Abstract
Recent evidence suggests that miR-520 family has an important role in regulating tumorigenesis and development of various types of solid cancers. However, as one of the most common cancers in the world, there is little known about the underlying regulatory mechanisms of miR-520 in colorectal cancer (CRC). In the present study, we investigated the expression of microRNA-520d-5p (miR-520d-5p) in CRC specimens and then explored its potential role and mechanism in CRC progression. We found that miR-520d-5p was markedly down-regulated in CRC clinical specimens compared with adjacent normal tissues by real-time PCR. Dual-luciferase assays confirmed that miR-520d-5p directly targeting CTHRC1 and SP1 transactivate miR-520d-5p by binding to its upstream promoter region. The biological functional experiments showed that ectopic re-expression of miR-520d-5p suppressed CRC cell proliferation, migration and invasion, whereas the inhibition of miR-520d-5p displayed an inverse effect in vitro and in vivo. Western blot shown that miR-520d-5p abrogated the epithelial-mesenchymal transition by inactivating the phosphorylation of Erk1/2. In conclusion, our findings indicate that miR-520d-5p is significantly down-expressed and involved in CRC progression and metastasis by targeting CTHRC1 and regulated by SP1, which provide new support for miR-520d-5p maybe as a novel anti-onco molecular target for the treatment of CRC in the future.
Keywords: Colorectal cancer, microRNA-520d-5p, CTHRC1, SP1, EMT, p-Erk1/2
Introduction
Colorectal cancer (CRC) is one of the most frequent malignant types of cancers and the fourth most common cause of cancer deaths worldwide [1,2]. Surgery with or without adjuvant radiation and chemotherapy treatments based on tumor stages have been recommended according to current guidelines. However, the clinical outcome and prognosis of colorectal cancer patients with advanced-stage disease remains poor. Like other solid cancers, the development of CRC is a multistep progression involving the activation of oncogenes and inactivation of tumor suppressor genes, which will affect all aspects of tumorigenicity of CRC, such as cell proliferation, apoptosis, invasion and metastasis [3]. The discovery of new molecular targets that regulate the initiation and progression of CRC will facilitate progress toward the eventual cure of this disease. However, because of the disease complexity, the specific molecular genetics and epigenetic alterations of CRC remain largely unknown.
MicroRNAs (miRNAs) are small (19-25 nucleotides), noncoding, regulatory RNAs that can negatively regulate gene expression by complementary base pairing with the 3’-untranslated region (UTR) of target messenger RNAs (mRNAs), leading to their degradation or repressing mRNA translation [4,5]. MiRNA deregulation is related to the pathogenesis of a wide range of diseases [6,7]. For cancer in particular, miRNAs can function as either oncogenes or tumor suppressors by pairing with the corresponding mRNA of the target anti-oncogene or oncogene in different types of cancer [8,9]. For example, miR-26a and miR-126 inhibit tumor growth and metastasis [10,11], but miR-181a and miR-224 are up-regulated in CRC tissue compared with adjacent non-tumor tissue [12,13]. It was reported that the miR-520 family acts as anti-onco-miRNAs by involvement in regulating the TGF-β and NF-κB signaling pathways in breast cancer [14]. As a member of the miR-520 family, miR-524 has been reported to act as a cancer suppressor in glioma [15]. However, little is known about the role of the miR-520 family in CRC.
CTHRC1 (collagen triple helix repeat containing 1) encodes a secreted glycoprotein with 12 repeats of the Gly-X-Y motif and can increase cell motility by limiting the deposition of collagen matrix and promoting cell migration [16]. CTHRC1 is usually up-regulated in many human solid tumors, including non-small-cell lung cancer [17], gastric cancer [18], hepatocellular cancer [19], breast cancer [20], pancreatic cancer [21] and CRC [22,23]. Meanwhile, CTHRC1 was confirmed that it it is associated with peritoneal carcinomatosis and may act as a new predictor of survival in our previous work by gene microarray [23]. Some recent studies have indicated that microRNA can regulate CTHRC1 at a post-transcriptional level. For example, miR-9 can inhibit Schwann cell migration by targeting CTHRC1 following sciatic nerve injury [24].
In this study, we showed that miR-520d-5p expression was negatively associated with CTHRC1 in CRC tissue and cell lines. By using gain-of and loss-of function studies, we confirmed the tumor suppressor function of miR-520d-5p, both in vitro and in vivo. Mechanically, miR-520d-5p directly targeted CTHRC1 and could be regulated by specificity protein 1 (SP1). These findings suggest that treatments involving miR-520d-5p have potential therapeutic implications for the cure of CRC.
Materials and methods
Clinical specimens and cell lines
Human CRC specimens were collected at Nanfang Hospital, Southern Medical University (Guangzhou, China), with written consent. Surgically removed tissues were immediately frozen in liquid nitrogen and stored at -80°C. Approval for these studies was obtained from the Southern Medical University Institutional Board. Seven colorectal cancer lines - HCT116, DLD-1, LS174T, HT29, SW480, SW620 and LoVo - were purchased from the American Type Culture Collection (ATCC) Cell Biology Collection and authenticated according to the recommendation. All cells were cultured in RMPI-1640 medium containing 10% FBS (Gibco) in a 5% CO2 container at 37°C.
MicroRNA mimics, siRNA transient transfection
MicroRNA mimics, miR-520d-5p inhibitor and SP1 siRNA were purchased from GenePharma (Suzhou, China). When cells covered 60% confluency, 100 pmol miR-520d-5p mimics or inhibitor or siRNA of SP1 were transfected by Lipofectamine2000 (Invitrogen) in Opti-MEM (Invitrogen, Carlsbad, CA, USA). The sequences of siRNAs were as follows: SP1 siRNA: sense: 5-GGAUGGUUCUGGUCAAAUATT-3 antisense: 5-UAUUUGACCAGAACCAUCCTT-3; miR-520d-5p mimics - sense: 5-CUACAAAGGGAAGCCCUUUC-3 antisense: 5-AAGGGCUUCCCUUUGUAGUU-3; negative control - sense: 5-UUCUCCGAACGUGUCACGUTT-3 antisense: 5-ACGUGACACGUUCGGAGAATT-3; miR-520d-5p inhibitor: 5-GAAAGGGCUUCCCUUUGUAG-3; inhibitor negative control: 5-CAGUACUUUUGUGUAGUACAA-3 After 24-48 h of transfection, the cells were harvested for further investigation.
RNA isolation and real-time RT-PCR
Total RNA was extracted from tissues or cells using Trizol regent (Invitrogen, Carlsbad, CA, USA). For miR-520d-5p detection, reverse transcription-polymerase chain reaction (RT-PCR) was performed using an All-in-OneTM miRNA quantitative RT-PCR (qRT-PCR) Detection Kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s instructions. A Reverse Transcriptase System and SYBR Green qPCR master mix (Takara, Dalian, China) was used to detect CTHRC1 and SP1 mRNA expression. U6 and GAPDH were used as references for miRNA and mRNA, respectively. Each sample was analyzed in triplicate. The 2-ΔΔCt method was used to quantify the relative levels of gene expression.
Western blotting
Total cell lysates were extracted using a RIPA lysis buffer and measured using a bicinchoninic acid (BCA) protein assay (Beyotime Institute of Biotechnology, Shanghai, China). Cell lysates (30 μg) were mixed with corresponding 5× loading buffer (Beyotime Institute of Biotechnology, Shanghai, China) and then denatured at 100°C for 10 min. Next, the lysate was separated on 10% SDS polyacrylamide gel, transferred to PVDF (polyvinylidene difluoride) membranes and blocked in 5% nonfat dry milk in Tris-buffered saline (TBS). Proteins were incubated with rabbit anti-CTHRC1 polyclonal antibody (1:1000, Proteintech, IL, USA), rabbit anti-SP1 polyclonal antibody (1:1000, Proteintech, IL, USA), rabbit anti-E-cadherin polyclonal antibody (1:1000, Cell Signaling, Danvers, MA, USA), rabbit anti-N-cadherin polyclonal antibody (1:500, Bioworld, MN, USA), rabbit anti-vimentin polyclonal antibody (1:500, Bioworld, MN, USA), rabbit anti-Erk1/2 polyclonal antibody (1:500, Bioworld, MN, USA) and rabbit anti-p-Erk1/2 polyclonal antibody (1:500, Bioworld, MN, USA) overnight at 4°C. The membranes were washed several times, followed by application of the appropriate second antibody. Protein bands were visualized using enhanced chemiluminescence (Beyotime Institute of Biotechnology, Shanghai, China).
Luciferase activity assay
To reveal the bind between miR-520d-5p and CTHRC1, the 3’UTR of CTHRC1 segment was amplified by polymerase chain reaction (PCR) and inserted into the pmirGLO vector. The mutant of the seed region of the putative miR-520d-5p binding sites in the CTHRC1 3’UTR was also developed using a QuikChange Site-Directed Mutagenesis Kit (Agilent, Roseville, CA, USA). Co-transfections of CTHRC1 3’UTR or Mut-CTHRC1 3’UTR plasmid with miR-520d-5p mimics were obtained using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA). Similarly, a 185 bp region containing the putative SP1-binding sites upstream of the miR-520d-5p putative transcription binding site and three mutant constructs were generated. Twenty-four hours after transfection of 40 pmol SP1 siRNA, the wt- and Mut-miR-520d-5p promoter vectors were transfected into the cells using Lipofectamine2000 (Invitrogen). Luciferase activity was measured 48 h after transfection by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Each assay was repeated in three independent experiments.
Plate clone formation assay
Cells (5×102/well) were seeded into six-well plates in triplicate. After incubation at 37°C for 14 days, the cells were washed twice with PBS and stained with 0.5% crystal violet solution. The number of colonies containing ≥ 50 cells was counted under a microscope using the formula: plate clone formation efficiency = (number of colonies/number of cells inoculated) ×100%.
CCK8 cell proliferation assay
Cells transfected with miRNA mimics/inhibitor or control siRNA (3×103/well) were seeded into a 96-well plate. On days 1, 2, 3, 4 and 5, 10 μl CCK8 solution (KeyGEN BioTECH, Nanjing, China) was added to each well, then the plate was incubated for 2 h at 37°C. Next, the absorbance was measured at 450 nm using a Vmax microplate spectrophotometer (Molecular Devices, Sunnyvale, CA). Each sample was assayed in triplicate. The data were plotted as means ± SD in three separate experiments.
Cell invasion and migration assay
A total of 2×105 cells were plated in the top chamber onto the Matrigel-coated membrane (24-well insert; pore size 8 μm; Corning Costar, NY, USA). Each well was coated freshly with Matrigel (60 μg; BD Bioscience, NJ, USA) before the invasion assay. Next, 600 μl 10% FBS media were added to the lower chamber. After incubation for 36 h, the cells that had not migrated through the pores were removed with a cotton swab. Cells that had migrated through the membrane and stuck to the lower membrane surface were fixed with methanol, stained with 0.5% crystal violet. Similar to the migration procedure, 2×105 cells were added to the upper chamber of the non-coated membrane (24-well insert; pore size 8 μm; Corning Costar, NY, USA), and 10% FBS medium in the lower chamber was used to induce the migration of the cells. After incubation for 24 h, the cells on the lower membrane were fixed with methanol and also stained with 0.5% crystal violet. The number of cells that had migrated through the membrane were counted under a light microscope (three random fields per well).
In vivo tumor growth assay
To conduct animal tumorigenesis assay, LoVo cells were infected with hsa-miR-520d-5p lentivirus (GenePharma, Suzhou, China). Four-to six-week-old BALB/c athymic nude mice were obtained from the Central Laboratory of Animal Science at Southern Medical University. All procedures involving animals and their care in this study were approved and performed by the Southern Medical University Institutional Animal Care and Use Committee. Each nude mouse was subcutaneously injected with 3×106 transfected LoVo cells. Ten days after injection, the tumor size was repeatedly measured with a vernier caliper every three days. The mice were killed on day 22 and the tumors were taken to perform H&E staining, immunohistochemistry (IHC), WB and qRT-PCR analyses. The tumor volume was calculated by the formula: V = long diameter × short diameter2 ×1/2.
Statistical analyses
All statistical data were analyzed using SPSS13.0 for Windows. All data were expressed as mean ± standard deviation (SD). Student’s t-test or analyses of variance were conducted to analyze these experiments in vitro or in vivo. Paired-samples t test was conducted to analyze the expression of miR-520d-5p in paired CRC specimens. Spearman’s correlation analyses were used to analyze the relationship between miR-520d-5p and CTHRC1 or SP1 mRNA expression. The data of colony formation assay, Transwell migration and invasion assay and relative luciferase activities were analyzed through One-way ANOVA and two-tailed independent-samples t test. The factorial design analysis of variance was applied to analyze the CCK8 assays. Variance analysis of repeated measurement data was adopted to analyze the in vivo tumor growth assay. The results were considered statistically significant when P < 0.05.
Results
MiR-520d-5p is significantly down-regulated in colorectal cancer compared with adjacent normal tissue
The expression of miR-520d-5p was detected in 42 paired CRC and adjacent noncancerous tissue biopsies by real-time PCR. As shown in Figure 1A, miR-520d-5p expression was down-regulated in 36 of 42 (85.7%) colorectal cancer tissues compared with their nontumorous counterpart from the same patients (Figure 1A). Moreover, Student’s t-tests showed that the expression of miR-520d-5p in CRC tissues was significantly lower than that in the paired noncancerous tissues (P = 0.004, Figure 1B). In addition, the relative expression of miR-520d-5p in low metastatic potential CRC cell lines SW480 and HCT116 was significantly higher than the expression in high metastatic CRC cell lines SW620 and LoVo (Figure 1C).
Figure 1.
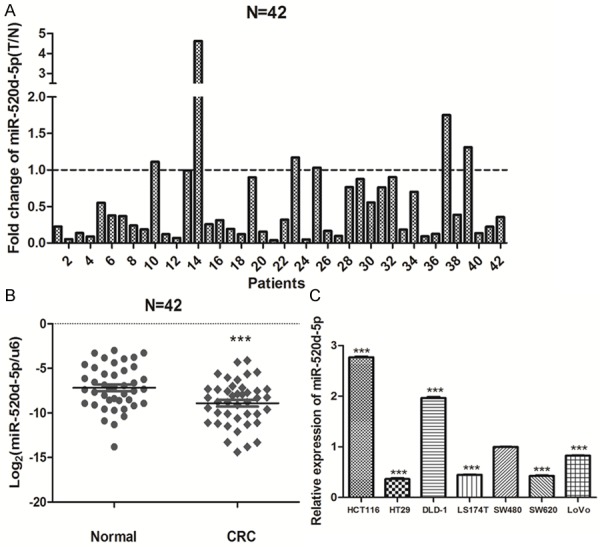
MiR-520d-5p is down-expressed in colorectal cancer. A. Real-time PCR analysis of miR-520d-5p in 42 paired human colorectal cancer tissues and the matched adjacent noncancerous normal tissues. B. Comparison of miR-520d-5p abundance in 42 paired CRC tissues (CRC) with the matched adjacent normal tissue (Normal). C. Real-time PCR analysis of miR-520d-5p in seven human colorectal cancer cell lines. Experiments were performed three times. Data, mean ± SD. **P < 0.01; ***P < 0.001.
MiR-520d-5p represses the proliferation, migration and invasion of CRC cells in vitro
To investigate the proliferation, migration and invasion ability of miR-520d-5p, we conducted transient transfection with mimics, inhibitor of miR-520d-5p and their respective NC counterpart. CCK8 assay was used to detect the effect of transfection of mimics or inhibitor. As shown in Figure 2A, the proliferation ability was markedly reduced in SW480, SW620 and LoVo cells when miR-520d-5p was over-expressed compared with NC groups. However, the miR-520d-5p inhibitor group notably increased the proliferation ability of HCT116 cells compared with the NC group (P < 0.05, Figure 2A). Moreover, the colony form test was performed to assess the long-term impact on cell growth of transfection. The transient expression of miR-520d-5p inhibited the colony formation; in contrast, repression of the miR-520d-5p augmented the colony formation of HCT116 cells. A one-way ANOVA test revealed that the colony numbers and colony formation rate differed markedly (P < 0.01, Figure 2B). Such evidence indicated that miR-520d-5p could impede the growth of CRC cells.
Figure 2.
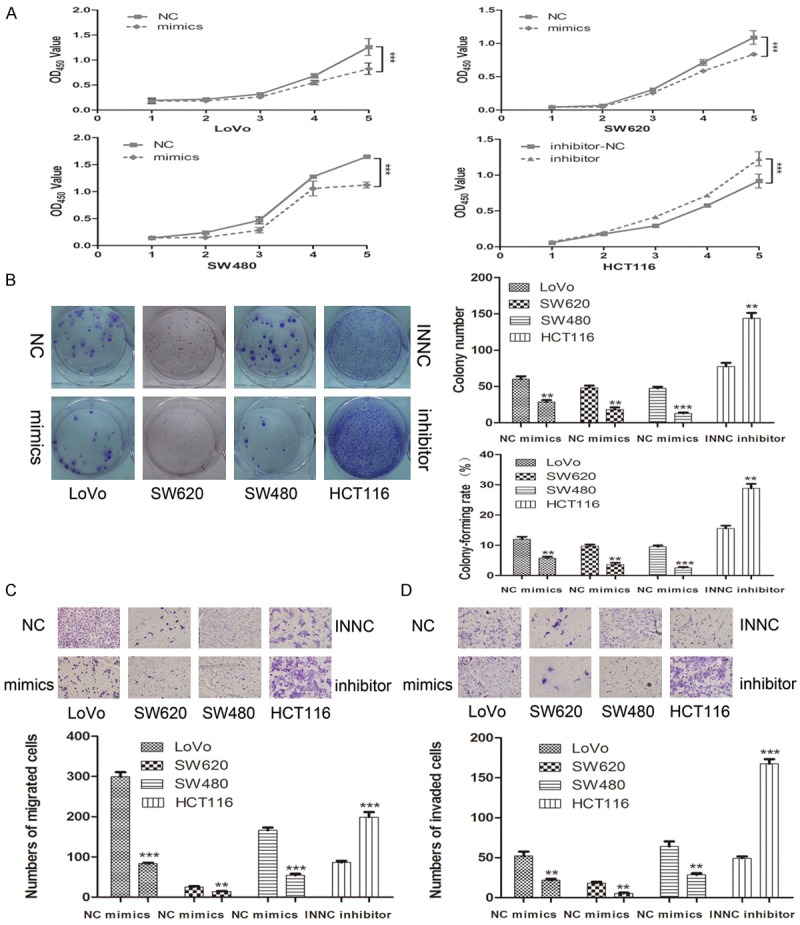
MIR-520d-5p suppresses CRC growth and metastasis in vitro. (A, B) Up-regulation of miR-520d-5p repressed CRC cell proliferation by CCK-8 assay (A) and colony formation assay (B); inhibition of miR-520d-5p exerted the opposite effects. (C, D) Impact of miR-520d-5p mimics and inhibitor on cell migration (C) and invasion (D) through a transwell chamber. Three independent experiments were performed. **P < 0.01; ***P < 0.001.
A transwell test was performed to research the different migration and invasion ability between the group of over-expressed or suppressed miR-520d-5p and their respective NC group. As shown in Figure 2C and 2D, ectopic expression of miR-520d-5p inhibited cell migration and invasion; however, silencing of miR-520d-5p in HCT116 cells promoted migration and invasion (P < 0.01). Therefore, miR-520d-5p was shown to dramatically decrease the migration and invasion ability of CRC cells.
CTHRC1 is a direct target of miR-520d-5p
To study the regulatory mechanism of miR-520d-5p in CRC progression and metastasis, we used five microRNA bioinformatic databases (miRBase, microRNAorg, miRWalk, TargetScan and PicTar) to determine potential study targets. Among these, we chose CTHRC1 as the next focus of our study because CTHRC1 has been reported to be associated with peritoneal carcinomatosis and may act as a new predictor of survival in our previous work by gene microarray in the CRC models of peritoneum metastasis [23]. Two putative binding sites were found using the TargetScan database; Figure 3A shows the more likely putative binding sequences.
Figure 3.
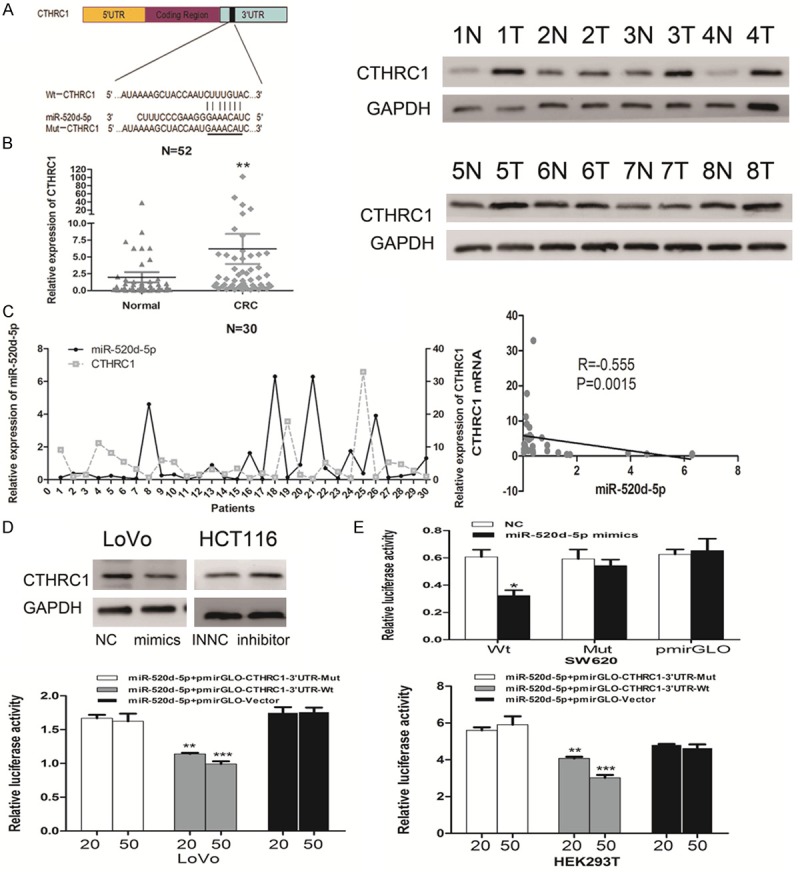
CTHRC1 is a direct target of miR-520d-5p. A. Predicted miR-520d-5p binding sequences in the 3’UTR of CTHRC1. The sequences of miR-520d-5p and wild-type (Wt) or mutant (Mut) putative target sites were compared in the 3’UTR of CTHRC1 mRNA. B. Expression of CTHRC1 in CRC tissues. (left) Real-time PCR analysis of CTHRC1 expression in 52 paired CRC tissues and the matched adjacent normal tissues. (right) Western blot analyses of CTHRC1 expression in eight paired CRC tissues. C. Inverse correlation between miR-520d-5p and CTHRC1 mRNA in 30 paired clinical tissue samples. D. The changes in CTHRC1 expression after transfection of miR-520d-5p mimics (left) and inhibitor (right) in CRC cell lines. E. Relative luciferase activity of the Wt-CTHRC1-3’UTR or Mut-CTHRC1-3’UTR or pmirGLO-vector after transfection with miR-520d-5p mimics and negative control in SW620 cells (top) or 20 and 50 pmol miR-520d-5p mimics in LoVo and HEK293 cells (bottom). Values, mean ± SD of three independent assays. *P < 0.05; **P < 0.01; ***P < 0.001.
The real-time PCR analysis of 52 paired CRC and adjacent noncancerous normal tissue specimens showed that the expression of CTHRC1 in CRC tissues was notably higher than in adjacent normal tissues. Consistently, similar results were found by Western blotting analyses (P = 0.0011, Figure 3B). Spearman correlation revealed that a significantly negative relationship existed between miR-520d-5p and CTHRC1 mRNA by real-time PCR analysis in 30 paired CRC and adjacent normal tissue biopsies (P = 0.0015, r = -0.555, Figure 3C).
In addition, a decreased expression of CTHRC1 was detected by Western blot analysis in LoVo cells transfected with miR-520d-5p mimics, compared with NC counterpart. In contrast, repressing miR-520d-5p strengthened CTHRC1 expression in HCT116 cells (Figure 3D).
Furthermore, to confirm the targeted binding between miR-520d-5p and the mRNA of CTHRC1, the entire 3’UTR of CTHRC1 mRNA was cloned into a pmirGLO vector. The dual-luciferase reporter assays revealed that miR-520d-5p mimics obviously decreased the relative luciferase activity of wild-type CTHRC1 3’UTR (Wt); however, no significant change was found in SW620 cells with mutant CTHRC1 3’UTR (Mut). Moreover, 50 pmol miR-520d-5p mimics exerted a lower relative luciferase activity than the 20 pmol group in LoVo and HEK293T cells (P < 0.05, Figure 3E). In summary, our findings revealed that miR-520d-5p reversely regulated CTHRC1 by directly targeting its mRNA 3’UTR.
Over-expression of miR-520d-5p suppresses CRC cell growth in vivo
To explore the effect of miR-520d-5p on CRC cell growth in vivo, the stable transfection cell lines LoVo/miR-520d-5p and LoVo/vector were injected subcutaneously into the nude mice. We found that the growth rate of LoVo/miR-520d-5p was evidently slower than LoVo/vector after a serial observation of 22 days. The tumors in LoVo/miR-520d-5p grew more slowly than those in LoVo/vector (P = 0.013, Figure 4A). When the mice were sacrificed, the tumor tissues were used to conduct H&E and Ki-67 (proliferative index) staining, real-time PCR and Western blot tests. Real-time PCR showed a higher expression of miR-520d-5p in LoVo/miR-520d-5p tumor than in LoVo/vector tumor. Meanwhile, Western blotting revealed a repressed CTHRC1 expression in LoVo/miR-520d-5p tumor compared with LoVo/vector tumor (P < 0.01, Figure 4B). IHC staining confirmed that the tumors of the LoVo/miR-520d-5p group displayed much lower Ki-67 indexes than the tumors from the LoVo/vector group (Figure 4C). The results showed that over-regulated miR-520d-5p repressed the CRC growth in vivo by inhibition of CTHRC1.
Figure 4.
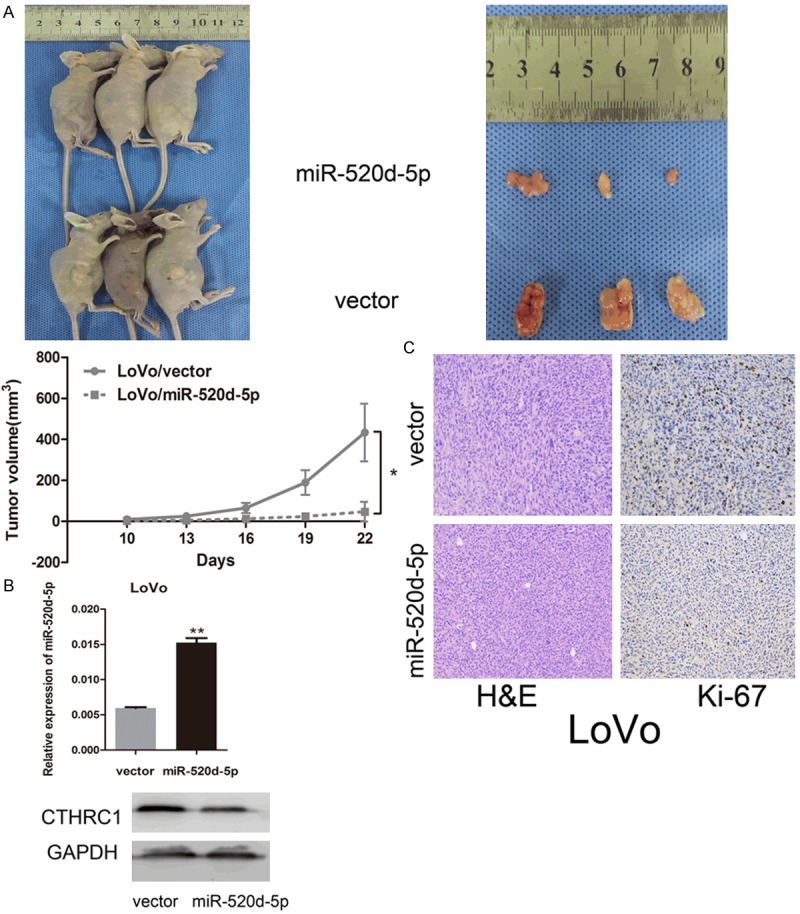
MIR-520d-5p suppresses the proliferation and tumorigenicity of colorectal cancer cells in vivo. A. Tumor xenograft model. Cells were injected into the hind limbs of nude mice (n=3). Data points are presented as the mean ± SD tumor volume. B. Real-time PCR analysis of miR-520d-5p (top) and Western blot analysis of CTHRC1 (bottom) in nude mice xenograft tumor tissue. C. Histopathology of xenograft tumors. H&E and IHC staining was performed on the tumor sections using antibody against Ki-67. *P < 0.05; **P < 0.01.
MiR-520d-5p abrogates EMT via inactivation of phosphorylation of Erk1/2
Previous studies showed that CTHRC1 could induce the expression of MMP9 via activation of Erk1/2 [25], thus promoting the epithelial mesenchymal transition (EMT) progression by cooperation with transcription factor Snail [26]. Therefore, we hypothesized that CTHRC1 might induce EMT by activating the phosphorylation of Erk1/2 in CRC. Western blotting was performed to investigate the potential role of miR-520d-5p in the signaling pathway. We found that re-expression of miR-520d-5p degraded the activation of phosphorylation Erk1/2 (p-Erk1/2), followed by an EMT progression reversion (mesenchymal epithelial transition): an enhanced E-cadherin expression and down-regulation of N-cadherin and vimentin. However, suppression of miR-520d-5p expression by transfection of miR-520d-5p inhibitor reinforced the phosphorylation of Erk1/2, with no marked change in total Erk1/2 expression level. Meanwhile, EMT marker molecules were induced in HCT116 cell lines (Figure 5).
Figure 5.
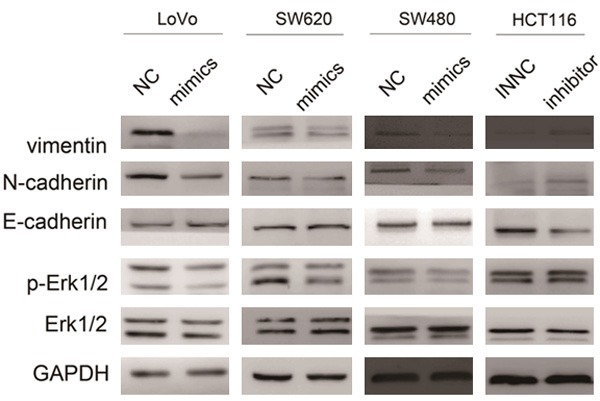
MIR-520d-5p abrogates the epithelial-mesenchymal transition (EMT) by inactivating the phosphorylation of Erk1/2. Western blot analyses of Erk1/2, p-Erk1/2, E-cadherin, N-cadherin and vimentin were conducted after transfection of miR-520d-5p mimics or inhibitor into the cells as indicated.
MiR-520d-5p is directly regulated by the transcription factor SP1
As a common gene, microRNA was transcribed as pri-miR by transcription factors and then spliced as mature microRNA. By Analyzing the 2 kp sequences of miR-520d upstream with some bioinformatics tools (Ensembl, UCSC, Transfac, TFSEARCH and MAPPER2.0), we found three SP1 putative binding sites (A, B, C were used to refer to the three sites) from -368 to -204 bp above the transcription start site (TSS) of miR-520d-5p (Figure 6A).
Figure 6.
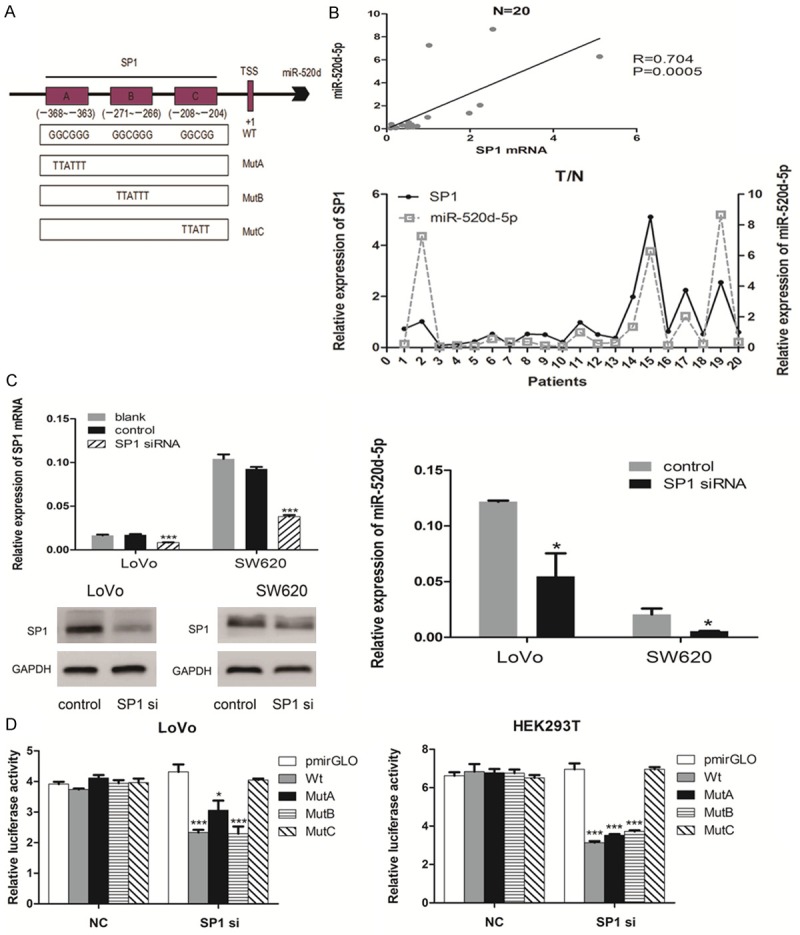
Specificity Protein 1 (SP1) transactivates miR-520d-5p by binding to its promoter. A. The promoter region structure of putative SP1 TFBS and the wild-type (Wt) and mutant (MutA, MutB and MutC) luciferase reporter vectors. B. A positive correlation existed between SP1 mRNA and miR-520d-5p. (top) Spearman correlation analysis showed a significantly positive relationship between SP1 mRNA and miR-520d-5p. (bottom) Real-time PCR analyses of the expression of SP1 mRNA and miR-520d-5p in 20 paired human colorectal cancer and adjacent normal tissue samples. C. Down-regulation of SP1 reduced the expression of miR-520d-5p. (left) Real-time PCR and Western blot analyses of the expression of SP1 after transfection with SP1 siRNA and negative control. (right) Real-time PCR analysis of the expression of miR-520d-5p after transfection. D. The relative luciferase activity of the indicated promoter vectors after transfection with SP1 siRNA or NC in LoVo (left) and HEK293T (right) cells. *P < 0.05; **P < 0.01; ***P < 0.001.
An evidently positive correlation was found between SP1 mRNA and miR-520d-5p by a real-time PCR analysis in 20 paired CRC and adjacent normal tissue specimens (P = 0.005, r = 0.704, Figure 6B). Moreover, qRT-PCR and Western blot analysis showed that the expression of miR-520d-5p was significantly reduced when SP1 was respectively inhibited by siRNA at mRNA and protein level (P < 0.05, Figure 6C).
A dual-luciferase reporter assay was performed to confirm the direct binding relationship between SP1 and the promoter of miR-520d-5p. The depletion of SP1 with siRNA suppressed the relative luciferase activity of miR-520d’s promoter wild-type vector (wt) compared with NC groups and blank-vector control (pmirGLO) groups in HEK293T and LoVo cell lines. The same result was observed when A or B was mutated alone (MutA or MutB), whereas no significant difference was found between C mutation (MutC) and NC groups (P < 0.05, Figure 6D). These experiments indicated that the C site may be the binding sequence of SP1 on miR-520d’s promoter.
Discussion
There is increasing evidence that microRNA plays a significant role in regulating proliferation, migration, invasion and metastasis in human solid carcinoma [8]. A recent study showed that miR-520c functions as a tumor suppressor in estrogen-receptor-negative breast cancer by targeting RELA and TGFBRII (transforming growth factor ß type II receptor) [27]. On the contrary, miR-520c was reported that it could promote tumor invasion and metastasis by targeting CD44 [28]. Moreover, a further study revealed that miR-520b was down-regulated in hepatocellular carcinoma tissue and could inhibit growth of hepatoma cells by targeting MEKK2 and Cyclin D1 [29,30]. Simultaneously, miR-520e can suppress growth of hepatoma cells by targeting the NF-κB-inducing kinase (NIK) [31]. From the evidence above, it can be concluded that the miR-520 family has a diverse and complicated effect on human solid tumors; this could be partly attributed to cancer heterogeneity. Current evidence indicates that miR-520d acts as a tumor suppressor: on the one hand, miR-520d-3p has been identified as a tumor suppressor upstream of EphA2 in ovarian cancer [32], while on the other hand, miR-520d can induce hepatoma cells to form normal liver tissues [33]. In the present study, we investigated the down-regulation in CRC tissues and cells, and provided new evidence that over-expression of miR-520d-5p can suppress the proliferation, migration, invasion, and ability of colony-formation in vitro and tumor growth in vivo.
CTHRC1 becomes dysregulated in many types of human solid tumors [22]. It can selectively activate the noncanonical Wnt pathway (Wnt/PCP) signal by the formation of a stable Cthrc1-Wnt-Fzd/Ror2 complex in CRC [34,35]. The downstream of the Wnt/PCP signal is some small GTPases, which consist of RhoA and Rac1. The activation of RhoA and Rac1 transforms the phenotype of the cell cytoskeleton, which in turn induces the EMT progression [36]. As is well known, TGF-β acts as a master regulator of the initiation and resolution of EMT within a variety of pathophysiological contexts [37]. In particular, CTHRC1 is regulated by TGF-β [16], and the expression level of CTHRC1 gradually increases with the stimulation of TGF-β1 in a time- and concentration-dependent manner [38,39]. In contrast, the phosphorylation of Smad2/3 is repressed by CTHRC1 in the progress of collagen deposition [40]. Seemingly contradictory, the role of CTHRC1 in EMT is complicated. Interestingly, a recent study has revealed that CTHRC1 can induce the expression of MMP9 via activation of Erk1/2 [25]. Furthermore, as a classical example of matrix metalloproteinases, MMP9 can cooperate with Snail to induce the EMT in cervical carcinoma [26]. Meanwhile, miR-520c increases the expression of MMP9 by directly targeting the 3’UTRs of mTOR and SIRT1 and activating the Ras/Raf/MEK/Erk signaling pathway [41]. In addition, miR-520e significantly decreases the phosphorylation of Erk1/2 in HepG2 cell by targeting the NF-κB-inducing kinase (NIK) [31]. Notably, miR-520d-3p mimics can abrogate the expression of MMP-9 in gastric cancer cells [42]. Taken together, the above studies indicate that miR-520d-5p may play a role in the EMT of CRC by inactivating the phosphorylation of Erk1/2 via directly targeting CTHRC1. We explored the expression relationship between miR-520d-5p and CTHRC1 in CRC tissues and cells and found a remarkably reverse correlation. Furthermore, dual-luciferase assays confirmed that CTHRC1 is the direct target of miR-520d-5p in CRC. Western blot analysis revealed that miR-520d-5p suppresses the phosphorylation of Erk1/2 and induces the secondary mesenchymal-epithelial transition (MET) progress through the inhibition of CTHRC1.
As with many common genes, mature microRNA was transcribed by RNA polymerase II, and then spliced by dicer enzyme. The transcription process is regulated by transcription factors [43]. For example, FoxD3 was reported that it can transactivate miR-137 in hepatocellular carcinoma [44]. Meanwhile, it was reported that Sox-2 can bind to the promoter of miR-200c to activate its expression at a transcriptional level in CRC [45]. Specificity Protein 1 (SP1), also known as GC-box, can encode a zinc finger protein transcription factor that binds to GC-rich motifs of many promoters. In terms of microRNA, SP1 can also activate or repress the expression of pri-miR at a transcriptional level; for example, it was reported that SP1 can regulate lung cancer progression by regulating miR-182 [46]. We confirmed that a positive correlation existed between SP1 and miR-520d-5p via real-time analysis of 20 paired clinical tissue specimens. Western blot tests further indicated that, to some degree, SP1 positively regulated miR-520d-5p. The dual-luciferase assay provided proof that SP1 transactivated miR-520d-5p through binding to its promoter region.
In conclusion, our study provides new evidence that miR-520d-5p is significantly down-expressed and associated with tumor growth and metastasis in CRC. MiR-520d-5p suppresses cell proliferation, migration, invasion by targeting CTHRC1. The SP1/miR-520d-5p/CTHRC1 regulatory axis is closely involved in CRC progression and may become a new therapeutic target.
Acknowledgements
We thank M.D. Wen-Jing Zhang for manuscript revision. This work was supported by grants from the Research Fund for the Doctoral Program of Higher Education of China (No. 20124433110010), Key Clinical Speciality Discipline Construction Program of China, Foundation for Distinguished Young Talents in Higher Education of Guangdong, China (No. 2013LYM_0006).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Herszényi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249–258. [PubMed] [Google Scholar]
- 3.Harris TJ, McCormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7:251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay S, Mitra R, Maulik U, Zhang MQ. Development of the human cancer microRNA network. Silence. 2010;1:6. doi: 10.1186/1758-907X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 9.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, Wei JW, Zhou HJ, Ren N, Ye QH, Dong QZ, Qin LX. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, Amadori D, Lu X, Xie D, Li QJ, Wang XF. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Z, Cui L, Mei Z, Liu M, Zhang D. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014;588:1773–1779. doi: 10.1016/j.febslet.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Liao WT, Li TT, Wang ZG, Wang SY, He MR, Ye YP, Qi L, Cui YM, Wu P, Jiao HL, Zhang C, Xie YJ, Wang JX, Ding YQ. microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res. 2013;19:4662–4672. doi: 10.1158/1078-0432.CCR-13-0244. [DOI] [PubMed] [Google Scholar]
- 14.Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T, Schneeweiss A, Sahin O, Wiemann S, Tschulena U. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31:4150–4163. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Zhang W, Yan W, Han L, Zhang K, Shi Z, Zhang J, Wang Y, Li Y, Yu S, Pu P, Jiang C, Jiang T, Kang C. The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis. 2012;33:2276–2282. doi: 10.1093/carcin/bgs261. [DOI] [PubMed] [Google Scholar]
- 16.Pyagay P, Heroult M, Wang Q, Lehnert W, Belden J, Liaw L, Friesel RE, Lindner V. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circ Res. 2005;96:261–268. doi: 10.1161/01.RES.0000154262.07264.12. [DOI] [PubMed] [Google Scholar]
- 17.Ke Z, He W, Lai Y, Guo X, Chen S, Li S, Wang Y, Wang L. Overexpression of Collagen Triple Helix Repeat Containing 1 (CTHRC1) is associated with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. Oncotarget. 2014;5:9410–9424. doi: 10.18632/oncotarget.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma MZ, Zhuang C, Yang XM, Zhang ZZ, Ma H, Zhang WM, You H, Qin W, Gu J, Yang S, Cao H, Zhang ZG. CTHRC1 acts as a prognostic factor and promotes invasiveness of gastrointestinal stromal tumors by activating Wnt/PCP-Rho signaling. Neoplasia. 2014;16:265–278. doi: 10.1016/j.neo.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YL, Wang TH, Hsu HC, Yuan RH, Jeng YM. Overexpression of CTHRC1 in hepatocellular carcinoma promotes tumor invasion and predicts poor prognosis. PLoS One. 2013;8:e70324. doi: 10.1371/journal.pone.0070324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JH, Baek TH, Yim HS, Kim KH, Jeong SH, Kang HB, Oh SS, Lee HG, Kim JW, Kim KD. Collagen triple helix repeat containing-1 (CTHRC1) expression in invasive ductal carcinoma of the breast: the impact on prognosis and correlation to clinicopathologic features. Pathol Oncol Res. 2013;19:731–737. doi: 10.1007/s12253-013-9636-y. [DOI] [PubMed] [Google Scholar]
- 21.Park EH, Kim S, Jo JY, Kim SJ, Hwang Y, Kim JM, Song SY, Lee DK, Koh SS. Collagen triple helix repeat containing-1 promotes pancreatic cancer progression by regulating migration and adhesion of tumor cells. Carcinogenesis. 2013;34:694–702. doi: 10.1093/carcin/bgs378. [DOI] [PubMed] [Google Scholar]
- 22.Tang L, Dai DL, Su M, Martinka M, Li G, Zhou Y. Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clin Cancer Res. 2006;12:3716–3722. doi: 10.1158/1078-0432.CCR-06-0030. [DOI] [PubMed] [Google Scholar]
- 23.Tan F, Liu F, Liu H, Hu Y, Liu D, Li G. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol. 2013;30:473. doi: 10.1007/s12032-013-0473-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Gao R, Hu W, Qian T, Wang N, Ding G, Ding F, Yu B, Gu X. MiR-9 inhibits Schwann cell migration by targeting Cthrc1 following sciatic nerve injury. J Cell Sci. 2014;127:967–976. doi: 10.1242/jcs.131672. [DOI] [PubMed] [Google Scholar]
- 25.Kim HC, Kim YS, Oh HW, Kim K, Oh SS, Kim JT, Kim BY, Lee SJ, Choe YK, Kim DH, Kim SH, Chae SW, Kim KD, Lee HG. Collagen Triple Helix Repeat Containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells. Oncotarget. 2014;5:519–529. doi: 10.18632/oncotarget.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin CY, Tsai PH, Kandaswami CC, Lee PP, Huang CJ, Hwang JJ, Lee MT. Matrix metalloproteinase-9 cooperates with transcription factor Snail to induce epithelial-mesenchymal transition. Cancer Sci. 2011;102:815–827. doi: 10.1111/j.1349-7006.2011.01861.x. [DOI] [PubMed] [Google Scholar]
- 27.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 28.Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2:361–369. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q, Cai N, Wang H, Liu F, Ye L, Zhang X. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner surviving through modulating miR-520b and HBXIP. Mol Cancer. 2014;13:128. doi: 10.1186/1476-4598-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Kong G, Zhang J, Wang T, Ye L, Zhang X. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One. 2012;7:e31450. doi: 10.1371/journal.pone.0031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-κB-inducing kinase (NIK) Oncogene. 2012;31:3607–3620. doi: 10.1038/onc.2011.523. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, Nicoloso MS, Wu SY, Almeida MI, Bottsford-Miller J, Pecot CV, Zand B, Matsuo K, Shahzad MM, Jennings NB, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Calin GA. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3:1302–1315. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuno S, Wang X, Shomori K, Hasegawa J, Miura N. Hsa-miR-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated process. Sci Rep. 2014;4:3852. doi: 10.1038/srep03852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Yamamoto S, Nishimura O, Misaki K, Nishita M, Minami Y, Yonemura S, Tarui H, Sasaki H. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Dev Cell. 2008;15:23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Kelley MW. Leading Wnt down a PCP path: Cthrc1 acts as a coreceptor in the Wnt-PCP pathway. Dev Cell. 2008;15:7–8. doi: 10.1016/j.devcel.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 2010;15:169–190. doi: 10.1007/s10911-010-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Cao J, Li M, Yu Y, Yang Y, Xiao X, Wu Z, Wang L, Tu Y, Chen H. Collagen triple helix repeat containing-1 inhibits transforming growth factor-β1-induced collagen type I expression in keloid. Br J Dermatol. 2011;164:1030–1036. doi: 10.1111/j.1365-2133.2011.10215.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang P, Wang YC, Chen XY, Shen ZY, Cao H, Zhang YJ, Yu J, Zhu JD, Lu YY, Fang JY. CTHRC1 is upregulated by promoter demethylation and transforming growth factor-β1 and may be associated with metastasis in human gastric cancer. Cancer Sci. 2012;103:1327–1333. doi: 10.1111/j.1349-7006.2012.02292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeClair RJ, Durmus T, Wang Q, Pyagay P, Terzic A, Lindner V. Cthrc1 is a novel inhibitor of transforming growth factor-beta signaling and neointimal lesion formation. Circ Res. 2007;100:826–833. doi: 10.1161/01.RES.0000260806.99307.72. [DOI] [PubMed] [Google Scholar]
- 41.Liu P, Wilson MJ. miR-520c and miR-373 target mTOR and SIRT1, activate the Ras/Raf/MEK/Erk pathway and NF-κB, with up-regulation of MMP9 in human fibrosarcoma cells. J Cell Physiol. 2012;227:867–876. doi: 10.1002/jcp.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Yuan W, Mei W, Yang K, Chen Z. MicroRNA-520d-3p inhibits gastric cancer cell proliferation, migration, and invasion by downregulating EphA2 expression. Mol Cell Biochem. 2014;396:295–305. doi: 10.1007/s11010-014-2164-6. [DOI] [PubMed] [Google Scholar]
- 43.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W, Yang YZ, Luo RZ, Zhang CZ, Yun JP. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget. 2014;5:5113–5124. doi: 10.18632/oncotarget.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu YX, Yuan L, Xue XL, Zhou M, Liu Y, Zhang C, Li JP, Zheng L, Hong M, Li XN. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin Cancer Res. 2014;20:2631–2642. doi: 10.1158/1078-0432.CCR-13-2348. [DOI] [PubMed] [Google Scholar]
- 46.Yang WB, Chen PH, Hsu T, Fu TF, Su WC, Liaw H, Chang WC, Hung JJ. Sp1-mediated microRNA- 182 expression regulates lung cancer progression. Oncotarget. 2014;5:740–753. doi: 10.18632/oncotarget.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]


