Abstract
Simian foamy viruses (SVF) are ubiquitous in nonhuman primates (NHP). SFV can be zoonotically transmitted to humans who either work with or live commensally with NHP. We analyzed the blood of 45 Bangladeshi performing monkey owners (an ethnic group called the Bedey) for SFV infection. Surprisingly, a PCR assay failed to detect SFV infection in any of these participants. This is in contrast to our previously reported infection rate of about 5% among Bangladeshi villagers.
TEXT
Humans can acquire new infectious agents via zoonotic transmission. Many of these zoonotic infections occur from nonhuman primate (NHP) viruses, such as the transmission of simian T-cell lymphotropic virus type 1 to humans (1). The global health toll from HIV, a retrovirus that emerged following the zoonotic transmission of a recombinant NHP virus, emphasizes the importance of studying the interplay of zoonotic viruses and their human and NHP hosts. Human movement and settlement conditions influence the ecological contexts in which infectious agent transmission occurs. This is readily evident in South and Southeast Asia, where the world's densest human populations are situated close to diverse NHP fauna and where cultural and religious traditions have encouraged tolerance of NHP. Simian foamy viruses (SFV), nearly ubiquitous in free-ranging primates, are the most frequently transmitted NHP retroviruses (2) and have been detected in different populations, including NHP bushmeat hunters in Africa (2), laboratory and zoo workers in North America (3), and temple workers and villagers in Asia (4).
A particularly intriguing human-primate interface available for examination is that of performing monkeys and their seminomadic owners (known as the Bedey in Bangladesh), who are found throughout South Asia. The Bedey travel seasonal circuits throughout South Asia, making a livelihood by staging performances with trained macaques, bears, and snakes. The Bedey, an ethnically homogenous group, are socially and economically marginalized. Despite their being part of a centuries-old tradition, little is known about the Bedey, their animal acquisition, husbandry, and/or training practices with macaques. Currently, an estimated 10,000 close-knit Bedey groups, each composed of 10 to 20 individuals, and a total of 5,000 to 10,000 macaques are involved in the performing monkey tradition in Bangladesh (5). Although only the men take the macaques into town for performances, both male and female Bedey take part in their training. Bedey receive bites, scratches, and mucosal splashes from a young age during their cohabitation with and training of these performing monkeys. We previously reported that 79% (30/38) of these macaques are PCR positive for SFV (6). We estimate that over the course of his or her lifetime, each Bedey is exposed to several hundred macaques. Each Bedey traveling group maintains about 12 macaques, with individual family groups averaging 2 working monkeys at any time; therefore, we expect that each Bedey is exposed to many SFV-positive animals during his or her lifetime.
In an extension of our previous work screening Bangladeshi residents for SFV infection (7), we recently tested the plasma of 45 Bedey (from six different groups/regions) for seroreactivity to SFVmac capsid protein (Gag) antigens. Only one Bedey, BGH251, tested potentially seropositive by Western blot assay (Fig. 1). We then performed PCR assays with primers homologous to highly conserved regions of SFV pol (8) and gag (9) which we have shown can amplify divergent strains of SFVmac in Bangladesh and other regions in South Asia, as well as SFV strains from other monkeys such as Macaca fascicularis (4). However, we could not detect the presence of provirus in the blood from BGH251. In contrast, in the villager group, 17 out of 269 individuals were seropositive for antibodies against SFV Gag (6.3%), with 12 of those being confirmed PCR positive for proviral sequences in blood (4.5%; Table 1). Occasionally, an individual tests negative serologically by Western blotting yet provirus is detectable by PCR (10). Therefore, we performed a nested PCR assay with SFV pol primers on all 45 Bedey blood samples. All were negative for SFV pol amplification, whereas our positive controls worked (data not shown). As a control, we amplified the genomic glyceraldehyde 3-phosphate dehydrogenase gene to ensure that the quality of the DNA samples from the Bedey were comparable to our villager DNA samples (data not shown). Previous quantitative PCR results indicated that we can detect one copy of SFV pol in 104 to 105 peripheral blood mononuclear cells (9). We cannot, however, rule out the presence of a much lower level of proviral DNA.
FIG 1.
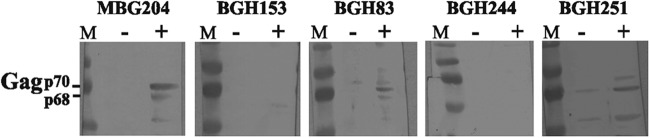
Western blot analysis. Lysates were generated from uninfected (−) Telo-RF macaque fibroblast cells (15) and cells infected with SFVmac (+). Each well was loaded with 15 μl of lysate generated from an equal number of cells, and proteins were separated with 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). Western blot assays were performed as previously described (6), with BGH sera at a 1:50 dilution and MBG sera at a 1:200 dilution in blocking buffer (5% dry milk powder, 0.05% Tween 20 in PBS). Goat anti-monkey IgG and goat monoclonal anti-human IgG were used at a 1:3,000 dilution as the secondary antibodies (Rockland Immunochemicals, Gilbertsville, PA). 3,3′,5,5′-Tetramethylbenzidine substrate (Thermo Fisher, Waltham, MA) was used to visualize proteins in accordance with the manufacturer's protocol. Lanes M contained molecular weight markers. MBG204 is an SFV-positive monkey. BGH153 and BGH83 are SFV-negative and -positive villagers, respectively. BGH244 is a negative Bedey, and BGH251 is potentially positive. Nonspecific bands were sometimes seen in control lanes (e.g., BGH83 and BGH251).
TABLE 1.
SFV infection of Bedey and villagers in Bangladesh
| Parameter | Bedey | Villagers |
|---|---|---|
| Total no. | 45 | 269 |
| No. of males | 34 | 114 |
| No. of females | 11 | 155 |
| No. ever bitten/total (%) | 40/45 (89) | 122/269 (45.4) |
| No. bitten more than once/total (%) | 29/45 (64) | 30/269 (11) |
| No. with scar/total (%) | 36/40 (90) | 106/122 (87) |
| No. WB+/total (%) | 1/45 (2.2) | 17/269 (6.3) |
| No. WB+/no. ever bitten (%) | 1/40 (2.5) | 17/122 (13.9) |
| No. PCR+ | 0a | 12a |
| No. PCR+/no. ever bitten (%) | 0/40 (0) | 12/122 (9.8) |
The PCR assay was performed with all Bedey samples but only with Western blot-positive (WB+) villager samples.
The low level of SFV infection found among the Bedey was unexpected, given that the Bedey have nearly constant exposure to macaques and are repeatedly bitten, often severely. Nearly 90% of the Bedey reported being bitten, and two-thirds of these reported multiple bites (Table 1). This is a much higher bite prevalence than that observed in village residents, who have only casual contact with macaques. As already stated, the prevalence of SFV infection in performing macaques is high, approximately 79% (6), ruling out a much lower infection prevalence in performing macaques than in village macaques as an explanation for these differences in zoonotic transmission. When controlling for bite history, i.e., only including individuals who had reported bites, we found a statistically significant difference between the apparent infection rates of these two groups (17/122 seropositive villagers versus 1/40 for the Bedey; P = 0.05 by Fisher's exact test). Provirus was detected (indicative of persistent infection) in 12/122 (9.8%) bitten villagers but in none of the 40 bitten Bedey (we expected approximately 4 if the same zoonotic infection rate applied). None of the Bedey subjects reported being related to other participants in this study and were sampled from five different nomadic traveling groups and one semipermanent village; therefore, we did not sample from just a few related family groups that might have possessed only SFV-negative animals. There was no statistically significant difference between the mean ages of the Bedey and villager participants (data not shown). Since our sample size is relatively small, we plan to sample more Bedey individuals in the future to confirm these data and conclusions.
One possible explanation for the difference in persistent infection between the villagers and performing monkey owners is that the latter group has a distinct genetic feature, for example, a polymorphism in an innate immune factor, that allows suppression of SFV infection. Human APOBEC3G has a role in suppressing the replication of SFV in zoonotically infected humans (11), as does TRIM5alpha and possibly other factors (12). Alternatively, since the Bedey are exposed to high levels of macaque SFV in saliva from a very young age, such exposure might stimulate the production of neutralizing antibodies in mucosal tissues, which could then suppress infection, as has been seen for HIV-1 (13, 14). We plan to examine these two possibilities in future experiments. Such studies could yield novel mechanistic information regarding specific host factors that render humans susceptible or resistant to SFV infection.
ACKNOWLEDGMENTS
We are grateful to the Bedey and Bangladeshi communities that continue to welcome our research team into their homes and for their cooperation. We thank Ted Gooley at the Fred Hutchinson Cancer Research Center for statistical analysis of the data.
This research was supported by funding from NIH-NIAID grants R01 AI078229 and R01 AI078229-03S1.
We have no competing interests to declare.
K.L.C. performed the pol PCR, analyzed the data, and wrote the manuscript. D.L.J., X.W., and K.S. performed experiments. G.A.E. and M.K.H. obtained field specimens and demographic data. M.M.F. facilitated field work in Bangladesh. L.J.-E. designed the study, obtained field samples and demographic data, and wrote the manuscript. M.L.L. designed the study and wrote the manuscript.
REFERENCES
- 1.Kazanji M, Mouinga-Ondémé A, Lekana-Douki-Etenna S, Caron M, Makuwa M, Mahieux R, Gessain A. 2015. Origin of HTLV-1 in hunters of nonhuman primates in Central Africa. J Infect Dis 211:361–365. doi: 10.1093/infdis/jiu464. [DOI] [PubMed] [Google Scholar]
- 2.Gessain A, Rua R, Betsem E, Turpin J, Mahieux R. 2013. HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology. Virology 435:187–199. doi: 10.1016/j.virol.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol 78:2780–2789. doi: 10.1128/JVI.78.6.2780-2789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones-Engel L, Steinkraus KA, Murray SM, Engel GA, Grant R, Aggimarangsee N, Lee BPY-H, May C, Schillaci MA, Somgird C, Sutthipat T, Vojtech L, Zhao J-Y, Linial ML. 2007. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging, Asian monkeys. J Virol 81:7330–7337. doi: 10.1128/JVI.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhtar S. 2013. Monkey performers in Bangladesh: monkey population, healthcare, human-monkey interaction and socio-economic condition of the monkey owners. M.S. thesis Jahangirnagar University, Savar, Bangladesh. [Google Scholar]
- 6.Feeroz M, Soliven K, Small C, Engel G, Pacheco M, Yee J, Wang X, Hasan K, Oh G, Levine K, Alam S, Craig K, Jackson D, Lee E-G, Barry P, Lerche N, Escalante A, Matsen F, Linial M, Jones-Engel L. 2013. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg Microbes Infect 2:e29. doi: 10.1038/emi.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel GA, Small CT, Soliven K, Feeroz MM, Wang X, Hasan K, Gunwha O, Alam S, Craig K, Jackson D, Matsen FA IV, Linial ML, Jones-Engel L. 2013. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infections among humans. Emerg Microbes Infect 2:e58. doi: 10.1038/emi.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweizer M, Neumann-Haefelin D. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577–582. doi: 10.1006/viro.1995.1120. [DOI] [PubMed] [Google Scholar]
- 9.Soliven K, Wang X, Small CT, Feeroz MM, Lee EG, Craig KL, Hasan K, Engel GA, Jones-Engel L, Matsen FA, Linial ML. 2013. Simian foamy virus infection of rhesus macaques in Bangladesh: relationship of latent proviruses and transcriptionally active viruses. J Virol 87:13628–13639. doi: 10.1128/JVI.01989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog 7:e1002306. doi: 10.1371/journal.ppat.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsen FA IV, Small CT, Soliven K, Engel GA, Feeroz MM, Wang X, Craig KL, Hasan MK, Emerman M, Linial ML, Jones-Engel L. 2014. A novel Bayesian method for detection of APOBEC3-mediated hypermutation and its application to zoonotic transmission of simian foamy viruses. PLoS Comput Biol 10:e1003493. doi: 10.1371/journal.pcbi.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rua R, Gessain A. 2015. Origin, evolution and innate immune control of simian foamy viruses in humans. Curr Opin Virol 10:47–55. doi: 10.1016/j.coviro.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seaton KE, Ballweber L, Lan A, Donathan M, Hughes S, Vojtech L, Moody MA, Liao HX, Haynes BF, Galloway CG, Richardson BA, Karim SA, Dezzutti CS, McElrath MJ, Tomaras GD, Hladik F. 2014. HIV-1 specific IgA detected in vaginal secretions of HIV uninfected women participating in a microbicide trial in Southern Africa are primarily directed toward gp120 and gp140 specificities. PLoS One 9:e101863. doi: 10.1371/journal.pone.0101863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi RY, Levinson P, Guthrie BL, Payne B, Bosire R, Liu AY, Hirbod T, Kiarie J, Overbaugh J, John Stewart G, Broliden K, Farquhar C. 2012. Cervicovaginal HIV-1 neutralizing IgA detected among HIV-1-exposed seronegative female partners in HIV-1-discordant Kenyan couples. AIDS 26:2155–2163. doi: 10.1097/QAD.0b013e328359b99b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchoff V, Wong S, St Jeor S, Pari GS. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch Virol 147:321–333. doi: 10.1007/s705-002-8322-9. [DOI] [PubMed] [Google Scholar]


