Abstract
Laribacter hongkongensis is a potential emerging pathogen associated with community-acquired gastroenteritis and traveler's diarrhea. We report the isolation of L. hongkongensis from the stool of a patient who had no history of travel outside the United States. The organism was identified by phenotypic tests, mass spectrometry, and gene sequencing.
CASE REPORT
A 17-year-old female with Down's syndrome and hypothyroidism presented with loose/watery stools with only one bowel movement per day. On examination, her temperature was afebrile (36.3°C). Surgical history included open heart surgery at age 3 months due to congestive heart failure and pulmonary hypertension. She had strabismus surgery and tonsils and adenoids removed in 1997. She owns a dog that lives in the house but does not sleep with her. The patient resides in southwest Texas (Houston metropolitan area), had never traveled outside Texas, and did not eat sushi or food from street vendors. It was reported that the young woman had a tendency to frequently put her hands and other objects in her mouth and that she attended classes at a school 3 days a week and did eat lunch there.
A stool sample was submitted for testing by the comprehensive stool analysis with parasitology x3 kit, which includes microbiological culture for aerobic, anaerobic, and yeast organisms, parasitology exam, and markers of digestion, absorption, and inflammation. The stool sample was collected on 11 August 2014 in Remel Cary-Blair transport medium, sodium acetate-acetic acid-formalin (SAF) transport medium, and a clean vial with no additives (Thermo Scientific, Lenexa, KS). The vials were received, cultured, and examined for ova and parasites on 13 August 2014. After 48 h of incubation at 35°C in ambient air, we observed a few small, nonfermenting organisms on Hektoen enteric agar (HEK) (Becton Dickinson, Cockeysville, MD), and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed on the isolate. The isolate was processed in a Bruker Daltonik MALDI-TOF MS with 1 μl of Bruker α-cyano matrix solution and subcultured to Trypticase soy II blood agar (TSA) (Becton Dickinson, Cockeysville, MD) for further testing. The spectrum was analyzed using a MALDI Biotyper RUO (for research use only) 3.1 (build 65), Reference Library version 3.1.2.0 software, and the MBT-BDAL 5627 MSP library (Bruker Daltonik, Bremen Germany), with the top score of 2.291 for Laribacter hongkongensis.
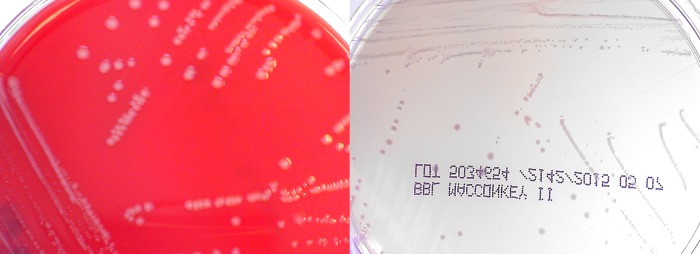
The organism grew on TSA as nonhemolytic gray colonies and on MacConkey agar (MAC) (Becton Dickinson, Cockeysville, MD) as smaller, non-lactose-fermenting colonies with a slight lavender appearance (Fig. 1). Gram stain showed slightly curved Gram-negative rods (Fig. 2). The organism was subsequently tested on the bioMérieux API 20E using APIWeb version 4.1 (API bioMérieux, Hazelwood, MO) and identified as Photobacterium damselae with 98.1% confidence based upon the profile number 201004. It produced cytochrome oxidase, urease, and arginine dihydrolase. It did not ferment any carbohydrates. The isolate was also tested on Vitek2 XL version 6.04 with GN Identification Knowledge Base 05.00.11 with low-discrimination identifications of Acinetobacter ursingii, Methylobacterium spp., and Acinetobacter lwoffii with positive reactions for tyrosine arylamidase, urease, and succinate alkalinization.
FIG 1.
Gray colonies grew on TSA, and lavender non-lactose-fermenting colonies on MAC.
FIG 2.
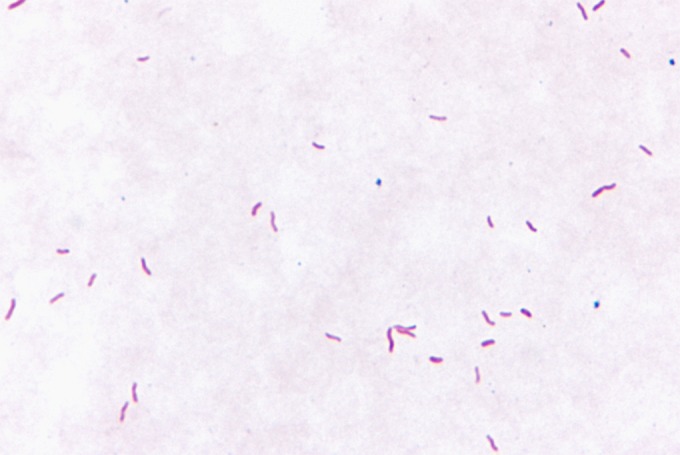
Gram stain showing slightly curved Gram-negative rods.
16S rRNA gene sequencing was performed at Quest Diagnostics Nichols Institute using a laboratory-developed assay, and the results were searched using the SmartGene Database. The sequence was determined to be genetically consistent with Laribacter hongkongensis with a match of 717 bp and an identity match of 100%, which corresponds to zero base pair mismatches. The 20 closest matches were all Laribacter hongkongensis.
Other microbiological findings from the patient's stool specimen included high quantities of Bacteroides fragilis group, Bifidobacterium spp., Clostridium spp., and Escherichia coli. Lesser quantities of Lactobacillus spp., Enterococcus spp., Saccharomyces cerevisiae/boulardii, and Streptococcus spp. were also identified in the specimen. Parasitology x3 exams were negative for ova and parasites by microscopic examination. The results for Giardia intestinalis and Cryptosporidium in enzyme immunoassays (Giardia/Cryptosporidium CHEK antigen test; TechLab, Blacksburg, VA) were negative.
The stool consistency was loose and watery. Microscopic evaluation revealed no white blood cells, red blood cells, or muscle fibers, and vegetable fibers were rare.
Chemistry findings revealed normal levels for elastase, fat stain, carbohydrates, lysozyme, lactoferrin, butyrate and total short-chain fatty acids, pH, and occult blood. The level of fecal secretory IgA was lower than the laboratory reference value (31.1 mg/dl; reference range, 51 to 204 mg/dl).
No treatment was given and the patient's loose stool resolved. Follow-up cultures were performed for the patient and household contacts 2 months after the initial episode and were negative for L. hongkongensis. No epidemiological investigation was performed, and it is presumed the patient acquired the organism from an unknown source outside the home.
In addition to this case, we identified four additional patients with L. hongkongensis from April 2014 to November 2014. Like our first case, all were tested and found negative for Salmonella spp., Shigella spp., Campylobacter spp., Vibrio spp., Aeromonas spp., Plesiomonas shigelloides, Edwardsiella tarda, Arcobacter butzleri, and parasites. The four additional cases included one from North Carolina who presented with soft stool and elevated lysozyme and three from outside the United States (United Kingdom, Australia, and Japan) for whom no history was available. All were identified as L. hongkongensis using MALDI-TOF MS, with top match scores ranging from 2.067 to 2.326. Unlike the isolate from the primary patient, all four were found on Campy CVA (cefoperazone, vancomycin, and amphotericin B) with 5% sheep blood agar (Becton Dickinson, Cockeysville, MD) after incubation for 72 h at 42°C under conditions supporting microaerophilic growth (Campy gas mixture of 5% O2, 10 CO2, and 85% N2). All four isolates grew as slightly spreading cytochrome oxidase-positive organisms on Campy CVA. All strains grew on TSA and MAC in ambient air at 35°C. Two of the strains were tested further for growth on several media and under different atmospheric conditions (Table 1). Interestingly, the isolates grew on Campy CVA but not on Campylobacter agar with 5 antimicrobics and 10% sheep blood, Blaser formula (Campy-BAP; Becton Dickinson, Cockeysville, MD). This most likely is due to the addition of trimethoprim and polymyxin B to the Blaser formula.
TABLE 1.
Growth characteristics of Laribacter hongkongensis
| Medium, type of atmosphere, temp (°C) | Growth of straina: |
|
|---|---|---|
| 1 | 2 | |
| TSA, ambient air | ||
| 35 | + | + |
| 42 | + | + |
| TSA, microaerophilic | ||
| 35 | + | + |
| 42 | + | + |
| MAC, ambient air | ||
| 35 | + | + |
| 42 | + | + |
| Campy CVA, ambient air | ||
| 35 | + | + |
| 42 | + | + |
| Campy CVA, microaerophilic | ||
| 35 | + | + |
| 42 | + | + |
| Campy BAP, ambient air | ||
| 35 | − | − |
| 42 | − | − |
| Campy BAP, microaerophilic | ||
| 35 | − | − |
| 42 | − | − |
−, no growth; +, good growth.
The same two isolates were tested on a MicroScan instrument with Biotype Server version 4.11 and LabPro Database version 4.11.1010.145 (MicroScan WalkAway-96 Plus; Beckman Coulter, Sacramento, CA), using the neg urine combo 51 panel, and both gave a biotype number of 42000006, with an interpretation of low-probability identification. The two strains were also tested in the Bruker Daltonics MBT-CA (FDA cleared) system and gave no identification. MALDI-TOF testing was not performed with the Vitek MS because it is not available in our laboratory. Of note, L. hongkongensis is not included in Vitek MS plus Saramis RUO or FDA-approved libraries. Conventional biochemical analysis of the two L. hongkongensis isolates showed both strains to be positive for motility, oxidase, catalase, nitrate reduction, arginine dihydrolase, malonate utilization, urease, and growth at 35°C and 42°C. Both strains were negative for DNase, starch, esculin and gelatin hydrolysis, reduction of NO2, ornithine and lysine decarboxylases, acetamide utilization, indole production, phenylalanine deaminase, growth in 6.5% NaCl, o-nitrophenyl-β-d-galactopyranoside (ONPG), 10% lactose, lecithinase, lipase, and pyrrolidonyl peptidase (PYR). In addition, both isolates were asaccharolytic and produced alkaline reactions in the oxidation-fermentation test with fructose, glucose, lactose, maltose, mannitol, sucrose, and xylose.
Laribacter hongkongensis is a facultative, anaerobic, motile, nonsporulating, nonfermentative, urease-positive, Gram-negative rod belonging to the family Neisseriaceae of the Betaproteobacteria. It was first isolated from the blood and empyemic pus of a 54-year-old Chinese man with alcoholic cirrhosis and bacterial thoracic empyema in Hong Kong (1). A case-control study showed the presence of an association between L. hongkongensis and community-acquired diarrhea, fish consumption, and travel (2). Intestinal samples from fish and frogs from retail markets were collected and cultured and found to contain L. hongkongensis with a prevalence of 16.3% from freshwater fish and 59.5% in edible frogs, suggesting a higher public health risk with the consumption of edible frogs (3). It also has been found in drinking water reservoirs in Hong Kong (4). L. hongkongensis has been reported in Asia, Europe, Africa, and Central America (2, 5–8). A review of the microbiological characteristics and pathogenicity of L. hongkongensis has been published by Raja and Ghosh (9).
We report the first validated L. hongkongensis isolate acquired in the United States. Based on the results from the culture media used in our laboratory, the Campy CVA agar appears to be a preferred medium for isolation of L. hongkongensis. In contrast, Woo et al. (2) and Lau et al. (10) isolated L. hongkongensis from stools on charcoal cefoperazone deoxycholate agar under microaerophilic conditions and Lau et al. (10) and Feng et al. (3) isolated L. hongkongensis on cefoperazone MacConkey agar in ambient air. Yuen et al. also reported growth of L. hongkongensis on MacConkey agar in a microaerophilic or anaerobic environment at 25 and 42°C but not at 4, 44, and 50°C (1). We were able to grow L. hongkongensis in ambient air at 35°C. We found the use of Campy CVA under either microaerophilic conditions or ambient air to be helpful for the cultivation of this organism because, unlike MacConkey agar, Campy CVA is also inhibitory to normal enteric flora.
Experiments that used adult zebrafish as an animal model linked L. hongkongensis to pathological changes in the liver (11). This was also observed in the two cases in which L. hongkongensis was isolated from blood cultures from cirrhotic patients, suggesting chronic liver disease as a possible risk factor for invasive L. hongkongensis infections. One fatal case of L. hongkongensis bacteremia has been reported in China in a patient with colonic carcinoma that metastasized to the liver (12).
While susceptibility testing was not performed on our isolates, a literature review was performed. The resistance of L. hongkongensis to antimicrobials is vast, as it is resistant to most β-lactams, including broad-spectrum penicillin and cephalosporin, due to a chromosomal class C β-lactamase (13). The emergence of multidrug-resistant L. hongkongensis was found in isolates from bighead carp that were resistant to at least three classes of antimicrobials. Two isolates from non-wide Chinese tiger frogs and one from Guenther's frog were resistant to 7 and 8 classes of antimicrobials, respectively (3). Tetracycline resistance was detected in 12.5% of human isolates and 4.2% of fish isolates (14). Naladixic acid resistance was observed in 13.4% of isolates from grass carp and 70.7% of isolates from Chinese tiger frogs. Thirty-two percent of the isolates from the Chinese tiger frogs were also resistant to ciprofloxacin (15).
Antibiotic treatment is not recommended for patients with self-limiting disease. However, in immunocompromised patients, antibiotic treatment may be necessary, and these findings show that it may be difficult to predict which antibiotic to use without susceptibility testing.
The failure to recognize L. hongkongensis from human stool samples is likely due to a combination of the organism not being tested for and misidentification by phenotypic methods. Microbiology laboratories that identify a Gram-negative rod with a profile number of 201004 or 200004 on bioMérieux API 20E or biotype number 42000006 on MicroScan combo panels should consider the organism suspicious for L. hongkongensis. Others have reported misidentification of Laribacter as A. lwoffii with a confidence level of 94% by Vitek 2 with the ID-GNB card (8). Laboratories are warned that current phenotypic methods are insufficient to identify L. hongkongensis, and we recommend that curved gram-negative rods recovered from stool samples that give phenotypic identifications as mentioned above be tested further by either the Bruker Daltonik Biotyper RUO MALDI-TOF MS or sequencing in order to confirm the identification of L. hongkongensis.
REFERENCES
- 1.Yuen KY, Woo PC, Teng JL, Leung KW, Wong MK, Lau SK. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel Gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J Clin Microbiol 39:4227–4232. doi: 10.1128/JCM.39.12.4227-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PC, Kuhnert AP, Burnens AP, Teng JL, Lau SK, Que TL, Yau HH, Yuen KY. 2003. Laribacter hongkongensis: a potential cause of infectious diarrhea. Diagn Microbial Infect Dis 47:551–556. doi: 10.1016/S0732-8893(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 3.Feng JL, Hu J, Lin JY, Liu S, Chowdhury N, Zhang O, Li JD, Shi L, Yamasaki S, Chen Q. 2012. The prevalence, antimicrobial resistance and PFGE profiles of Laribacter hongkongensis in retail freshwater fish and edible frogs of southern China. Food Microbiol 32:118–123. doi: 10.1016/j.fm.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Lau SK, Woo PC, Fan RY, Ma SS, Hui WT, Au SY, Chan LL, Chan JY, Lau AT, Leung KY, Pun TC, She HH, Wong CY, Wong LL, Yen KY. 2007. Isolation of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis, from drinking water reservoirs in Hong Kong. J Appl Microbiol 103:507–515. doi: 10.1111/j.1365-2672.2006.03263.x. [DOI] [PubMed] [Google Scholar]
- 5.Woo PC, Lau SK, Teng JL, Que TL, Yung RW, Luk WK, Lai RW, Hui WT, Wong SS, Yau HH, Yuen KY. 2004. Association of Laribacter hongkongensis in community-acquired human gastroenteritis with travel and with eating fish: a multicentre case-control study. Lancet 363:1941–1947. doi: 10.1016/S0140-6736(04)16407-6. [DOI] [PubMed] [Google Scholar]
- 6.Woo PC, Lau SK, Teng JL, Yuen KY. 2005. Current status and future directions for Laribacter hongkongensis, a novel bacterium associated with gastroenteritis and traveller's diarrhoea. Curr Opin Infect Dis 18:413–419. doi: 10.1097/01.qco.0000180162.76648.c9. [DOI] [PubMed] [Google Scholar]
- 7.Ni XP, Ren SH, Sun JR, Xiang HQ, Gao Y, Kong QX, Cha J, Pan JC, Yu H, Li HM. 2007. Laribacter hongkongensis isolated from a patient with community-acquired gastroenteritis in Hangzhou City. J Clin Microbiol 45:255–256. doi: 10.1128/JCM.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DS, Wi YM, Choi JY, Peck KR, Song JH, Ko KS. 2011. Bacteremia caused by Laribacter hongkongensis misidentified as Acinetobacter lwoffii: report of the first case in Korea. J Korean Med Sci 26:679–681. doi: 10.3346/jkms.2011.26.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raja MK, Ghosh AR. 2014. Laribacter hongkongensis: an emerging pathogen of infectious diarrhea. Folia Microbiol 59:341–347. doi: 10.1007/s12223-013-0299-6. [DOI] [PubMed] [Google Scholar]
- 10.Lau SK, Woo PC, Hui WT, Li MW, Teng JL, Que TL, Luk WK, Lai RW, Yung RW, Yuen KY. 2003. Use of cefoperazone MacConkey agar for selective isolation of Laribacter hongkongensis. J Clin Microbiol 41:4839–4841. doi: 10.1128/JCM.41.10.4839-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie J, He J-B, Shi J-W, Xiao Q, Li L, Woo PCY. 2014. An adult zebrafish model for Laribacter hongkongensis infection: Koch's postulates fulfilled. Emerg Microbes Infect 3:e73. doi: 10.1038/emi.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cindy WS, Curreem SO, Cheung I, Tang BS, Leung KW, Lau SK, Woo PC. 2014. A novel MLST sequence type discovered in the first fatal case of Laribacter hongkongensis bacteremia clusters with the sequence types of other human isolates. Emerg Microbes Infect 3:e41. doi: 10.1038/emi.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau SK, Ho PL, Li MW, Tsoi HW, Yung RW, Woo PC, Yuen KY. 2005. Cloning and characterization of a chromosomal class C β-lactamase and its regulatory gene in Laribacter hongkongensis. Antimicrob Agents Chemother 49:1957–1964. doi: 10.1128/AAC.49.5.1957-1964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau SK, Wong GK, Li MW, Woo PC, Yuen KY. 2008. Distribution and molecular characterization of tetracycline resistance in Laribacter hongkongensis. J Antimicrob Chemother 61:488–497. doi: 10.1093/jac/dkm539. [DOI] [PubMed] [Google Scholar]
- 15.Chen DQ, Yang L, Luo YT, Mao MJ, Lin YP, Wu AW. 2013. Prevalence and characterization of quinolone resistance in Laribacter hongkongensis from grass carp and Chinese tiger frog. J Med Microbiol 62:1559–1564. doi: 10.1099/jmm.0.059451-0. [DOI] [PubMed] [Google Scholar]