Abstract
Successful sequencing of the Clostridium difficile genome requires high-quality genomic DNA (gDNA) as the starting material. gDNA extraction using conventional methods is laborious. We describe here an optimized method for the simple extraction of C. difficile gDNA using the QIAamp DNA minikit, which yielded high-quality sequence reads on the Illumina MiSeq platform.
TEXT
Advances in the epidemiology and pathogenesis of Clostridium difficile infection have been progressing at a rapid pace with the advent of high-throughput whole-genome sequencing (WGS) (1–6). Obtaining high-quality, large-molecular-weight genomic DNA (gDNA) is the first step toward a successful WGS run, as it will have an impact on the quality of the sequence reads (7, 8; for submission requirements, see http://mendel.stanford.edu/SSC/howto.html). A simple method for gDNA extraction from C. difficile has not been described. The use of a phenol-chloroform-isoamyl alcohol extraction method yields high-quality gDNA, but it is time consuming and technically demanding (10). In this report, we describe an optimized protocol for extraction of gDNA from C. difficile that we developed using the QIAamp DNA minikit (Qiagen, Germantown, MD).
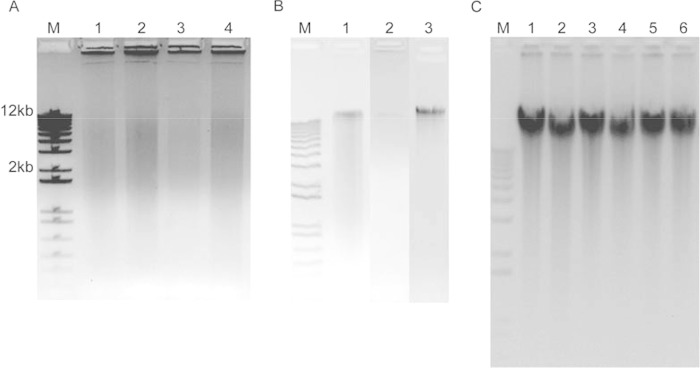
The gDNA from C. difficile was first extracted using each of the three existing protocols in the QIAamp handbook (11), namely (i) the Gram-positive bacteria (GP) protocol, (ii) the bacterial plate cultures (PC) protocol, and (iii) the bacterial suspension cultures (SC) protocol. For the GP and SC protocols, bacterial density in Trypticase soy broth (TSB) was adjusted to a 0.5 to 1 McFarland standard, using colonies from brucella blood agar, and incubated overnight at 35°C under anaerobic conditions. For the PC protocol, a single colony was suspended in 180 μl of ATL buffer and vortexed. As shown in Fig. 1A, no distinct bands were observed with the GP protocol. A smear was seen on the gel, suggesting shearing of the DNA. Similarly, no gDNA was obtained using the PC protocol (data not shown). Isolation of gDNA using the SC protocol yielded a very faint large-molecular-weight band. However, significant smearing was present (Fig. 1B), and the amount of gDNA obtained was low and variable, ranging from 0.5 to 2.5 μg.
FIG 1.
Gel electrophoresis results for genomic DNA (gDNA) extracted from Clostridium difficile using three different QIAamp protocols. Gram-positive protocol (A), lanes 1 to 4: gDNA extracts from isolates 38A2 (lane 1 and 2) and 69B1 (lane 3 and 4). Each lane shows an independent extraction. Bacterial suspension protocol (B), lanes 1 to 3: gDNA extracts from isolate 38A2 obtained in three independent extractions. Optimized protocol (C), lanes 1 to 6: gDNA extracts from isolates 127A4, 161A1, 208A2, 208A3, 224C1, and 224A3, respectively. M, molecular-weight marker.
Given that the SC protocol was the most promising for the extraction of gDNA from C. difficile, we conducted a series of experiments to optimize this protocol through experimentation with different variables, such as culture inoculum, culture volume, culture incubation duration, degree of homogenization of the bacterial pellet in the kit reagent, duration of lysis with proteinase K, and frequency of vortexing during the lysis step (data not shown). In the final version of the optimized protocol, colonies from the brucella agar were inoculated into 12 ml of TSB broth, and the optical density was adjusted to a McFarland standard of 3 to 4. Cultures were incubated anaerobically at 35°C for 48 h. The cultures were then sedimented at 3,000 × g for 15 min. The supernatant was gently decanted without disturbing the sediment. Any residual broth was gently aspirated and discarded. A total of 180 μl of ATL buffer was added, ensuring that the sediment was completely resuspended by mixing the contents using a micropipette. Care was taken not to form excessive frothing during resuspension of the pellet. The suspension was transferred to a 1.5-ml microcentrifuge tube, and subsequent steps were performed according to the instructions in the handbook, Protocol: DNA purification from tissues, starting at step 3 with some modifications. The tube was incubated at 56°C for 1 h, and the content was vortexed for 15 s at 10-min intervals instead of 2 to 3 times as indicated in the handbook. The use of RNase A was omitted in our protocol. The DNA was eluted from the QIAamp minispin column in 50 μl of buffer AE in two steps, achieving a final volume of 100 μl. As shown in Fig. 1C, the optimized protocol yielded large-fragment gDNA of more than 12 kbp in size. The bands appeared sharp with minimal smearing, suggesting the absence of degradation or shearing of gDNA during the extraction procedure. The quantity of gDNA obtained was consistently above 10 μg, with an A260/A280 ratio between 1.80 and 1.85 (Table 1).
TABLE 1.
Recovered quantity of gDNA and A260/A280 absorbance ratio for gDNA extracted using the optimized protocol
| Strain no. | Quantity (μg) | A260/A280 ratio |
|---|---|---|
| 127A4 | 18.7 | 1.81 |
| 161A1 | 15.8 | 1.81 |
| 208A2 | 17.2 | 1.81 |
| 208A3 | 13.1 | 1.80 |
| 224C1 | 15.6 | 1.81 |
| 224A3 | 14.6 | 1.85 |
To test the quality of gDNA obtained with the optimized protocol for downstream WGS, we performed WGS on six isolates using the standard Illumina Trueseq DNA HT sample prep protocol and the MiSeq reagent kit v3 for 300-bp paired-end reads on the Illumina MiSeq platform (Illumina, San Diego, CA) (12, 13). Adapters and barcodes were trimmed per the default setting in the Illumina experiment manager, generating 301-bp reads. The quality of the unprocessed reads was assessed using FastQC (Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom [http://www.bioinformatics.babraham.ac.uk/projects/fastqc/]) (15). Result showed that all reads achieved a sequence length of 301 bp for all samples without any overrepresented sequences (data not shown). The median per-base sequence quality was more than 34 (Illumina 1.9 encoding) from base-pair position 1 to 299.
This study shows that none of the three existing protocols in the handbook yielded satisfactory gDNA extracts. In contrast, the optimized protocol consistently produced high-quality and high-quantity gDNA. The total estimated amount of gDNA generally exceeded 10 μg, which is more than sufficient for paired-end sequencing on most next-generation sequencing platforms (7, 8; see also http://mendel.stanford.edu/SSC/howto.html). The optimized protocol also provides an environmentally friendly alternative to phenol-chloroform extraction, with the hands-on time for processing a batch of 12 samples of approximately 3 h. Analysis of the WGS sequence data obtained on the Illumina MiSeq platform confirmed that the gDNA was of adequate quality, with all reads achieving the desired length and satisfactory quality scores across all bases. There were no overrepresented sequences, indicating the absence of adaptor contamination and the adequate size of the gDNA for generation of inserts with a length sufficient for 300-bp paired-end sequencing (15, 16).
There are several plausible explanations for the high-quality and high-quantity gDNA yield from the modifications in the optimized protocol. First, a higher inoculum, consisting of a 3 to 4 McFarland standard, in a larger culture volume (12 ml) means more bacteria were used for extraction. Given that TSB does not enhance the growth of C. difficile (data not shown), incubating the bacteria in TSB broth likely facilitated the downstream resuspension of C. difficile in the lysis buffer. Further, the broth culture likely kept the bacteria in a vegetative state versus spores, which may have aided with lysis. Additional steps that may have contributed to the effective lysis of cells include thorough homogenization of the pellet and frequent vortexing at 10-min intervals during the lysis step.
In conclusion, we described a simple and rapid protocol for the QIAamp DNA minikit to extract high-quality and high-quantity gDNA from C. difficile for downstream WGS application.
ACKNOWLEDGMENT
This study was supported by an investigator-initiated grant from Merck & Co. to N.B.
REFERENCES
- 1.Darling AE, Worden P, Chapman TA, Roy Chowdhury P, Charles IG, Djordjevic SP. 2014. The genome of Clostridium difficile 5.3. Gut Pathog 6:4. doi: 10.1186/1757-4749-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre DW, Griffiths D, Vaughan A, Golubchik T, Acharya M, O'Connor L, Crook DW, Walker AS, Peto TE. 2013. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS One 8:e78445. doi: 10.1371/journal.pone.0078445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim SK, Stuart RL, Mackin KE, Carter GP, Kotsanas D, Francis MJ, Easton M, Dimovski K, Elliott B, Riley TV, Hogg G, Paul E, Korman TM, Seemann T, Stinear TP, Lyras D, Jenkin GA. 2014. Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis 58:1723–1730. doi: 10.1093/cid/ciu203. [DOI] [PubMed] [Google Scholar]
- 4.Eyre DW, Babakhani F, Griffiths D, Seddon J, Del Ojo Elias C, Gorbach SL, Peto TE, Crook DW, Walker AS. 2014. Whole-genome sequencing demonstrates that fidaxomicin is superior to vancomycin for preventing reinfection and relapse of infection with Clostridium difficile. J Infect Dis 209:1446–1451. doi: 10.1093/infdis/jit598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, Griffiths D, Vaughan A, O'Connor L, Golubchik T, Batty EM, Piazza P, Wilson DJ, Bowden R, Donnelly PJ, Dingle KE, Wilcox M, Walker AS, Crook DW, Peto TE, Harding RM. 2012. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 13:R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broad Institute of MIT and Harvard. 2015. Sample quantity and quality specifications. Broad Institute, Cambridge, MA: https://www.broadinstitute.org/scientific-community/science/platforms/genomics/sample-quality-and-quantity-specifications. [Google Scholar]
- 8.Wellcome Trust Sanger Institute. DNA requirements for sequencing. Wellcome Trust Sanger Institute, Hinxton, United Kingdom: https://www.sanger.ac.uk/resources/technologies/dna_requirements_for_sequencing_june2010.pdf. [Google Scholar]
- 9.Reference deleted.
- 10.Bouillaut L, McBride SM, Sorg JA. 2011. Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol 9:Unit 9A.2. doi: 10.1002/9780471729259.mc09a02s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiagen. 2012. QIAamp DNA mini and blood mini handbook, 3rd ed, June 2012 Qiagen, Germantown, MD. [Google Scholar]
- 12.Illumina. 2012. TruSeq sample preparation guide, July 2012 Illumina, San Diego, CA. [Google Scholar]
- 13.Illumina. 2014. MiSeq reagent kit v3-PGS reagent preparation guide, revision A, June 2014 Illumina, San Diego, CA. [Google Scholar]
- 14.Reference deleted.
- 15.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. 2014. Three-stage quality control strategies for DNA resequencing data. Brief Bioinform 15:879–889. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]