Abstract
The Uppsala University Chlamydia trachomatis multilocus sequence type (MLST) database (http://mlstdb.bmc.uu.se) is based on five target regions (non-housekeeping genes) and the ompA gene. Each target has various numbers of alleles—hctB, 89; CT058, 51; CT144, 30; CT172, 38; and pbpB, 35—derived from 13 studies. Our aims were to perform an overall analysis of all C. trachomatis MLST sequence types (STs) in the database, examine STs with global spread, and evaluate the phylogenetic capability by using the five targets. A total of 415 STs were recognized from 2,089 specimens. The addition of 49 ompA gene variants created 459 profiles. ST variation and their geographical distribution were characterized using eBURST and minimum spanning tree analyses. There were 609 samples from men having sex with men (MSM), with 4 predominating STs detected in this group, comprising 63% of MSM cases. Four other STs predominated among 1,383 heterosexual cases comprising, 31% of this group. The diversity index in ocular trachoma cases was significantly lower than in sexually transmitted chlamydia infections. Predominating STs were identified in 12 available C. trachomatis whole genomes which were compared to 22 C. trachomatis full genomes without predominating STs. No specific gene in the 12 genomes with predominating STs could be linked to successful spread of certain STs. Phylogenetic analysis showed that MLST targets provide a tree similar to trees based on whole-genome analysis. The presented MLST scheme identified C. trachomatis strains with global spread. It provides a tool for epidemiological investigations and is useful for phylogenetic analyses.
INTRODUCTION
Chlamydia trachomatis is one of the most common sexually transmitted infections (STIs) worldwide (1), and besides urogenital infections, it also causes lymphogranuloma venereum (LGV), which is a rare but more invasive sexually transmitted disease. In addition, C. trachomatis causes the eye infection trachoma, which is the major infectious cause of preventable blindness worldwide. Severe sequelae from urogenital chlamydia infections include ectopic pregnancy and infertility (2). In spite of testing, treatment, partner notification, and counseling, huge public health efforts have not been able to control urogenital chlamydia infections. Current knowledge about the role of repeated infections and transmission in sexual networks is still limited and needs to be extended to achieve a reduction in the rate of infections.
In this context, it is important to have adequate tools for genotyping to understand the epidemiology of chlamydia infections. Traditional typing of C. trachomatis was based on serotyping of the major outer membrane protein (MOMP) and, later on, genotyping of the ompA gene, which encodes MOMP. However, neither MOMP nor ompA provides sufficient discriminatory power for epidemiological purposes (3). In most countries, almost half of all urogenital chlamydia infections are of serotype E, and within this serotype the ompA E/Bour genotype predominates (4–8). Therefore, other typing methods were developed, such as multilocus variable-number tandem-repeat analysis (MLVA) (8, 9) and multilocus sequence typing (MLST) (10–12).
MLST relies on PCR amplification and DNA sequencing of several genomic loci. There are three such schemes described for C. trachomatis. Two of them are based on housekeeping genes, have a resolution similar to that of ompA sequencing, and are suitable for evolutionary studies (11, 12). The third scheme was developed by Klint et al. (10); it is intended for short-term clinical epidemiology and outbreak investigations and is based on five highly variable genomic loci (hctB [CT046], CT058, CT144, CT172, and pbpB [CT682]). The MLST scheme has been slightly modified to facilitate rational processing with only 1% loss of discriminatory capacity (13), and the current setup can be found at the Uppsala University C. trachomatis MLST database (http://mlstdb.bmc.uu.se).
Since its development in 2007, the scheme has been applied to a variety of clinical specimens, including samples from cases of urogenital chlamydia (7, 13–21), LGV (22, 23), and trachoma (24). It has been useful for different purposes: (i) identification of clonal spread of LGV (22) and the new variant of C. trachomatis (16, 17); (ii) differentiation of strains within the predominating serotype, E (7, 17); (iii) differentiation of strains infecting men who have sex with men (MSM) and heterosexuals (15, 19); (iv) investigation of the role of tissue tropism (21); and (v) molecular epidemiology and antibiotic treatment of trachoma (24). At present, our database comprises 415 unique sequence types (STs) derived from 2,089 C. trachomatis specimens.
The aims of the current study were to (i) perform an overall analysis of all C. trachomatis MLST STs in the current database, (ii) identify and examine STs with global spread, and (iii) evaluate the use of the MLST targets for phylogenetic analysis.
MATERIALS AND METHODS
Specimens.
A total of 2,089 entries were obtained from 13 studies with different objectives and are available at the Uppsala University C. trachomatis MLST database (http://mlstdb.bmc.uu.se).
Study populations.
Samples from studies with specific objectives to investigate chlamydia infections in MSM are here designated samples from the compiled MSM population (15, 19, 22, 23). Samples from studies including clinics for STI, gynecology, and family planning (10, 13–21) and screening among high school students (7) are here designated samples from heterosexuals. However, a minor proportion of samples from heterosexuals may originate from MSM. In addition, one study comprised samples from trachoma patients (24).
PCR amplification, sequencing, and genotyping.
The currently used protocol for PCR amplification, sequencing, and genotyping of the five highly variable target regions included in this MLST scheme is available at the database above. Study sites could also use their own amplification and sequencing protocols as long as the trimmed sequences of the loci were complete from primer to primer. In addition to the five MLST targets, ompA sequence data were also included.
D.
The diversity index (D) of a typing method refers to the probability that two unrelated strains sampled from the test population will be placed into different typing groups. The D was determined for entries in the MLST database using Hunter and Gaston's modification of Simpson's diversity index (25). A cluster was defined as a group of STs differing by not more than one locus from another ST.
eBURST analysis.
eBURST v.3 (http://eburst.mlst.net/; Department of Infectious Disease Epidemiology, Imperial College London, United Kingdom) software was used to identify founders among the sequence types. A founder was defined as the ST with the largest number of single-locus variants in a group. An ST that appears to have diversified to produce multiple single-locus variants is called a subgroup founder. A list of all STs was inserted into the single data set function at the eBURST website, and the number of loci was set to 5 (ompA not included). The analysis was computed generating groups and predicted founders, and for each larger group a diagram was drawn.
MST analysis.
BioNumerics software (version 7.0; Applied Maths, Sint-Martens-Latem, Belgium) was used to construct a minimum spanning tree (MST) of all entries in the database. The sequence data for all 2,089 samples (ompA not included) was entered into a database within the BioNumerics software. As the algorithm, we used the predefined template “MST for categorical data” plug-in, which uses the categorical coefficient to calculate the similarity matrix. This calculates a standard MST with single- and double-locus variants.
Whole-genome analysis.
C. trachomatis genomes were downloaded from GenBank (6 draft genomes and 122 complete genomes and plasmids) and from the Ribosomal Multilocus Sequence Typing (rMLST) database (http://pubmlst.org/rmlst/, containing 397 genomes). MLST was performed on all genomes using the approach by Larsen et al. (26), with source code kindly provided by the authors and using our own MLST scheme. We selected a subset of genomes for further investigation. One to three representative full genomes were chosen for each of 5 founder STs (see Results), for a total of 12 genomes. The number of genomes for each ST depended on the number of genomes including this particular ST. Complete genomes were selected preferably instead of draft genomes. We also selected 22 genomes with other STs, in order to have a large number of profiles represented for comparative analyses. See Table S1 in the supplemental material for a list of genomes in the subset. For the genomes lacking annotations, coding sequences were predicted using Prodigal version 2.6 (27).
Identifying clusters of genes.
The program OrthoMCL version 2.0 (28) was used to cluster genes based on all-against-all blastp searches. As the genomes of C. trachomatis are highly conserved between strains, a high level of similarity is expected within groups. The blast comparison was performed without filtering for sequences with low information content, to avoid artificially short matches of nearly identical sequences. We required at least 90% sequence identity within groups and used an inflation index of 1.5 for the clustering. Members of the groups containing only sequences from genomes with founder STs were searched against the NCBI nucleotide database using blastp and standard settings. Typical best matches for each group, with 100% sequence identity in the aligned area when not otherwise stated, are listed in Table S2 in the supplemental material.
Phylogenetic analyses.
Two data sets based on the MLST genes were analyzed and compared with the phylogenomic analysis in the work of Harris et al. (29). The first data set consisted of the MLST genes from the same strains as in the work of Harris et al. (referred to here as the Harris et al. data set). The second data set also included all unique STs in the MLST database (http://mlstdb.bmc.uu.se; referred to here as the high-resolution [hr]-CT-MLST data set). For both data sets the individual genes were aligned by eye using Seaview (30). Potential recombinations between strains within genes were detected using RDP4 at default settings. To reduce the number of ambiguously aligned regions and number of gap positions, the poly(G) and intron regions of CT172, and the repetitive elements of hctB were removed. CT144 was removed from further analysis since it included a large amount of potential recombinations in both data sets. A phylogenetic tree was constructed for each gene separately using RAxML (31) with the GTR plus gamma model. Support values were estimated with 100 rapid bootstraps. The trees were rooted using the L strains as the outgroup. Sequences in clades separated from the rest by long branches (>0.05) were excluded to reduce long-branch attraction, and the phylogenetic analysis was redone. For the Harris et al. data set, conflicts between gene trees that had bootstrap support (BS) above 70 (in both gene trees) were identified and resolved by removing the least number of gene sequences. The same was done for the hr-CT-MLST data set, but using a cutoff of 65 to include apparent conflicts that were not identified using the cutoff of 70. The gene alignments were then concatenated and analyzed with RAxML. The GTR model and gamma distribution were used with parameters estimated separately for each gene. Support values were estimated using 10,000 rapid bootstraps. For the hr-CT-MLST data set, this analysis is referred to as “no conflict.”
Ethics.
Ethic permissions were obtained for each individual study. The present study includes no information that could identify individual patients.
RESULTS
The 2,089 entries in our database comprised 13 studies from 16 countries: the Netherlands (39%), Sweden (16%), Norway (12%), Suriname (8%), Tunisia (5%), China (4%), the United States (4%), Argentina (4%), Gambia and Senegal (4%), and France, Chile, Denmark, Spain, Australia, and Germany (all ≤1%). The collection of clinical specimens spanned more than 19 years (1992 to 2011), and reference strains were isolated from 1958 onwards. Overall, 609 (29%) of the database samples originated from MSM (75% of samples from rectum), 1,383 (66%) urogenital samples were assumed to be from heterosexuals, and 75 (4%) ocular samples were from trachoma patients. In addition, 22 C. trachomatis reference strains representing 15 genovars were included.
The database comprised 415 unique STs derived from 2,089 C. trachomatis specimens. Polymorphism of target regions was reflected in various numbers of alleles: hctB, 89; CT058, 51; CT144, 30; CT172, 38; and pbpB, 35. With the addition of 49 ompA gene variants, a total of 459 STs existed in December 2013.
High genetic variation, but with few STs predominating.
The frequency distribution of STs ranged from 1 to 140. Most STs were represented by only a few samples; thus, 255 STs were found in a single sample (singletons) and 122 STs in 2 to 9 samples. All these uncommon STs represented by 1 to 9 samples comprised 33% of the total database. Thirty-one STs were medium sized, with 10 to 43 samples each, and constituted 25% of the database. The distribution of STs and ompA genovars is shown in Fig. 1A.
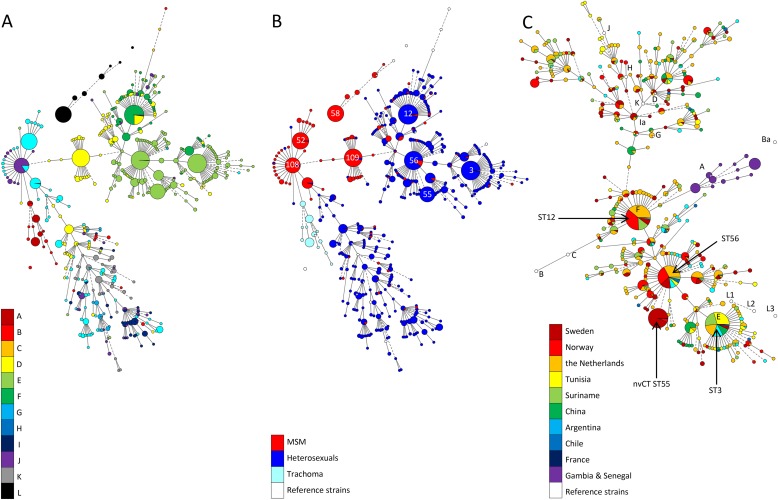
FIG 1.
(A and B) Minimum spanning tree analysis of the entire database based on the five MLST target regions visualized by ompA genovar distribution (A) and by sexual behavior (B). Sphere sizes indicate the numbers of samples in each sphere. Solid branches show single-locus variants, and dashed branches show double-locus variants. The most common STs are indicated by number. (C) Minimum spanning tree showing the geographical ST distribution of 1383 heterosexual samples, 75 trachoma samples (Gambia and Senegal), and 15 reference strains in the MLST database. Sphere size is proportional to the number of samples of each sequence type. Solid branches show single-locus variants, dashed branches show double-locus variants, and dotted branches show triple-locus variants.
There were 8 STs that dominated the database, with numbers of samples ranging from 83 to 140, comprising a total of 868 samples (42% of the database). These 8 STs were widely distributed in the different countries. Analysis with eBURST identified 3 of the 8 predominating profiles as founders and another 3 as subgroup founders. MST analysis placed these 6 large STs centrally in the tree (Fig. 1B). The two large STs that were nonfounders were LGV2b (ST58) and the new variant C. trachomatis (ST55), which both have shown clonal spread in Europe in the last decade.
Four of the 8 predominating STs were strongly associated with MSM (ST52 [n = 98], ST58 [n = 97], ST108 [n = 83], and ST109 [n = 115]), with 97 to 100% of the samples coming from MSM (Fig. 1B). In the other four large clusters (ST3 [n = 128], ST12 [n = 140], ST55 [n = 84], and ST56 [n = 123]), samples from mostly heterosexuals comprised >90% of the samples. Studies including urogenital samples from heterosexuals were performed in 8 countries (Sweden, Norway, the Netherlands, Tunisia, Suriname, Argentina, Chile, and China). Of these, ST3 (genovar E) was found in all 8 countries, ST56 (genovar E) in 7 countries, and ST12 (genovars F, D, and J) in 5 countries. The geographical distribution of specimens from heterosexuals is illustrated in Fig. 1C. It shows that some STs are common and are spread on different continents on the globe.
The diversity index (D) was 0.975 (95% confidence interval [CI], 0.974 to 0.976) based on all 2089 entries in the database. The Ds for different clusters are shown in Table 1. The D was smaller in samples from MSM than from heterosexuals: 0.893 (0.887 to 0.899) and 0.968 (0.966 to 0.970), respectively. The three predominating non-LGV STs in MSM (ST52, ST108, and ST109) comprised 47% of all non-LGV cases from MSM, while the four STs predominating among heterosexuals (ST3, ST12, ST55, and ST56) comprised 31% of all cases from this group. In Fig. 1B it is also shown that STs in MSM are predominantly confined to four clusters, while STs in heterosexuals are less clustered.
TABLE 1.
Diversity indices for MSM, heterosexuals, five predominating clusters,a and the complete database
| Diversity index (95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| All MSM (n = 609) | MSM-associated cluster |
All heterosexuals (n = 1,383) | Heterosexual-associated cluster |
Complete database (n = 2,089) | |||
| Cluster 52/108b (n = 247) | Cluster 109 (n = 140) | Cluster 3 (n = 180) | Cluster 12 (n = 247) | Cluster 56 (n = 247) | |||
| 0.893 (0.887–0.899) | 0.70 (0.69–0.72) | 0.32 (0.27–0.38) | 0.968 (0.966–0.970) | 0.49 (0.44–0.54) | 0.66 (0.63–0.69) | 0.72 (0.69–0.75) | 0.975 (0.974–0.976) |
A cluster was defined as a group of STs allowing single locus variance of one step from the founder.
Cluster 52/108 is a composite of two predominating STs since they differ only in the hctB locus (Fig. 1B), where ST108 is 60 bp shorter than ST52 and has three point mutations.
The present work is a compilation of 13 studies with different design and sampling strategies, and the study populations are not representative for the general sexually active population in the 16 countries. To compare the ompA genotype distribution in our study with the ompA distribution in previous studies, we performed ompA sequencing of the 1,383 samples assumed to be from heterosexuals, with the following results: E, 45% (n = 617); F, 16% (n = 217); D, 13% (n = 173); G, 10% (n = 136); K, 5.8% (n = 80); I, 5.4% (n = 75); J, 3.9% (n = 54); H, 2.3% (n = 32); and B, 0.01% (n = 10).
Differences between C. trachomatis typing results from MSM and heterosexuals are summarized in Table 2.
TABLE 2.
Differences between MSM and heterosexuals in C. trachomatis genotypes and population structures
| Parameter | MSM populationa | Heterosexual population |
|---|---|---|
| Most predominant ompA genovars | D (26%, n = 160) | D (12%, n = 170); P < 0.001 for the five serotypes |
| E (5%, n = 29) | E (45%, n = 617) | |
| G (29%, n = 179) | G (10%, n = 134) | |
| F (4%, n = 22) | F (16%, n = 215) | |
| J (14%, n = 85) | J (4%, n = 53) | |
| No. clustersb | A few large clusters | Multiple clusters of various size |
| Overlap with clusters in the other risk groupc | Almost absent | A small proportion |
| Subpopulations within clusters | No | Ethnicityd |
| Geographic variation within clusters | No | Yese |
The D for 75 trachoma specimens was lower (0.772 [0.742 to 0.803]) than for urogenital samples (0.968 [0.966 to 0.970]). The number of ompA variants was 10, and the number of STs using the five target regions in MLST was 12, while a combination of ompA and MLST resulted in 19 STs.
Whole-genome analysis of strains with predominating MLST STs.
The distribution pattern of STs clearly shows that a few strains have been successful in global spread in different populations. Concurrently, many strains are single-locus variants to the founders and subgroup founders identified by eBURST analysis and seem to be genetically closely related to the predominating strains. Twelve C. trachomatis genomes containing any of the predominating STs (ST3, ST12, ST52, ST56, and ST108), here designated founders, were compared with 22 other available C. trachomatis genomes to investigate if specific genes or proteins could be identified in the strains with founder STs. No genome containing ST109 was yet available in GenBank.
After OrthoMCL analysis, where the proteins of the 34 selected genomes were clustered into groups of orthologues, 17 groups of proteins were identified as unique for strains with any of the founder STs. However, no group contained proteins from all genomes with founder STs. There were 13 groups containing protein sequences from more than one genome. Four of these groups consisted of sequences that are fragments of longer proteins. Most of the groups are similar to hypothetical proteins; only three of these are from C. trachomatis, while the others are from eukaryotic organisms (see Table S2 in the supplemental material). The analysis could not identify any specific gene that was common for strains with predominating STs but missing in other strains.
Phylogenetic analysis.
With regard to the Harris et al. data set, B/TZ1A828/OT was on a long branch in pbpB, indicating highly divergent sequences, and was therefore excluded for this gene. Three cases with conflicts between gene trees were identified. The placement of L1 1322-p2 was inconsistent between CT058 and CT172 and was removed from CT172. D/SotonD6 and G11222 clustered differently in CT058 and pbpB and were removed from pbpB.
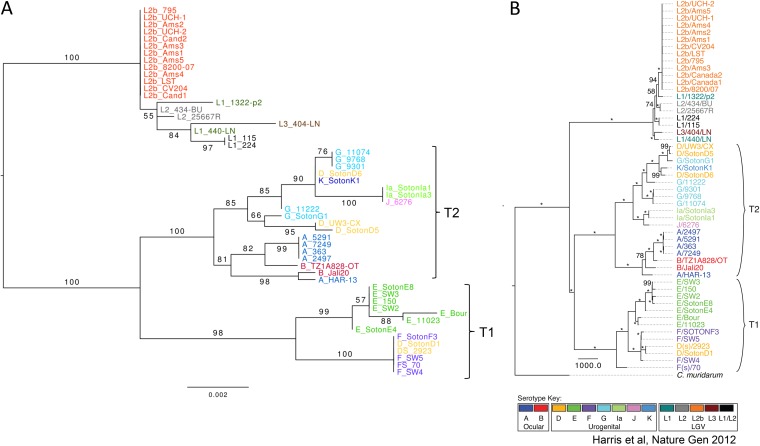
The concatenated tree was largely in agreement with the full genome tree of Harris et al. (29), but with less resolution and lower bootstrap support values (Fig. 2). There are, however, some differences supported by bootstrap values of >70. In the Harris et al. data set, F(s)/70 is the sister to a clade including E, F, and D strains, while it is nested within that clade in the MLST tree. A/HAR-13 and B/Jali20 form a clade for the MLST genes, while they constitute the basal grade to a clade dominated by A and B strains in the Harris et al. data set. There are several differences in the clade composing G, D, K, Ia, and J strains. In the clade with L strains, the MLST tree has a clade with L1/224, L1/115, L1/440/LN, and L1/404/LN, while these are part of a basal grade in the phylogeny of Harris et al. (29).
FIG 2.
Comparison of phylogenetic trees obtained from 52 strains by maximum likelihood analysis after removal of recombination events. (A) MLST concatemer of five targets; (B) whole-genome sequences (reproduced from reference 29 by permission from Macmillan Publishers Ltd.). Numbers on nodes represent percentage of bootstrap replicates supporting each clade. Asterisks indicate 100% support. The tree in panel A was rooted based on the previous study whose results are shown in panel B.
The hr-CT-MLST data set resulted in a tree that has essentially the same topology as obtained when using the Harris et al. data set but with considerably more branches (see Fig. S1 in the supplemental material).
DISCUSSION
This report is a unique summary of the most frequently used high-resolution genotyping scheme for C. trachomatis to date with the database including more than 2,000 samples from 16 countries worldwide. The distribution of STs clearly shows that a few strains have been successful in spreading in many countries among different populations and thus appears to predominate in both space and time. Different STs predominate among MSM and heterosexual individuals. Currently, multiple strains are single-locus variants of the founders and subgroup founders identified by eBURST analysis and seem to be genetically closely related to the predominating strains. However, the traits that contribute to successful spread may not be directly linked to the 5 MLST targets or the ompA gene, but these targets may be biomarkers for other genes or traits in strains that are successfully spread worldwide. Our comparison of whole genomes from strains with and without founder STs could not identify any gene that was common for strains with predominating STs but missing in other strains.
The MLST scheme has been used in study populations and provides high resolution for differentiation of C. trachomatis strains (32). A major concern about a typing method is its reliability. Since the 5 MLST targets are not housekeeping genes, but were selected from the most divergent genes in the few genomes available in 2005, it has been questioned if the targets are stable. In previous studies the stability of the included target regions has been demonstrated in laboratory experiments (33) as well as in longitudinal studies of the new variant of C. trachomatis and lymphogranuloma venereum (16, 17, 22, 34). The longitudinal studies also indicated that ompA was more unstable, which is in agreement with recent whole-genome studies that have shown extensive recombination in ompA (29, 35, 36). Our MLST has therefore proved suitable for use in epidemiological studies. Identification of founders in eBURST analysis showed that ST12 was detected in a reference strain collected as early as 1960 (F/IC-cal3), a common and widespread genotype. This further indicates the stability of the MLST targets.
This MLST scheme has shown up to a 5-fold-higher resolution than ompA (7). The D was 0.975 (95% CI, 0.974 to 0.976) when calculated on all entries in the database. In previous studies of sexually transmitted chlamydia infections, the D varied between 0.84 and 0.97 depending on the degree of epidemiological relatedness between samples in the study populations (7, 13, 17, 37). By calculating an overall D for all 2,089 samples, epidemiological relatedness was eliminated. The obtained index was well above 0.95, an “ideal” cutoff value for a molecular typing method (38). The lower D for the predominating clusters in Table 1 is explained by the inclusion of some epidemiologically related samples, especially among MSM who may be connected via extended international sexual networks (15, 19).
Regarding trachoma, the MLST has been used only in a limited number of samples where many cases were epidemiologically linked, explaining why a low D (0.77) was obtained. The number of ompA variants was almost equal to the number of STs based on the five MLST targets (24). This is in contrast to the case with sexually transmitted chlamydia infections, for which up to 5-fold more STs have been found compared to ompA variants (7).
As most of the 13 studies included STI clinics, these samples were not representative for samples from the general population. However, the distribution of ompA genotypes among the 1,383 assumed heterosexual samples is similar to that in other studies (3) and indicates that our study reflects strain distribution in most countries where genotyping of C. trachomatis has been performed.
Previous ompA typing studies have shown that specific genotypes are frequently found in rectal samples from MSM (39–42), and tissue tropism has been suggested as an explanation (35, 43–46), mainly based on genetic and cell biological findings. However, recent MLST data show that differing sexual network structures and distributions of C. trachomatis strains in MSM versus heterosexual networks may be an important reason for the findings of certain genotypes in rectum (21). This also explains the findings in our data set of mostly different STs in MSM and heterosexuals.
Maximum likelihood analysis showed that obtained trees were similar to trees obtained from whole-genome analysis by Harris et al. (29), but bootstrap support values were lower. The phylogeny based on the MLST targets is also in agreement with the four identified lineages in whole-genome analysis by Joseph et al. (36). Analyses of MLST targets are easier to perform than obtaining whole genome sequences and thereafter performing analyses. Using MLST may therefore facilitate phylogenetic studies. We also applied phylogenetic analysis to the entire hr-CT-MLST data set and confirmed the results of the limited data set, but it also gave rise to more clades. A limitation is the occurrence of recombinations that disturb the phylogenetic analysis. Therefore, identified recombinations were removed which influenced the tree topology, but the major structure remained intact.
The MLST target genes used in this study were originally selected because of their sequence variation, which was thought to provide high resolution and to be suitable for short-term investigations. Considering the conserved nature of the C. trachomatis genomes, we expected that this MLST system would be saturated with a limited number of STs. However, in each new study population, several new STs have been identified. The number of detected alleles provides a theoretical number of 8.8 × 109 STs, assuming no linkage between targets, indicating that only a fraction of possible STs have been identified. More refined genetic analysis is needed to elucidate why a few STs predominate in space and time. To provide a better understanding of evolutionary changes for C. trachomatis and link it to short-term variations, it would be rewarding to combine our MLST system with data from a housekeeping-based MLST. Furthermore, the role of the host response to different infecting strains is not well understood.
The strength of this study is the size of the data set. Although the geographic representation is patchy and the separate studies have different objectives, the results provide an overall picture of strain distribution for C. trachomatis in time and space and in populations with different sexual behavior that has not been observed previously.
In summary, the presented MLST system provides high resolution for C. trachomatis strains, is useful for epidemiological investigations, has identified a few predominating strains that have spread successfully in several countries, and is beneficial for phylogenetic analysis.
Supplementary Material
ACKNOWLEDGMENTS
The study was financed by local funds from Uppsala University Hospital, Sweden.
Additional members of The Chlamydia trachomatis MLST Study Group include Bertille de Barbeyrac and Olivia Peuchant (University of Bordeaux, Bordeaux, France), Reinier Bom (Municipal Health Service Amsterdam, Amsterdam, the Netherlands), Hans Fredlund (Örebro University Hospital, Örebro, Sweden), Houda Gharsallah and Adnene Hammami (Habib Bourguiba University, Sfax, Tunisia), Charlotte A Gaydos (Johns Hopkins University, Baltimore, MD), David Mabey (London School of Hygiene and Tropical Medicine, London, United Kingdom), Servaas A Morré (VU University Medical Center, Amsterdam, the Netherlands), Marcelo Rodríguez Fermepin (University of Buenos Aires, Buenos Aires, Argentina), Julius Schachter (University of California, San Francisco, CA), Henry JC de Vries (Municipal Health Service Amsterdam, Amsterdam, the Netherlands), Qianqiu Wang (Institute of Dermatology, CAMS National Center for STD Control, Nanjing, China), and Fuquan Long (Shanghai Dermatology Hospital, China).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00249-15.
REFERENCES
- 1.World Health Organization. 2012. Global incidence and prevalence of selected curable sexually transmitted infections—2008. World Health Organization, Geneva, Switzerland: http://www.who.int/reproductivehealth/publications/rtis/2008_STI_estimates.pdf. [Google Scholar]
- 2.Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. 2010. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis 201(Suppl 2):S134–S155. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- 3.Pedersen LN, Herrmann B, Møller JK. 2009. Typing Chlamydia trachomatis: from egg yolk to nanotechnology. FEMS Immunol Med Microbiol 55:120–130. doi: 10.1111/j.1574-695X.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 4.Jurstrand M, Falk L, Fredlund H, Lindberg M, Olcen P, Andersson S, Persson K, Albert J, Bäckman A. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J Clin Microbiol 39:3915–3919. doi: 10.1128/JCM.39.11.3915-3919.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lysén M, Österlund A, Rubin C-J, Persson T, Persson I, Herrmann B. 2004. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J Clin Microbiol 42:1641–1647. doi: 10.1128/JCM.42.4.1641-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jónsdóttir K, Kristjńsson M, HjaltalÍn Olafsson J, Steingrmsson O. 2003. The molecular epidemiology of genital Chlamydia trachomatis in the greater Reykjavik area, Iceland. Sex Transm Dis 30:249–256. doi: 10.1097/00007435-200303000-00015. [DOI] [PubMed] [Google Scholar]
- 7.Gravningen K, Christerson L, Furberg A-S, Simonsen GS, Ödman K, Ståhlsten A, Herrmann B. 2012. Multilocus sequence typing of genital Chlamydia trachomatis in Norway reveals multiple new sequence types and a large genetic diversity. PLoS One 7(3):e34452. doi: 10.1371/journal.pone.0034452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peuchant O, Le Roy C, Herrmann B, Clerc M, Bébéar C, de Barbeyrac B. 2012. MLVA subtyping of genovar E Chlamydia trachomatis individualizes the Swedish variant and anorectal isolates from men who have sex with men. PLoS One 7(2):e31538. doi: 10.1371/journal.pone.0031538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen LN, Pødenphant L, Møller JK. 2008. Highly discriminative genotyping of Chlamydia trachomatis using omp1 and a set of variable number tandem repeats. Clin Microbiol Infect 14:644–652. doi: 10.1111/j.1469-0691.2008.02011.x. [DOI] [PubMed] [Google Scholar]
- 10.Klint M, Fuxelius H-H, Goldkuhl RR, Skarin H, Rutemark C, Andersson SGE, Persson K, Herrmann B. 2007. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J Clin Microbiol 45:1410–1414. doi: 10.1128/JCM.02301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean D, Bruno WJ, Wan R, Gomes JP, Devignot S, Mehari T, de Vries HJC, Morré SA, Myers G, Read TD, Spratt BG. 2009. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis 15:1385–1394. doi: 10.3201/eid1509.090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannekoek Y, Morelli G, Kusecek B, Morré SA, Ossewaarde JM, Langerak AA, van der Ende A. 2008. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol 8:42. doi: 10.1186/1471-2180-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bom RJM, Christerson L, Schim van der Loeff MF, Coutinho RA, Herrmann B, Bruisten SM. 2011. Evaluation of high-resolution typing methods for Chlamydia trachomatis in samples from heterosexual couples. J Clin Microbiol 49:2844–2853. doi: 10.1128/JCM.00128-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christerson L, de Vries HJC, Klint M, Herrmann B, Morré SA. 2011. Multilocus sequence typing of urogenital Chlamydia trachomatis from patients with different degrees of clinical symptoms. Sex Transm Dis 38:490–494. doi: 10.1097/OLQ.0b013e31820b8be0. [DOI] [PubMed] [Google Scholar]
- 15.Christerson L, Bom RJM, Bruisten SM, Yass R, Hardick J, Bratt G, Gaydos CA, Morré SA, Herrmann B. 2012. Chlamydia trachomatis strains show specific clustering for men who have sex with men compared to heterosexual populations in Sweden, the Netherlands, and the United States. J Clin Microbiol 50:3548–3555. doi: 10.1128/JCM.01713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann B, Törner A, Low N, Klint M, Nilsson A, Velicko I, Söderblom T, Blaxhult A. 2008. Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg Infect Dis 14:1462–1465. doi: 10.3201/eid1409.080153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurstrand M, Christerson L, Klint M, Fredlund H, Unemo M, Herrmann B. 2010. Characterisation of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex Transm Infect 86:56–60. doi: 10.1136/sti.2009.037572. [DOI] [PubMed] [Google Scholar]
- 18.Bom RJM, van der Helm JJ, Bruisten SM, Grünberg AW, Sabajo LOA, Schim van der Loeff MF, de Vries HJC. 2013. The role of Surinamese migrants in the transmission of Chlamydia trachomatis between Paramaribo, Suriname and Amsterdam, The Netherlands. PLoS One 8(11):e77977. doi: 10.1371/journal.pone.0077977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bom RJM, van der Helm JJ, Schim van der Loeff MF, van Rooijen MS, Heijman T, Matser A, de Vries HJC, Bruisten S. 2013. Distinct transmission networks of Chlamydia trachomatis in men who have sex with men and heterosexual adults in Amsterdam, The Netherlands. PLoS One 8(1):e53869. doi: 10.1371/journal.pone.0053869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Helm JJ, Bom RJM, Grünberg AW, Bruisten SM, Schim van der Loeff MF, Sabajo LOA, de Vries HJC. 2013. Urogenital Chlamydia trachomatis infections among ethnic groups in Paramaribo, Suriname; determinants and ethnic sexual mixing patterns. PLoS One 8(7):e68698. doi: 10.1371/journal.pone.0068698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Versteeg B, van Rooijen MS, Schim van der Loeff MF, de Vries HJC, Bruisten SM. 2014. No indication for tissue tropism in urogenital and anorectal Chlamydia trachomatis infections using high-resolution multilocus sequence typing. BMC Infect Dis 14:464. doi: 10.1186/1471-2334-14-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christerson L, de Vries HJC, de Barbeyrac B, Gaydos CA, Henrich B, Hoffmann S, Schachter J, Thorvaldsen J, Vall-Mayans M, Klint M, Herrmann B, Morré SA. 2010. Typing of lymphogranuloma venereum Chlamydia trachomatis strains. Emerg Infect Dis 16:1777–1779. doi: 10.3201/eid1611.100379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vall-Mayans M, Isaksson J, Caballero E, Sallés B, Herrmann B. 2014. Bubonic lymphogranuloma venereum with multidrug treatment failure. Int J STD AIDS 25:306–308. doi: 10.1177/0956462413501158. [DOI] [PubMed] [Google Scholar]
- 24.Harding-Esch EM, Christerson L, Grannas K, Molina S, Roberts CH, Holland MJ, Andreasen AA, Sillah A, Sarr B, Jeffries D, Grundy C, Bailey RL, Mabey DC, Herrmann B. 2010. Multi-locus sequence typing: a useful tool for trachoma molecular epidemiology, p 55–58. In Paavonen J, Saikku P, Starnbach M, Stary A, Stephens RS, Timms P, Wyrick PB (ed), Chlamydial infections. Proceedings of the 12th International Symposium on Human Chlamydial Infections; Salzburg International Chlamydia Symposium San Francisco, CA. [Google Scholar]
- 25.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ. 2011. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics 35:6.12.1–6.12.19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris SR, Clarke IN, Seth-Smith HMB, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morré SA, Speksnijder A, Bébéar CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44:413–419, S1. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vries HJ, Schim van der Loeff MF, Bruisten SM. 2015. High resolution typing of Chlamydia trachomatis: epidemiological and clinical uses. Curr Opin Infect Dis 28:61–71. doi: 10.1097/QCO.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 33.Labiran C, Clarke IN, Cutcliffe LT, Wang Y, Skilton RJ, Persson K, Bjartling C, Herrmann B, Christerson L, Marsh P. 2012. Genotyping markers used for multi locus VNTR analysis with ompA (MLVA-ompA) and multi sequence typing (MST) retain stability in Chlamydia trachomatis. Front Cell Infect Microbiol 2:68. doi: 10.3389/fcimb.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klint M, Hadad R, Christerson L, Loré B, Anagrius C, Österlund A, Larsson I, Sylvan S, Fredlund H, Unemo M, Herrmann B. 2011. Prevalence trends in Sweden for the new variant of Chlamydia trachomatis. Clin Microbiol Infect 17:683–689. doi: 10.1111/j.1469-0691.2010.03305.x. [DOI] [PubMed] [Google Scholar]
- 35.Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78:2544–2553. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joseph SJ, Didelot X, Rothschild J, de Vries HJC, Morré SA, Read TD, Dean D. 2012. Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol 29:3933–3946. doi: 10.1093/molbev/mss198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikryannikova LN, Shkarupeta MM, Shitikov EA, Il'ina EN, Govorun VM. 2010. Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol Med Microbiol 59:188–196. doi: 10.1111/j.1574-695X.2010.00678.x. [DOI] [PubMed] [Google Scholar]
- 38.Van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):S1–S46. [DOI] [PubMed] [Google Scholar]
- 39.Barnes RC, Rompalo AM, Stamm WE. 1987. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J Infect Dis 156:953–958. doi: 10.1093/infdis/156.6.953. [DOI] [PubMed] [Google Scholar]
- 40.Bax CJ, Quint KD, Peters RPH, Ouburg S, Oostvogel PM, Mutsaers JA, Dörr PJ, Schmidt S, Jansen C, van Leeuwen AP, Quint WG, Trimbos JB, Meijer CJ, Morré SA. 2011. Analyses of multiple-site and concurrent Chlamydia trachomatis serovar infections, and serovar tissue tropism for urogenital versus rectal specimens in male and female patients. Sex Transm Infect 87:503–507. doi: 10.1136/sti.2010.048173. [DOI] [PubMed] [Google Scholar]
- 41.Klint M, Löfdahl M, Ek C, Airell A, Berglund T, Herrmann B. 2006. Lymphogranuloma venereum prevalence in Sweden among men who have sex with men and characterization of Chlamydia trachomatis ompA genotypes. J Clin Microbiol 44:4066–4071. doi: 10.1128/JCM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morré S, Rozendaal L, Valkengoed I van, Boeke A, Voorst Vader PC, van Schirm J, de Blok S, van Den Hoek JA, van Doornum GJ, Meijer CJ, van Den Brule AJ. 2000. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J Clin Microbiol 38:2292–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdelsamed H, Peters J, Byrne GI. 2013. Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol 8:1129–1146. doi: 10.2217/fmb.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almeida F, Borges V, Ferreira R, Borrego MJ, Gomes JP, Mota LJ. 2012. Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates. J Bacteriol 194:6574–6585. doi: 10.1128/JB.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunes A, Borrego MJ, Gomes JP. 2013. Genomic features beyond Chlamydia trachomatis phenotypes: what do we think we know? Infect Genet Evol 16:392–400. doi: 10.1016/j.meegid.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Borges V, Nunes A, Ferreira R, Borrego MJ, Gomes JP. 2012. Directional evolution of Chlamydia trachomatis towards niche-specific adaptation. J Bacteriol 194:6143–6153. doi: 10.1128/JB.01291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.