4 cases with L. donaldsonii | ||||
| 1, 35/M; BAL fluid | CML, allo-HSCT; 2.5, 1.13 | Pneumonia, neutropenic fever | 99.6 (1,403/1,409) to Z49724 | FEP, AZM, MEM; recovered |
| 12, 43/F; BAL fluid | ALL; 0, 0 | Pneumonia, neutropenic fever | 99.6 (1,403/1,409) to Z49724 | CIP, AZM, SXT; recovered |
| 33, 72/F; BAL fluid | CLL; 21.6-leukemic, 0.65 | Pneumonia, neutropenic fever | 99.6 (1,403/1,409) to Z49724 | MEM, LZD, CIP; recovered |
| 26, 66/M; BAL fluid | CLL; 15.8-leukemic, 0.4 | Pneumonia, neutropenic fever | 98.6 (1,389/1,409) to Z49724 | FEP, CIP, LZD; recovered |
| 3 cases with L. micdadei | ||||
| 9, 54/M; BAL fluid | ALL, allo-HSCT; 8.8, 6.59 | Pneumonia | 100 (1,405/1,405) to AF227162 | CIP, LZD; recovered |
| 23, 57/M; BAL fluid | Large-cell lymphoma; 2.9, 0.7 | Pneumonia, neutropenic fever | 100 (1,443/1,443) to AF227162 | FEP, MEM, VAN; recovered |
| 25, 71/F; BAL fluid | CLL; 11.8-leukemic, 0.8 | Pneumonia-severe, neutropenic fever, coagulopathy | 100 (1,442/1,442) to AF227162 | FEP, MEM, VAN; recovered |
| 8 cases with other Legionella species | ||||
| 3, 65/F; BAL fluid | Myeloma, auto-HSCT; 2.2, 1.15 | Pneumonia, resp. failure, neutropenic fever | 99.4 (1,387/1,396) to L. bozemanae, Z49718 | SXT, DOX; death @ 23 days |
| 11, 59/M; BAL fluid | Lung; 10.5, 8.67 | Pneumonia (postobstructive, severe) | 99.7 (1,404/1,408) to L. feeleii, X73406 | LVX; hospice, death |
| 8, 65/M; BAL fluid | ALL; 0.4, 0.22 | Pneumonia (severe), neutropenic fever | 99.4 (1,415/1,424) to L. gormanii, Z32639 | AMK, MEM, VAN, AZM; death @ 14 days |
| 31, 59/M; BAL fluid | AML; leukemic, 3.0 | Pneumonia | 100 (1,455/1,455) to L. longbeachae, FN650140 | FEP, TGC, CIP; hospice, death |
| 27, 81/F; BAL fluid | Myelodysplastic syndrome; 2.0, 0.6 | Pneumonia (low grade) | 99.9 (1,442/1,443) to L. maceachernii, NR_041790 | FEP, LZD; death @ 4 days, unrelated |
| 20, 50/M; BAL fluid | CML, allo-HSCT, GVHD; 4.9, 4.12 | Pneumonia | 99.5 (1,396/1,403) to L. parisiensis, Z49731 | LVX, DOX; recovered |
| 2, 28/M; BAL fluid | Acute leukemia, allo-HSCT, GVHD; 5.5, 4.4 | Pneumonia | 99.5 (1,445/1,452) to L. sainthelensi, X73399 | CRO, AZM, GAT; recovered |
| 13, 38/F; BAL fluid | CLL; 3.1, 0.42 | Pneumonia, neutropenic fever | 99.9 (1,360/1,361) to Legionella sp. strain D5382, JN380990 | LVX, DOX; recovered |
F, female; BAL, bronchoalveolar lavage; M, male.
HSCT, hematopoietic stem cell transplant; WBC, white blood cell count; ANC, absolute neutrophil count; AML, acute myelogenous leukemia; allo-HSCT, allogeneic HSCT; auto-HSCT, autologous HSCT; CML, chronic myelogenous leukemia; ALL, acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; GVHD, graft-versus-host disease.
FEP, cefepime; AZM, azithromycin; VAN, vancomycin; LVX, levofloxacin; MXF, moxifloxacin; MEM, meropenem; (+), positive; CIP, ciprofloxacin; CRO, ceftriaxone; (−), negative; TZP, piperacillin-tazobactam; LZD, linezolid; SXT, trimethoprim-sulfamethoxazole; DFA, direct fluorescent assay; TGC, tigecycline; DOX, doxycycline; GAT, gatifloxacin.
resp., respiratory.
Phily-1, strain Philadelphia-1.
All patients except one, who was found to be an asymptomatic carrier, had clinical signs, symptoms, and radiographic images that were consistent with pneumonia. Twenty-four of the 33 patients recovered with therapy, while nine patients died, with the Legionella pneumonia likely the immediate cause of death in six of them. Nine patients had evidence of respiratory failure that required mechanical ventilation on presentation or shortly after hospitalization. The antibiotics most commonly given were the broad class of fluoroquinolones (24 cases), consistent with current recommendations (9), with or without addition of azithromycin, doxycycline, cephalosporins, and/or carbapenems as part of empirical coverage for neutropenic pneumonia.
Culture and microbiological features.
Thirty-two of the 33 Legionella strains were isolated from respiratory specimens obtained through bronchoscopy, while one was isolated from a blood culture. With near-full-length 16S gene sequencing (∼1,460 bp), all strains except one showed 99.4% to 100% match with at least one reference sequence or species in GenBank to allow a confident species assignment. The match details are also shown in Table 1.
The most common species was L. pneumophila subsp. pneumophila, with 15 strains. These strains matched with many reference strains (genomes) that showed minor differences of one to two nucleotides in their 16S gene sequences. Fourteen of these 15 strains were serogrouped, with 10 being serogroup 1, 2 being serogroup 6, and one each from serogroups 3 and 5. Thus, serogroup 1 accounted for 10 of the total 32 typed strains in this study. The two patients who likely acquired their infection far from Houston had this subspecies (one serogroup 6 and one not typed).
There were 3 strains of L. pneumophila subsp. fraseri or subsp. pascullei. These two subspecies had identical 16S gene sequences and were not further differentiated. However, they differed from L. pneumophila subsp. pneumophila by 12 nucleotides in the 1,500-bp 16S gene to allow confident separation. One of these three strains was typed as being serogroup 6.
The other 15 strains of Legionella species included 4 strains of L. donaldsonii, 3 strains of L. micdadei, and one each of L. bozemanae, L. feeleii, L. gormanii, L. longbeachae, L. maceachernii, L. parisiensis, L. sainthelensi, and Legionella sp. D5382. It is notable that the strains isolated from six patients with allogeneic HSCT included two L. pneumophila subsp. pneumophila serogroup 1 and one each of L. donaldsonii, L. micdadei, L. parisiensis, and L. sainthelensi.
During the study years, direct fluorescent antibody staining with microscopy was also applied to all bronchoscopy-derived specimens; positive specimens rarely matched the culture results (data not shown), with only one specimen being positive with both methods. This strain, L. pneumophila subsp. pneumophila serogroup 3, was isolated from a 61-year-old man with lymphoma who had no countable blood neutrophils (Table 1, case 21). Thus, despite polyvalent antibodies of the fluorescent assay that covered all the Legionella species and serogroups in this study except L. donaldsonii, this direct assay failed to detect 28 strains in the specimens. The urine Legionella antigen test was not used consistently; in two patients with infection due to L. pneumophila subsp. pneumophila serogroup 1, one was positive (case 28) and one negative (case 32).
The above results suggest that diverse Legionella species may cause pneumonia in patients with cancer in the Houston area, and culture recovery from bronchoscopy-derived respiratory specimens likely provides the most reliable, sensitive, and specific diagnosis. Selected cases are presented below to illustrate unusual features or the first report of an organism in human infection, along with brief discussions.
Four cases of L. donaldsonii infection.
L. donaldsonii is a tentative name used in the GenBank accession for a clinical isolate (Glasgow 86/35785, without clinical details) (11). The isolation of this organism from the environment was reported recently in Taiwan (14). The four cases in this series (cases 1, 12, 26, and 33) all involved patients with refractory leukemia. They occurred in 2002, 2010, 2014, and 2014, respectively, with acquisition of infection likely occurring in the greater Houston area.
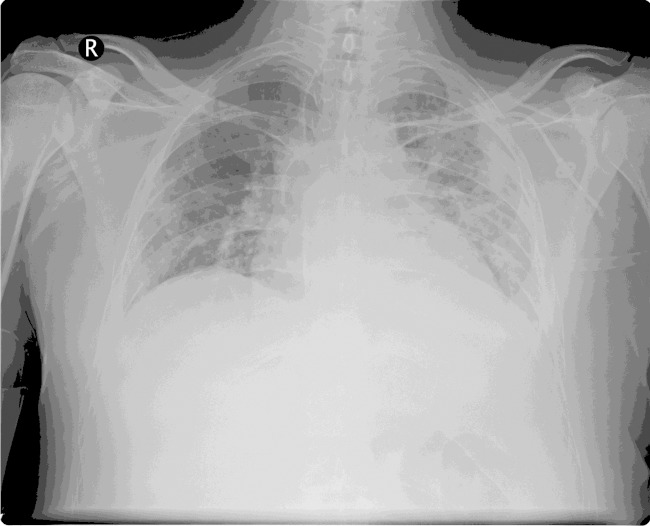
Case 1 (Table 1) was a 35-year-old man who received an allogeneic HSCT for chronic myelogenous leukemia. The patient had suffered from graft-versus-host disease that required methylprednisolone and tacrolimus therapy. Approximately 2 months following HSCT, he was admitted to the hospital, complaining of subjective fevers and shortness of breath. On chest examination, there were inspiratory crackles and dullness to percussion. His labs showed pancytopenia. A chest radiograph showed new bilateral pulmonary opacities with pleural effusions (Fig. 1), and a CT also revealed peripheral lung nodules. A bronchoscopy was performed, and BAL fluid cultures grew L. donaldsonii without other pathogens. The patient was treated with cefepime, azithromycin, and meropenem until recovery during the 46-day hospitalization. Intubation with mechanical ventilation was also required.
FIG 1.
Chest radiograph of L. donaldsonii pneumonia in a 35-year-old man with a hematopoietic stem cell transplant.
Case 12 was a 43-year-old woman being treated for leukemia, with zero blood neutrophils per microliter. She presented to the emergency department with fever (39°C) and tachycardia (150 beats/min). The admission chest radiograph showed pulmonary opacities most consistent with pneumonia. A chest CT performed later also showed worsening nodular opacities in the left upper lobe compared to an earlier CT. The BAL fluid cultures grew L. donaldsonii without other pathogens. The patient was treated empirically and then specifically during the 45-day hospitalization. She recovered.
Case 33 was a 72-year-old woman with a history of chronic lymphocytic leukemia (CLL) for 7 years. While being treated for relapsed leukemia with neutropenia, she presented to the emergency room with fever (39.1°C), mild nonproductive cough, and rhinorrhea. Chest radiograph and CT scan revealed the new development of left lower lobe opacities. A BAL fluid sample was obtained 2 days later, which grew L. donaldsonii. A rhinovirus was also detected in the nasal wash sample. The patient was hospitalized for 6 days and treated with meropenem, ciprofloxacin, and linezolid. She also recovered.
Case 26 involved a 66-year-old man with CLL for 3 years. Within the 2-month period after the initiation of chemotherapy, the patient experienced two episodes of fever and possible pneumonia with admission to a community hospital, but the etiology was not ascertained. This admission began with the presentation of fever (39°C), dry cough, and dyspnea in the emergency room. Chest radiography and CT scan showed left lower lobe consolidation and bilateral multifocal opacities. A bronchoscopy was performed the next day, and the culture grew an L. donaldsonii-like organism without other pathogens. The patient was hospitalized for 7 days and treated with cefepime, ciprofloxacin, and linezolid. He recovered.
Three strains from cases 1 (in 2002), 12 (strain MDA4726 in 2010), and 33 (in 2014) showed almost identical 16S gene sequences (zero to one mismatch of 1,444 bp). They all matched 99.6% with L. donaldsonii (GenBank accession no. Z49724, 1,403 of 1,409 bp) (Table 1). They matched second with L. feeleii (GenBank accession no. X73406, 99.3% [1,439/1,449]). The fourth strain (MDA6655 from case 26 in 2014) matched best with both L. donaldsonii and Legionella tunisiensis at 98.6% (1,389/1,409 and 1,449/1,470, respectively). This strain was thus considered L. donaldsonii-like and potentially novel. Notably, L. tunisiensis is a newly described species in 2012 that was isolated from environmental water with hitherto unknown clinical significance (15). The 16S sequences of strains MDA4726 and MDA6655 showed considerable difference, matching at 98.7% (1,460/1,479). These two sequences were deposited to the GenBank in view of the clinical significance of these organisms, sequence quality, and/or novelty.
Case of Legionella sp. D5382 infection.
Strain D5382 is a Legionella organism without species status, and its 16S gene sequence was directly deposited to GenBank from Australia (GenBank accession no. JN380990, 1,363 bp, R. M. Ratcliff, 2011). No infection by this strain was known prior to this study. The strain recovered in this study matched best (99.9% [1,360/1,361]) with D5382, and the next-level matches showed a difference of nearly 20 nucleotides with several Legionella anisa strains in GenBank (∼98.7%). The patient was a 38-year-old female with CLL (case 13) who presented to the emergency department with fever (39.3°C) and hypotension (87/51 mm Hg). Her admission chest radiograph showed a large left upper lobe consolidation; a CT confirmed the air space opacities in the left upper lobe, with additional bilateral ground glass nodular opacities. On review of prior radiographs, it was noted that subtle evidence of a developing pneumonia likely started 2 months earlier. A BAL fluid culture on the day of admission grew Legionella sp. strain D5382. The patient was treated empirically with ciprofloxacin, piperacillin-tazobactam, and linezolid. She was discharged on levofloxacin, and her condition improved gradually over 7 weeks.
Case of L. parisiensis infection.
L. parisiensis, initially isolated in Paris from a cooling tower in 1985 (16), has been reported to cause pneumonia in two cases, in a 34-year-old French woman who had undergone liver transplant and immunosuppressive therapy (17), and in a German patient with immunosuppression (lack of clinical details) (18). The present case thus represented the third confirmed infection and likely the first in the United States. The patient was a 50-year-old male with chronic myelogenous leukemia post-allogeneic HSCT for 5 months (case 20). He had experienced severe graft-versus-host disease, for which he received tacrolimus and high-dose prednisone. He presented to the emergency department with a 2-day history of fever, productive cough with yellow sputum, left-sided pleuritic chest pain, and dyspnea. On admission, his temperature was 39.4°C, and his blood was pancytopenic. The admission chest imaging showed the new development of multifocal pneumonia, along with nodules involving the left lower lobe and left parahilar region. A BAL fluid sample obtained on hospital day 2 grew L. parisiensis. He was treated with levofloxacin, piperacillin-tazobactam, and vancomycin. His condition improved further after discharge with additional antimicrobial therapy.
Bacteremia with L. pneumophila subsp. fraseri or subsp. pascullei.
Cases of L. pneumophila bacteremia were reported a few times decades ago (19–21), but the bacterial strains were rarely further investigated for additional features due to technical limits and their uncertain significance. Similarly, since the initial descriptions of L. pneumophila subsp. fraseri and subsp. pascullei (22), there have been limited studies of these organisms and clinical reports. Thus, the case of bacteremia caused by L. pneumophila subsp. fraseri or subsp. pascullei in this series may be instructive.
The patient was a 74-year-old male nonsmoker post-chemotherapy for metastatic renal cell carcinoma (case 5). He presented in May 2007 with fever, cough, dyspnea, profound generalized weakness, and episodes of confusion. Bilateral lung crackles were noted on physical examination, along with pulmonary opacities on chest radiography, acute respiratory acidosis on arterial blood gas analysis, and coagulopathy in view of positive tests for D-dimers and prolonged thrombin time. His white blood cell count rose from 8,300 × 106/liter at baseline to 12,900 × 106/liter, with 97% neutrophils. In the emergency room, blood cultures in a Bactec bottle and an Isolator tube were drawn, and he was treated empirically with intravenous cefepime and levofloxacin and admitted for intensive care.
While the Bactec bottle remained negative for 7 days, the Isolator tube culture, upon spread of the blood sediments on agar plates and incubation for ∼90 h, grew 75 colonies of a Gram-negative rod on the two chocolate agar plates but not on the two sheep blood agar plates. The fastidiousness of this organism, along with a slender Gram-negative microscopic morphology, prompted the use of 16S gene sequencing, which led to the identification of L. pneumophila. The pneumonia was refractory to antimicrobial therapy, and hemoptysis also developed. At 11 days after admission, a bronchoscopy was performed, which revealed a diffuse alveolar hemorrhage, but cultures of the BAL fluid specimen were negative. The condition of the patient eventually improved, and he was discharged after 50 days of hospitalization. Further 16S sequencing of the strain later led to the subspecies assignment. The strain did require cysteine to grow.
The blood source, sole growth on chocolate agar, and large number of colonies made this culture unusual for Legionella species. This case led us to include a BCYE agar plate in exchange for a sheep blood agar plate in our Isolator blood cultures starting in September 2009; so far, no Legionella organism has been isolated this way, but this medium has improved the recovery of Methylobacterium radiotolerans, another environmental Gram-negative bacillus (23).
Case of asymptomatic carriage.
Airway colonization or carriage of Legionella species has been rarely reported (24–27). Thus, the present case of an asymptomatic carrier is noteworthy. The patient (case 18) was a 56-year-old male ex-smoker with metastatic esophageal cancer who had undergone chemoradiation therapy. A restaging CT showed patchy pulmonary opacities in the right upper lobe that correlated with the radiation treatment history to suggest radiation effects. Prior to surgery, a bronchoscopy was performed for staging, and the bronchial washings (without BAL in view of the absence of significant lung pathology) grew L. pneumophila subsp. fraseri or subsp. pascullei serogroup 6. The patient had no complaints to suggest a respiratory tract infection, nor did repeat chest imaging reveal significant abnormalities. As a precaution, he was empirically treated with levofloxacin as an outpatient.
DISCUSSION
By using 16S gene sequencing in this study, we determined the Legionella species with confidence. We noted 12 Legionella species or subspecies in 33 cases. These results suggest that diverse Legionella species may cause infection in patients with cancer in the Houston area. We attribute the recovery of diverse species to our inclusive cultures, the timely lavage of the infection site, and the increased vulnerability of our cancer patient population to opportunistic pathogens, particularly those with hematologic malignancies (27 of the 33 patients).
In the general population, most Legionella infections are caused by L. pneumophila; for instance, a multinational study of community-acquired Legionnaires' disease identified 508 culture-confirmed cases, in which L. pneumophila was responsible for 91.5% of the cases, and among these, serogroup 1 accounted for 84.2% (28). In the environment, however, Legionella species are diverse, as revealed in a French study (29). Another large-scale French study compared clinical and environmental Legionella isolates and noted that L. pneumophila serogroup 1 accounted for 28% of the environmental isolates, compared to 95% of the clinical isolates (30). Therefore, the clinical dominance of L. pneumophila, among the large number of Legionella species described so far, is a reflection of the higher pathogenicity of this species.
In contrast, our data showed that the 31 infections that were likely acquired in the greater Houston area and its vicinity involved 13 strains of L. pneumophila subsp. pneumophila and 18 strains of other species or subspecies. In particular, serogroup 1 of L. pneumophila, with or without a subspecies designation, accounted for 10 of the 31 strains, or 32.3%, a frequency similar to the 28% in the French environmental strains (30), as noted above. This dominance of non-pneumophila Legionella species should reflect the presence of these diverse Legionella species in the Houston area environment and the vulnerability of our cancer patient population to these organisms. Despite the lack of data on the environmental distribution of Legionella species, the Houston area is known for warm temperatures, ample rain, many water bodies, and frequent standing water. During the 13 study years, we did notice yearly fluctuation of the isolation of these Legionella strains, and the reason behind this is under investigation. Rainfall is a reported risk factor for legionellosis (31, 32). We previously noted that the clinical recovery of rapidly growing mycobacteria, also of an environmental origin, followed the seasonal rise and fall of rainfall in Houston (33). Thus, future studies of Legionella species in the Houston area environment may be revealing.
The four cases of L. donaldsonii pneumonia and the one due to Legionella sp. D5382 are likely the first reports of human infection with these organisms. All five patients had underlying leukemia with leukopenia and/or neutropenia. Upon antimicrobial treatment, they all recovered, although two patients required long hospitalizations. These infections also hint that L. donaldsonii is likely not rare in the Houston area environment, similar to L. micdadei, a known species that infected three of our patients. L. donaldsonii clusters with L. feeleii (11), and they are relatively distant from other species. Our findings suggest that the species designation L. donaldsonii may be warranted, pending additional phenotypic studies of our strains and other strains.
It has been reported that among recipients of HSCT or solid organ transplant, L. pneumophila, L. micdadei, and L. bozemanae, in descending order, are the most commonly isolated Legionella species (8, 34–36). Our six patients with allogeneic HSCT were infected by five different Legionella species, including L. pneumophila subsp. pneumophila serogroup 1, L. micdadei, L. donaldsonii, L. parisiensis, and L. sainthelensi. This finding thus adds L. donaldsonii, L. parisiensis, and L. sainthelensi to the list of opportunistic pathogens.
ACKNOWLEDGMENTS
This work was supported in part by a University Cancer Foundation grant from the University of Texas MD Anderson Cancer Center and the National Institutes of Health grant P30 CA016672 to our Institutional DNA Analysis Core Facility.
We thank the staff at our DNA Analysis Core Facility and molecular diagnostic laboratory for DNA sequencing, the staff of the microbiology laboratory for cultures, the clinical staff for taking care of the patients, and Mylene Truong for critical review of the radiologic aspects of the cases and article.
We declare no conflicts of interest.
REFERENCES
- 1.McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 2.Brenner DJ, Steigerwalt AG, McDade JE. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med 90:656–658. doi: 10.7326/0003-4819-90-4-656. [DOI] [PubMed] [Google Scholar]
- 3.Edelstein PH. 2011. Legionella, p 770–785. In Versalovic J, Carroll K, Funke G, Jorgensen JH, Landry ML, Warnock D (ed), Manual of clinical microbiology, 10th ed, American Society for Microbiology, Washington, DC. [Google Scholar]
- 4.Fields B, Benson R, Besser R. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelstein PH, Roy CR. 2015. Legionnaires' disease and Pontiac fever, p 2933–2944. In Bennett JE, Dolin R, Blaser MJ (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 8th ed Saunders, Philadelphia, PA. [Google Scholar]
- 6.Stout J, Yu V. 1997. Legionellosis. N Engl J Med 337:682–687. [DOI] [PubMed] [Google Scholar]
- 7.Muder RR, Yu VL. 2002. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis 35:990–998. doi: 10.1086/342884. [DOI] [PubMed] [Google Scholar]
- 8.Chow J, Yu VL. 1998. Legionella: a major opportunistic pathogen in transplant recipients. Semin Respir Infect 13:132–139. [PubMed] [Google Scholar]
- 9.Roig J, Rello J. 2003. Legionnaires' disease: a rational approach to therapy. J Antimicrob Chemother 51:1119–1129. doi: 10.1093/jac/dkg191. [DOI] [PubMed] [Google Scholar]
- 10.She RC, Billetdeaux E, Phansalkar AR, Petti CA. 2007. Limited applicability of direct fluorescent-antibody testing for Bordetella spp. and Legionella spp. specimens for the clinical microbiology laboratory. J Clin Microbiol 45:2212–2214. doi: 10.1128/JCM.00548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hookey JV, Saunders NA, Fry NK, Birtles RJ, Harrison TG. 1996. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol 46:526–531. doi: 10.1099/00207713-46-2-526. [DOI] [Google Scholar]
- 12.Ratcliff RM. 2013. Sequenced-based identification of Legionella, p 57–72. In Buchrieser C, Hubert H (ed), Legionella: methods and protocols, methods in molecular biology, vol 954 Springer Science+Business Media, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13.Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra RR. 2002. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol 118:796–801. doi: 10.1309/HN44-XQYM-JMAQ-2EDL. [DOI] [PubMed] [Google Scholar]
- 14.Huang SW, Hsu BM, Ma PH, Chien KT. 2009. Legionella prevalence in wastewater treatment plants of Taiwan. Water Sci Technol 60:1303–1310. doi: 10.2166/wst.2009.410. [DOI] [PubMed] [Google Scholar]
- 15.Campocasso A, Boughalmi M, Fournous G, Raoult D, La Scola B. 2012. Legionella tunisiensis sp. nov. and Legionella massiliensis sp. nov., isolated from environmental water samples. Int J Syst Evol Microbiol 62:3003–3006. doi: 10.1099/ijs.0.037853-0. [DOI] [PubMed] [Google Scholar]
- 16.Brenner DJ, Steigerwalt AG, Gorman GW, Wilkinson HW, Bibb WF, Hackel M, Tyndall RL, Campbell J, Feeley JC, Thacker WL, Skaliy P, Martin WT, Brake BJ, Fields BS, Maceachern HV, Corcoran LK. 1985. Ten new species of Legionella. Int J Syst Bacteriol 35:50–59. doi: 10.1099/00207713-35-1-50. [DOI] [Google Scholar]
- 17.Lo Presti F, Riffard S, Vandenesch F, Reyrolle M, Ronco E, Ichai P, Etienne J. 1997. The first clinical isolate of Legionella parisiensis, from a liver transplant patient with pneumonia. J Clin Microbiol 35:1706–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igel L, Helig J, Luck P. 2004. Isolation and characterization of a nonfluorescent strain of Legionella parisiensis. J Clin Microbiol 42:2877–2878. doi: 10.1128/JCM.42.6.2877-2878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chester B, Poulos EG, Demaray MJ, Albin E, Prilucik T. 1983. Isolation of Legionella pneumophila serogroup 1 from blood with nonsupplemented blood culture bottles. J Clin Microbiol 17:195–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rihs D, Yu VL, Zurvaleff JJ, Goetz A, Muder RR. 1985. Isolation of Legionella pneumophila from blood with Bactec system: a prospective study yielding positive results. J Clin Microbiol 22:422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin RS, Marrie TJ, Best L, Sumarah RK, Peppard R. 1984. Isolation of Legionella pneumophila from the blood of a patient with Legionnaires' disease. Can Med Assoc J 131:1085–1087. [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner DJ, Steigerwalt AG, Epple P, Bibb WF, McKinney RM, Starnes RW, Colville JM, Selander RK, Edelstein PH, Moss CW. 1988. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J Clin Microbiol 26:1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Tarrand JJ, Han XY. 2015. Microbiologic and clinical features of four cases of catheter-related infection by Methylobacterium radiotolerans. J Clin Microbiol 53:1375–1379. doi: 10.1128/JCM.03416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridge J, Edelstein PH. 1983. Oropharyngeal colonization with Legionella pneumophila. J Clin Microbiol 18:1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukunaga H, Akagi K, Yabuuchi E. 1990. Asymptomatic infection of Legionella pneumophila in four cases with pulmonary diseases. Nihon Saikingaku Zasshi 45:833–840. (In Japanese.) doi: 10.3412/jsb.45.833. [DOI] [PubMed] [Google Scholar]
- 26.Marrie TJ, Bezanson G, Haldane DJ, Burbridge S. 1992. Colonisation of the respiratory tract with Legionella pneumophila for 63 days before the onset of pneumonia. J Infect 24:81–86. doi: 10.1016/0163-4453(92)91094-R. [DOI] [PubMed] [Google Scholar]
- 27.Pedro-Botet ML, Sabrià M, Sopena N, Garc ía, Núñez M, Morera J, Reynaga E. 2002. Environmental legionellosis and oropharyngeal colonization by Legionella in immunosuppressed patients. Infect Control Hosp Epidemiol 23:279–281. doi: 10.1086/502051. [DOI] [PubMed] [Google Scholar]
- 28.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired pneumonia: an international collaborative survey. J Infect Dis 186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 29.Parthuisot N, West NJ, Lebaron P, Baudart J. 2010. High diversity and abundance of Legionella spp. in a pristine river and impact of seasonal and anthropogenic effects. Appl Environ Microbiol 76:8201–8210. doi: 10.1128/AEM.00188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doleans A, Aurell H, Reyrolle M, Lina G, Freney J, Vandenesch F, Etienne J, Jarraud S. 2004. Clinical and environmental distributions of Legionella strains in France are different. J Clin Microbiol 42:458–460. doi: 10.1128/JCM.42.1.458-460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hicks LA, Rose CE Jr, Fields BS, Drees ML, Engel JP, Jenkins PR, Rouse BS, Blythe D, Khalifah AP, Feikin DR, Whitney CG. 2007. Increased rainfall is associated with increased risk for legionellosis. Epidemiol Infect 135:811–817. doi: 10.1017/S0950268806007552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Vidal C, Labori M, Viasus D, Simonetti A, Garcia-Somoza D, Dorca J, Gudiol F, Carratalà J. 2013. Rainfall is a risk factor for sporadic cases of Legionella pneumophila pneumonia. PLoS One 8:e61036. doi: 10.1371/journal.pone.0061036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han XY. 2008. Seasonality of clinical isolation of rapidly growing mycobacteria. Epidemiol Infect 136:1189–1191. doi: 10.1017/S095026880700982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwebke JR, Hackman R, Bowden R. 1990. Pneumonia due to Legionella micdadei in bone marrow transplant recipients. Clin Infect Dis 12:824–828. doi: 10.1093/clinids/12.5.824. [DOI] [PubMed] [Google Scholar]
- 35.Ernst A, Gordon FD, Hayek J, Silvestri RC, Koziel H. 1998. Lung abscess complicating Legionella micdadei pneumonia in an adult liver transplant recipient: case report and review. Transplantation 65:130–134. doi: 10.1097/00007890-199801150-00025. [DOI] [PubMed] [Google Scholar]
- 36.Knirsch CA, Jakob K, Schoonmaker D, Kiehlbauch JA, Wong SJ, Della-Latta P, Whittier S, Layton M, Scully B. 2000. An outbreak of Legionella micdadei pneumonia in transplant patients: evaluation, molecular epidemiology, and control. Am J Med 108:290–295. doi: 10.1016/S0002-9343(99)00459-3. [DOI] [PubMed] [Google Scholar]