Abstract
Pancreatic cancer (PC) is one of the most malignant tumors. Rho GTPases can affect several types of human cancers, including PC. In this study, we investigated the role of Ras homolog family member T1 (RHOT1), a new member of Rho GTPases in PC. IHC results showed that RHOT1 was expressed significantly higher in PC tissues than paracancerous tissues (P<0.01) and SMAD family member 4 (SMAD4) was expressed lower in PC tissues (P<0.01). RHOT1 was widely expressed in PC cell lines analyzed by reverse transcription PCR (RT-PCR), real-time quantitative PCR (RT-qPCR) and western blotting (WB). SiRNA-RHOT1 significantly suppressed the proliferation and migration of SW1990 cells. Moreover, SMAD4 was identified as an effector of RHOT1. Our findings suggest that RHOT1 can regulate cell migration and proliferation by suppressing the expression of SMAD4 in PC, which may provide a novel sight to explore the mechanism and therapeutic strategy for PC.
Keywords: RHOT1, SMAD4, proliferation, migration, pancreatic cancer
Introduction
Pancreatic cancer (PC) is one of the most malignant tumors and ranks as the fourth leading cause of cancer-related deaths [1]. In 2011, a total of 44,030 new PC cases and 37,660 PC-related deaths occurred in the United States [2]. Due to its late diagnosis, highly aggressive behavior and ineffective treatments, the 5-year survival rate of PC is less than 5% [3]. Therefore, more accurate and reliable biological markers need to be identified to facilitate earlier detection and better drug targets. Furthermore, therapeutic strategies should be developed for disease treatment. The key to attaining these objectives is to understand the molecular mechanisms related to the pathogenesis of PC.
Multiple key genetic mutations and environmental factors contribute to tumor initiation and progression, some proteins including Ras homolog family (Rho) GTPases have been identified as crucial players in the proliferation, invasion and survival of various cancer cells [4-6]. RhoA, a Rho family member, plays important roles in the morphology, migration speed and invasion ability of cancer cells [7]. The depletion of RhoB, another Rho family member, may lead to a decrease in migration and invasion of prostate cancer cells [8]. In this study, we focused on the Ras homolog family member T1 (RHOT1), also named mitochondrial Rho (MIRO-1), which is a new member of Rho GTPases and was reported to be involved in mitochondrial transport [9], lymphocyte migration and polarity [10]. Furthermore, mutations in the sequestosome 1 (SQSTM1) gene of Paget’s disease were associated with gene expression of RHOT1 which may contribute to the over activity of osteoclasts [11]. However, the role of RHOT1 in cancer is still elusive.
In the current work, we attempt to shed light on the molecular mechanism of RHOT1 in PC. First, the expression level of RHOT1 was confirmed in PC tissues and PC cell lines. Then, RHOT1 down-regulation assay was performed to detect proliferation and migration in the PC cell line. Finally, SMAD family member 4 (SMAD4) was identified as one of the main effectors of RHOT1. Our study provides new insights into the role of RHOT1 in PC and suggests RHOT1 may be a potential target for the diagnosis and treatment of PC.
Materials and methods
Samples collection and immunohistochemistry (IHC)
All volunteers for this study were recruited with informed consent. Moreover, this study was approved by the committee on ethics of biomedicine research at Shanghai East Hospital, Tongji University. A total of 221 paraffin-embedded pancreatic cancer tissues and paracancerous tissues were selected from Shanghai Eastern Hospital and Biobank Center of National Engineering Center for Biochip at Shanghai. For IHC staining, the tissue sections were first dewaxed in xylene and then rehydrated with graded alcohol solutions. Antigen retrieval was performed by microwave heating the sections in sodium citrate buffer (pH 6.0) for 10 minutes. Afterwards, the sections were incubated with antibodies against RHOT1 (1:15) or SMAD4 (1:15) respectively overnight. The sections were washed with TBST and incubated with peroxidase-conjugated affinipure goat anti-rabbit antibody for 1 hour. Finally, the sections were incubated with 3, 3’-diaminobenzidine (DAB) chromogen and counterstained with hematoxylin (Sigma, USA). Stained sections were visualized and photographed using a Leica microscope (Leica Microsystems, Germany). The IHC scoring systems was used as previously described [12]. Two pathologists scored the sections in a double-blind manner.
Cell culture
Human PC cell lines AsPC-1, BxPC-3, CFPAC-1, PANC-1 and SW1990 were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. Cell lines Capan-2 and JF305 were obtained from the Tumor Marker Research Center, Beijing, China. AsPC-1, BxPC-3, Capan-2, JF305 and SW1990 were all cultured in RPMI 1640 (GIBCO, USA). PANC-1 and CFPAC-1 were cultivated in high glucose DMEM (GIBCO, USA) and IMDM (GIBCO, USA), respectively. All media were supplemented with 10% fetal bovine serum (GIBCO, USA), and 1% penicillin-streptomycin (GIBCO, USA). The cell lines were incubated at 37°C with 5% CO2 and sub-cultured when the cells reached 80%-90% confluence. Otherwise, the media were replaced every 2 days.
RNA extraction, reverse transcription PCR and quantitative PCR
Total RNA from PC cells was extracted using Trizol reagent (Invitrogen, USA) under RNase-free conditions, according to the manufacturer’s instructions. All qualified RNA were reverse transcribed using the first strand cDNA synthesis kit (CWBIO, CHINA). Reverse transcription PCR (RT-PCR) with Taq MasterMix (CWBIO, CHINA) and real-time quantitative PCR (RT-qPCR) with SYBR Green Master Mix (TOYOBO, JAPAN) assays were conducted to detect the mRNA levels of target genes. All primers and annealing temperatures are listed in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene. Each sample was performed in triplicate. Relative expression quantification analysis relied on the classical delta-delta-Ct method [13].
Table 1.
Primers used for RT-PCR and RT-qPCR analyses
| Gene | Forward primer (5’-3’) | Ta (°C)a | Amplicon size (bp) |
|---|---|---|---|
|
| |||
| Reverse primer (5’-3’) | |||
| RHOT1 | F: GGGAGGAACCTCTTCTGGA | 60 | 105 |
| R: ATGAAGAAAGACGTGCGGAT | |||
| GAPDH | F: TGCACCACCAACTGCTTAGC | 60 | 87 |
| R: GGCATGGACTGTGGTCATGAG | |||
| SMAD4 | F: GCTGCTGGAATTGGTGTTGATG | 60 | 108 |
| R: AGGTGTTTCTTTGATGCTCTGTCT | |||
| P53 | F: TCAACAAGATGTTTTGCCAACTG | 60 | 118 |
| R: ATGTGCTGTGACTGCTTGTAGATG | |||
| P16 | F: CCCTCGTGCTGATGCTACTG | 60 | 72 |
| R: CATCATGACCTGGTCTTCTAGGAA | |||
| Kras | F: ACAGAAGTGGAGGATGCTTT | 60 | 100 |
| R: TTTCACACAGCCAGGAGTCTT |
annealing temperature for PCR amplification.
Western blotting (WB)
Total protein from human PC cells was extracted, quantified, and subjected to 10% SDS-PAGE under denaturing conditions. The samples were transferred to polyvinylidene fluoride membranes (Millipore, USA). Membranes were blocked for 1 h with TBS-T buffer (with 10% non-fat dried milk). Afterwards, membranes were incubated overnight at 4°C with primary antibodies against RHOT1 (1:1000), β-actin (1:2000), or SMAD4 (1:500). Then, membranes were washed 3 times with PBS-T buffer, followed by incubation in goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (1:5000; Life Science, USA) for 1 h at room temperature. After washing with PBS-T 3 times, immunoreactive proteins were detected using an eECL Western Blot kit (CWBIO, CHINA).
Small interfering RNA (siRNA) assay
In order to choose the best effector, three candidate siRNA pairs for human RHOT1 gene and a negative control (NC) were designed and chemically synthesized by Biotend Biotechnology Co., Ltd (Shanghai). All siRNA sequences are shown in Table 2. SW1990 cells were transfected with a candidate siRNA (final concentration 50 nM) or a negative control using Lipofectamine 2000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s instructions. Then, RT-qPCR and WB were used to analyze the knock-down efficiency at various time points.
Table 2.
Sequence of oligonucleotides used in this study
| Oligonucleotides | Sequence (5’-3’) |
|---|---|
| siRNA1-RHOT1-S | GCACUACUGAAUUAAAUCAdTdT |
| siRNA1-RHOT1-AS | UGAUUUAAUUCAGUAGUGCdTdT |
| siRNA2-RHOT1-S | CACGACUUAUUUAGAUGUAdTdT |
| siRNA2-RHOT1-AS | UACAUCUAAAUAAGUCGUGdTdT |
| siRNA3-RHOT1-S | CAACACUUUAUGGACAGCAdTdT |
| siRNA3-RHOT1-AS | UGCUGUCCAUAAAGUGUUGdTdT |
| siRNA-NC-S | UUCUCCGAACGUGUCACGUdTdT |
| siRNA-NC-AS | ACGUGACACGUUCGGAGAAdTdT |
Cell proliferation and migration assays
To assess the role of RHOT1 in cell proliferation, SW1990 cells were plated in 96-well plates at a density of 1×105 cells in each well and silenced with siRNAs. The number of cells was evaluated with a Cell Counting Kit-8 (DOJINDO, CHINA) according to the manufacture’s protocol at 24, 48, 72 and 96 hours after siRNA transfection. The optical density (OD) at 450 nm was measured using Multiskan MK3 (Themo, Finland). The scratch assay was performed to assess the effect of RHOT1 on cell migration. Scratch wounds were created on the confluent cell monolayer 24 hours after transfection. To visualize wound healing, images were taken at 0, 10 and 20 hours after scratching.
Statistical analysis
For statistical analysis, each experiment was repeated at least three times. Data analysis was performed using a two-tailed Student’s t-test. P<0.05 was considered statistically significant. All analyses were carried out using SPSS19.0 software (San Rafael, USA).
Results
Clinical characteristics of PC patients
The characteristics of PC patients enrolled in this study are summarized in Figure 1A. There were a total of 221 PC patients including 139 males (63%) and 82 females (37%). The median age of the participants was 61 years (range 41-85 years). The tumor size ranged from 1 cm to 8 cm. According to the seventh edition of the American Joint Committee on Cancer (AJCC) staging system, 13 (6%) cases were in stage I; 186 (84%) cases were in stage II; and 22 cases (10%) were in stage III.
Figure 1.
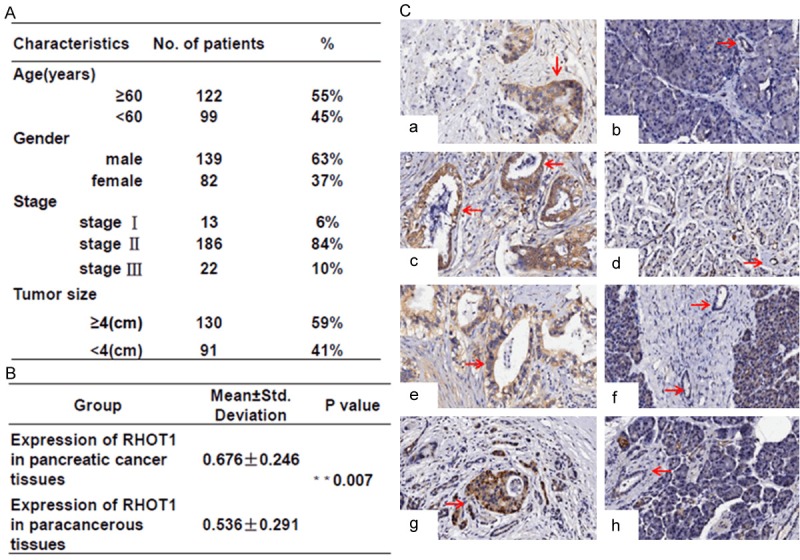
The expression levels of RHOT1 in PC tissues. A. Characteristics of the 221 patients with PC. B. Samples paired T test was performed to analyze different expression of RHOT1 between cancer tissues and paracancerous tissues. **P<0.01, statistically significant. C. Immunohistochemical staining of RHOT1 in four patients. a, c, e and g showed the positive cytoplasmic expression of RHOT1 in cancer tissues; b, d, f and h showed the negative cytoplasmic expression of RHOT1 in paracancerous tissues. All the four patients were in stage II. No. 1, 2, 4 patients are males, No.3 is female. Survival time of all these patients was less than 15 months after surgery.
The expression levels of RHOT1 in pancreatic cancer tissues and cell lines
According to the IHC results, RHOT1 is mainly expressed in pancreatic ductal epithelial cells and islet cells (data not shown). Moreover, the expression level of RHOT1 in pancreatic ductal epithelial cells in PC was significantly higher (P=0.007) than in paracancerous tissues (Figure 2B, 2C). At the cellular level, RT-PCR, RT-qPCR and WB confirmed RHOT1 expression in all PC cell lines, including AsPC-1, BxPC-3, Capan-2, CFPAC-1, JF305, PANC-1 and SW1990 (Figure 2A-C). Nevertheless, the expression intensities of RHOT1 in multiple cell lines were significantly different. RHOT1 demonstrated significantly high expression in PANC-1, and the expression was relatively lower in capan-2 (Figure 2B).
Figure 2.
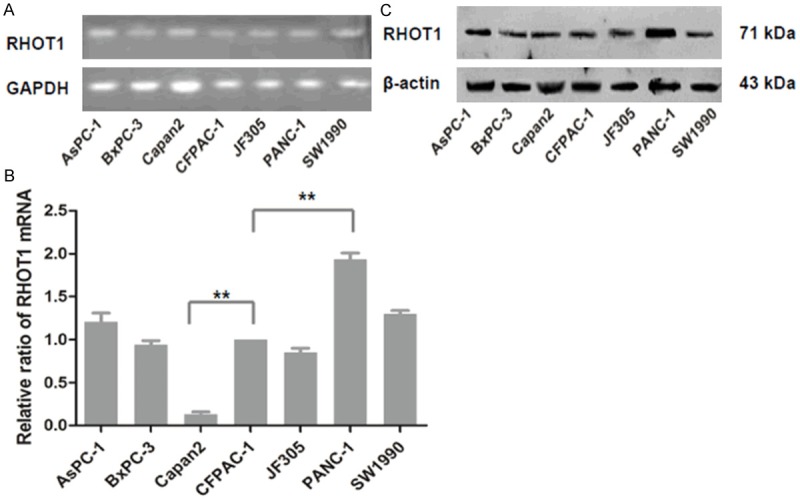
Expression of RHOT1 in PC cell lines. A. The mRNA expression of RHOT1 could be observed among seven PC cell lines by RT-PCR. B. Relative RHOT1 expression levels of mRNA were measured by RT-qPCR, and normalized to the expression level of RHOT1 in CFPAC-1. All data are shown as mean ± SD. **P<0.01. C. Expression of RHOT1 protein in seven PC cell lines was detected by western blotting and normalized to that of β-actin.
Knockdown of RHOT1 in SW1990 inhibits cell migration and proliferation
To examine the knock-down efficiency of candidate siRNAs, mRNA and protein were isolated from SW1990 cells to test the expression level of RHOT1. Compared with other treated groups, candidate siRNA1-RHOT1 could significantly depress the mRNA and protein expression of RHOT1 in untreated NC groups (Figure 3A-C). To detect whether RHOT1 was associated with cell behavior, we analyzed the effect of interfering with RHOT1 expression on the proliferation and migration of SW1990. Transfection of siRNA-RHOT1 resulted in a significant reduction of cell proliferation and migration compared with the negative control or untreated cells (Figure 4A-C). These results demonstrated that RHOT1 could regulate the behaviors of PC cells in vitro.
Figure 3.

Effect of RHOT1 knockdown. A. Red fluorescence-labeled SW1990 cells reflect positive effect of transfection. The image was taken at 200x magnification. B. The mRNA levels were quantified using real-time PCR and the ratio of siRNA-RHOT1 to untreated group was determined. GAPDH was used as a housekeeping control. All data are shown as mean ± SD. **P<0.01. C. Protein level of RHOT1 was decreased in SW1990 cells transfected with siRNA1-RHOT1.
Figure 4.
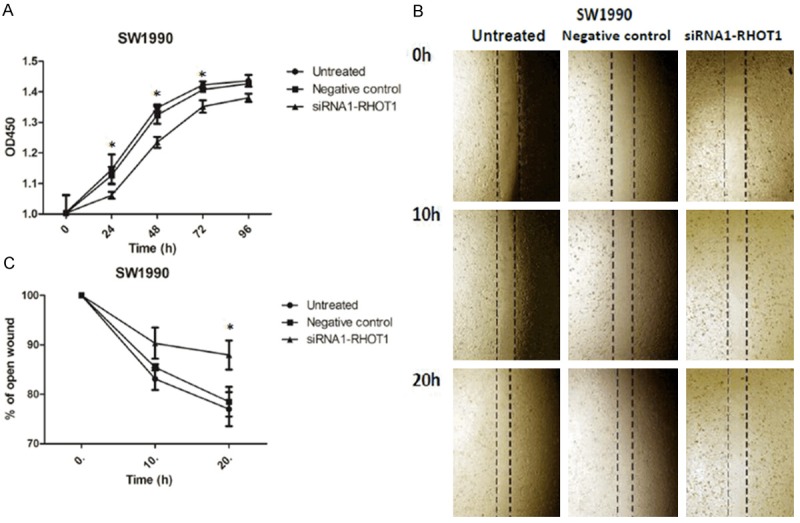
Knocking-down of RHOT1 inhibits pancreatic cancer cell proliferation and migration in vitro. A. Knocking down RHOT1 decreased SW1990 cells proliferation. Proliferation tested by CCK8 assay. The OD 450 nm was assessed at 0, 24, 48, 72 and 96 hours. *P<0.05. B, C. SW1990 cells were transfected or not with 50 nmol/L of siRNA-RHOT1 for 24 h, and wounds were made. Magnification for identification of migration is ×40. The relative ratio of wound closure per field is shown. Data are shown as mean ± SD. *P<0.05.
SMAD4 was an effector to the negative regulation of RHOT1
Previous studies showed that genetic mutations were generally observed in carcinogenesis. Activation of Kras [4] and inactivation of SMAD4 [14], P16 [15] or p53 [16] is associated with PC. The detailed relationship among RHOT1 and these genes was analyzed by knocking down RHOT1. As shown in Figure 5A, 5B, the mRNA level of SMAD4 was significantly increased while suppressing the expression of RHOT1. Moreover, WB showed that the expression of the SMAD4 protein increased significantly in the siRNA1-RHOT1 group when compared to the untreated control or siRNA-NC groups (Figure 5C). We then performed IHC to confirm the expression of SMAD4 in PC and paracancerous tissues. SMAD4 was significantly decreased (P=0.017) in cancer tissues (Figure 6A, 6B). While collecting RHOT1 and SMAD4 expression data from the same sample, we found that the percent of patients with lower SMAD4 and higher RHOT1 was more than 50% (Figure 6C).
Figure 5.

SMAD4 is a potential effector of RHOT1. A. Relative RHOT1, SMAD4, P53, P16 and KRAS expression levels of mRNA were measured by RT-qPCR, normalized to the expression level of RHOT1, **P<0.01. B, C. Both RT-qPCR and western bloting result showed that expression of SMAD4 increased while RHOT1 decreased after transfection, β-actin served as control.
Figure 6.

The expression levels of SMAD4 in PC tissues. A. Samples paired T test was performed to analyze different expression of SMAD4 between cancer tissues and paracancerous tissues. *P<0.05, statistically significant. B. Expression data of RHOT1 and SMAD4 from the same sample were collected; the percent of patients was shown. C. RHOT1 and SMAD4 expressed oppositely in the same patients. a, c, e and g showed the expression of RHOT1 or SMAD4 in cancer tissues; b, d, f, and h showed the expression of RHOT1 or SMAD4 in paracancerous tissues.
Discussion
The involvement of Rho GTPases in cancer is complex, with evidence supporting roles for these molecules as tumor suppressors or promoters [17,18]. To date, there have not been many reports on the role of RHOT1 in cancers. In this study, we focused on the expression and function of RHOT1 in PC. Our data have shown that the expression of RHOT1 is higher in PC tissues than in paracancerous tissues (Figure 1A-C). Furthermore, analysis of RHOT1 in seven PC cell lines (AsPC-1, BxPC-3, Capan-2, CFPAC-1, JF305, PANC-1 and SW1990) showed that RHOT1 is expressed in all PC cell lines as well (Figure 2A-C). For functional analysis, targeted RHOT1- siRNA could significantly suppress the proliferation and migration of cancer cells (SW1990) (Figure 4A-C). Finally, we discovered that RHOT1 may regulate the expression of SMAD4 to influence the behaviors of tumor cell (Figure 5A-C). These observations suggested that RHOT1 may act as a potential oncogene in PC. To the best of our knowledge, this was the first evidence on the molecular mechanism of RHOT1 in PC.
Whole-exome and whole-genome sequencing and copy number analysis revealed that mutations in Kras, p53, p16 and SMAD4 are the most common genetic alterations in PC tissues [19,20]. Because of their importance in PC, these 4 genes were investigated as potential targets affected by RHOT1. The qRT-PCR result showed that only the mRNA expression of SMAD4 increased significantly when we interfered with RHOT1 expression (Figure 5). SMAD4 has been identified as a tumor suppressor gene that is deleted, mutated or inactivated in many human tumors [21,22]. In recent years, it has been demonstrated that SMAD4 can function as an oncogene or as tumor suppressor in the initiation and progression of PC via the transforming growth factor-β (TGF-β) signaling pathway [23] and Wnt/β-Catenin signaling pathway [24]. The TGF-β pathway is one of the most crucial and frequently activated pathways in pancreatic cancer and is mechanistically linked to the regulation of SMAD4 [25]. SMAD4 is a key mediator of the TGF-β pathway and is involved in the control of cell proliferation, differentiation and apoptosis [26]. In the canonical TGF-β pathway, SMAD2 and 3 are phosphorylated by the type I TGF-β receptor, after which they form transcriptional complexes with SMAD4 and are translocated to the nucleus where they activate or repress the transcription of target genes [25]. In our research, knocking down RHOT1 can significantly increase SMAD4 expression (Figure 5A-C). This result suggested that SMAD4 may be one downstream effector protein of RHOT1. Although it was unclear whether RHOT1 directly or indirectly binds to SMAD4 during regulation of gene expression, our finding suggested that RHOT1 may regulate the proliferation and migration of PC cells via SMAD4-dependent TGF-β signaling.
The immunolocalization of RHOT1 in NIH3T3 and COS7 cells indicated that it is localized at the outer mitochondrial membrane [9]; therefore, the function of RHOT1 may be closely related to mitochondria. In neurons, RHOT1 can regulate mitochondrial trafficking [27] and calcium homeostasis [28]. More importantly, two Parkinson’s disease proteins, the Ser/Thr kinase PTEN induced putative kinase 1 (PINK1) and ubiquitin ligase Parkinson disease 2 (Park2), have been found to control the phosphorylation, ubiquitination and degradation of RHOT1 to arrest mitochondrial motility [29,30]. In murine models with neuron-specific loss of RHOT1, the mouse had progressive neurological deficits mirroring human motor neuron disease [31]. These studies suggest that RHOT1 has a striking association with the neuron disease that is characterized by the dysfunction of mitochondria. Moreover, expression of RHOT1 is also involved in the pagetic osteoclast phenotype [32], spinal bone mineral density [33], lymphocyte migration and polarity [34], mesenchymal stem cell rescue [10], and embryonic development [35,36]. In these biological processes, the main role of RHOT1 at the subcellular (organelle) level is mitochondrial trafficking [37]. Because of the key role of mitochondrial dysfunction in tumorigenesis, RHOT1 may also be involved in the proliferation, migration and invasion of cancers cells [38]. Nevertheless, the function of RHOT1 in cancer has been poorly understood. In this study, we attempted to investigate the molecular mechanism of RHOT1 in PC. The down-regulation of RHOT1 can significantly suppress the proliferation and migration of cancer cells (Figure 4). This finding is in line with a previous study demonstrating that over-expression of RHOT1 could increase the cell apoptotic rate [9]. Furthermore, in epithelial cancer cells, RHOT1 could serve as a linkage between mitochondria and microtubules to control the number of anterior-localized mitochondria, which determine the invasive abilities between cells [39]. More importantly, mitochondrial function determines tumor cell survival in pancreatic cancer [40]. Based on these clues, we speculated that RHOT1 may also regulate these cell biological behaviors by controlling the mitochondrial function in PC.
Our previous study showed that a low cytoplasmic expression level of RhoT1 was significantly associated with reduced survival [41]. In recent studies, we attempted to reconfirm this result with a larger number of samples. However, the expression of RHOT1 was higher in PC tissues than in paracancerous tissues in 221 PC patients. In molecular and cellular experiments, down-regulation of RHOT1 could significantly suppress the proliferation and migration of cancer cells. This conclusion supported the hypothesis that RHOT1 functions as an oncogene with higher expression in PC tissue.
Despite continuous improvements in therapies, pancreatic cancer remains one of the most clinically challenging diseases. The genetic landscape of PC demonstrates comprehensive activating mutations of Rho GTPases and inactivating mutations of SMAD4. Here, we demonstrate that RHOT1, a new member of Rho GTPases, is expressed widely in pancreatic cancer cell lines and promotes cell proliferation and migration via SMAD4 regulation. These findings provide new insights into specific gene functions in PC. RHOT1 may be a potential target in the diagnosis and therapy for PC patients.
Acknowledgements
This work was supported by the Outstanding Leaders Training Program of Pudong new area health system, Shanghai, China (No. PWR-12012-01) and the Science and Technology Commission of Shanghai Municipality, China (No.13ZR1433800).
Disclosure of conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2013. Ann Oncol. 2013;24:792–800. doi: 10.1093/annonc/mdt010. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 4.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Grise F, Bidaud A, Moreau V. Rho GTPases in hepatocellular carcinoma. Biochim Biophys Acta. 2009;1795:137–151. doi: 10.1016/j.bbcan.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Cheng KW, Agarwal R, Mills GB. Ras-superfamily GTP-ases in ovarian cancer. Cancer Treat Res. 2009;149:229–240. doi: 10.1007/978-0-387-98094-2_11. [DOI] [PubMed] [Google Scholar]
- 7.Ridley AJ. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc. 2013;251:242–249. doi: 10.1111/jmi.12025. [DOI] [PubMed] [Google Scholar]
- 8.Alfano D, Ragno P, Stoppelli MP, Ridley AJ. RhoB regulates uPAR signalling. J Cell Sci. 2012;125:2369–2380. doi: 10.1242/jcs.091579. [DOI] [PubMed] [Google Scholar]
- 9.Fransson A, Ruusala A, Aspenström P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- 10.Morlino G, Barreiro O, Baixauli F, Robles-Valero J, González-Granado JM, Villa-Bellosta R, Cuenca J, Sánchez-Sorzano CO, Veiga E, Martín-Cófreces NB, Sánchez-Madrid F. Miro-1 links mitochondria and microtubule Dynein motors to control lymphocyte migration and polarity. Mol Cell Biol. 2014;34:1412–1426. doi: 10.1128/MCB.01177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinck R, Laberge G, Bisson M, McManus S, Michou L, Brown JP, Roux S. Alternative splicing in osteoclasts and Paget’s disease of bone. BMC Med Genet. 2014;15:98. doi: 10.1186/s12881-014-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H, Li Q, He C, Li F, Sheng H, Shen X, Zhang X, Zhu S, Chen H, Chen X, Yang C, Gao H. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J Cancer Res. 2014;4:537–544. [PMC free article] [PubMed] [Google Scholar]
- 13.Meijerink J, Mandigers C, van de Locht L, Tönnissen E, Goodsaid F, Raemaekers J. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J Mol Diagn. 2001;3:55–61. doi: 10.1016/S1525-1578(10)60652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackford A, Serrano OK, Wolfgang CL, Parmigiani G, Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Eshleman JR, Goggins M, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Cameron JL, Olino K, Schulick R, Winter J, Herman JM, Laheru D, Klein AP, Vogelstein B, Kinzler KW, Velculescu VE, Hruban RH. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15:4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin T, Wei H, Gou S, Shi P, Yang Z, Zhao G, Wang C. Cancer Stem-Like Cells Enriched in Panc-1 Spheres Possess Increased Migration Ability and Resistance to Gemcitabine. Int J Mol Sci. 2011;12:1595–1604. doi: 10.3390/ijms12031595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsey C, Balakrishnan V, O’Dell MR, Huang JL, Newman L, Whitney-Miller CL, Hezel AF, Land H. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep. 2014;7:1143–1155. doi: 10.1016/j.celrep.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arpaia E, Blaser H, Quintela-Fandino M, Duncan G, Leong HS, Ablack A, Nambiar SC, Lind EF, Silvester J, Fleming CK, Rufini A, Tusche MW, Brüstle A, Ohashi PS, Lewis JD, Mak TW. The interaction between caveolin-1 and Rho-GTPases promotes metastasis by controlling the expression of alpha5-integrin and the activation of Src, Ras and Erk. Oncogene. 2012;31:884–896. doi: 10.1038/onc.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Zhu Y, Zhang G, Liu N, Sun L, Liu M, Qiu M, Luo D, Tang Q, Liao Z, Zheng Y, Bi F. A distinct role of RhoB in gastric cancer suppression. Int J Cancer. 2011;128:1057–1068. doi: 10.1002/ijc.25445. [DOI] [PubMed] [Google Scholar]
- 19.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N Australian Pancreatic Cancer Genome Initiative. Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, Hruban RH. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 2012;18:4257–4265. doi: 10.1158/1078-0432.CCR-12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 22.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, Deng C, Lu SL, Wang XJ. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia X, Wu W, Huang C, Cen G, Jiang T, Cao J, Huang K, Qiu Z. SMAD4 and its role in pancreatic cancer. Tumour Biol. 2015;36:111–9. doi: 10.1007/s13277-014-2883-z. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, Cui J, Du Y, Wei D, Huang S, Xie K. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 25.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;9:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 26.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 27.Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnóczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, Takahashi R, Hattori N, Imai Y, Lu B. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen TT, Oh SS, Weaver D, Lewandowska A, Maxfield D, Schuler MH, Smith NK, Macfarlane J, Saunders G, Palmer CA, Debattisti V, Koshiba T, Pulst S, Feldman EL, Hajnóczky G, Shaw JM. Loss of Miro1-directed mitochondrial movement results in a novel murine model for neuron disease. Proc Natl Acad Sci U S A. 2014;111:E3631–3640. doi: 10.1073/pnas.1402449111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Danilchanka O, Mekalanos JJ. Vibrio cholerae T3SS Effector VopE Modulates Mitochondrial Dynamics and Innate Immune Signaling by Targeting MiroGTPasesVibrio cholerae T3SS Effector VopE Modulates Mitochondrial Dynamics and Innate Immune Signaling by Targeting MiroGTPases. Cell Host Microbe. 2014;16:581–591. doi: 10.1016/j.chom.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alam I, Sun Q, Koller DL, Liu L, Liu Y, Edenberg HJ, Foroud T, Turner CH. Genes influencing spinal bone mineral density in inbred F344, LEW, COP, and DA rats. Funct Integr Genomics. 2010;10:63–72. doi: 10.1007/s10142-009-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klinck R, Laberge G, Bisson M, McManus S, Michou L, Brown JP, Roux S. Alternative splicing in osteoclasts and Paget’s disease of bone. BMC Med Genet. 2014;15:98. doi: 10.1186/s12881-014-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmad T, Mukherjee S, Pattnaik B, Kumar M, Singh S, Kumar M, Rehman R, Tiwari BK, Jha KA, Barhanpurkar AP, Wani MR, Roy SS, Mabalirajan U, Ghosh B, Agrawal A. Miro1 regulates intercellular mitochondrial transport enhances mesenchymal stem cell rescue efficacy. EMBO J. 2014;33:994–1010. doi: 10.1002/embj.201386030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y, Kono T. Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol Reprod. 2008;78:568–576. doi: 10.1095/biolreprod.107.064634. [DOI] [PubMed] [Google Scholar]
- 37.Yamaoka S, Leaver CJ. EMB2473/MIRO1, an Arabidopsis MiroGTPase, is required for embryogenesis and influences mitochondrial morphology in pollen. Plant Cell. 2008;20:589–601. doi: 10.1105/tpc.107.055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang H, He C, Geng S, Sheng H, Shen X, Zhang X, Li H, Zhu S, Chen X, Yang C, Gao H. RhoT1 and Smad4 Are Correlated with Lymph Node Metastasis and Overall Survival in Pancreatic Cancer. PLoS One. 2012;7:e42234. doi: 10.1371/journal.pone.0042234. [DOI] [PMC free article] [PubMed] [Google Scholar]


