ABSTRACT
Latent membrane protein 1 (LMP1) of Epstein-Barr virus (EBV) induces constitutive signaling in EBV-infected cells to ensure the survival of the latently infected cells. LMP1 is localized to lipid raft domains to induce signaling. In the present study, a genome-wide screen based on bimolecular fluorescence complementation (BiFC) was performed to identify LMP1-binding proteins. Several actin cytoskeleton-associated proteins were identified in the screen. Overexpression of these proteins affected LMP1-induced signaling. BiFC between the identified proteins and LMP1 was localized to lipid raft domains and was dependent on LMP1-induced signaling. Proximity biotinylation assays with LMP1 induced biotinylation of the actin-associated proteins, which were shifted in molecular mass. Together, the findings of this study suggest that the association of LMP1 with lipid rafts is mediated at least in part through interactions with the actin cytoskeleton.
IMPORTANCE LMP1 signaling requires oligomerization, lipid raft partitioning, and binding to cellular adaptors. The current study utilized a genome-wide screen to identify several actin-associated proteins as candidate LMP1-binding proteins. The interaction between LMP1 and these proteins was localized to lipid rafts and dependent on LMP1 signaling. This suggests that the association of LMP1 with lipid rafts is mediated through interactions with actin-associated proteins.
INTRODUCTION
Epstein-Barr virus (EBV) is a DNA tumor virus and an etiologic agent of infectious mononucleosis (1, 2). Current models suggest that lytic replication occurs in epithelial cells and that latent infection occurs primarily in B lymphocytes. Latent infection of B lymphocytes with EBV induces a number of cellular changes that reprogram the latently infected cells to establish a subset of memory B cells that contain the viral genome and persist for the life of the infected host. In vitro infection of peripheral blood mononuclear cells with EBV is sufficient to establish latently infected, immortalized lymphocyte cell lines (LCLs). In addition, latent infection is associated with human cancer (3–9). Nearly all patients with endemic cases of Burkitt's lymphoma and nasopharyngeal carcinoma contain EBV, and a significant number of patients with Hodgkin's lymphoma and gastric carcinoma contain EBV. Finally, in the presence of immunodeficiency, EBV induces lymphoproliferative diseases.
During lytic replication, EBV expresses the cadre of herpesvirus genes required to replicate the viral DNA and assemble virus particles de novo. During latency, a number of EBV nuclear antigens (EBNAs), latent membrane proteins (LMPs), and regulatory RNAs are expressed (1, 2). These regulate the latently infected cells to alter the cellular environment to favor the survival of the infected cells and the maintenance of the viral episome. LMP1 is required for the establishment of latency in vitro and is considered the oncogene of EBV since it can induce the phenotypic transformation of rodent fibroblasts (10–15). Fibroblasts that express LMP1 grow in an anchorage-independent fashion and can overcome contact inhibition. A number of signaling pathways induced by LMP1, including the c-Jun N-terminal kinase (JNK), phosphatidylinositol 3-kinase (PI3K), extracellular signal-regulated kinase (ERK), and both canonical and noncanonical nuclear factor kappa B (NF-κB) pathways, have been defined, and induction of these signaling pathways results in altered gene expression in LMP1 cells (13, 16–22). Activation of these signaling pathways by LMP1 has been associated with specific cellular phenotypes. Both ERK signaling and PI3K signaling have been correlated with rodent fibroblast transformation (13, 23, 24). Activation of PI3K is associated with increased motility and invasion of epithelial cells (25). Inhibition of NF-κB signaling in LCLs induces apoptosis (26).
Signaling of LMP1 is induced through three activities; oligomerization, lipid raft partitioning, and adaptor binding. The 6-pass transmembrane domain of LMP1 oligomerizes within the membrane without ligand binding, and LMP1 is constitutively present in the cholesterol-rich, lipid raft domains of the membrane. Signaling of LMP1 is induced through the binding of proteins in the tumor necrosis factor receptor (TNFR) pathway to the C-terminal cytoplasmic domain of LMP1. C-terminal activating region 1 (CTAR1) binds TNFR-associated factors (TRAFs) and functions similarly to CD40 signaling. CTAR2 also induces binding to TRAFs through TRADD and RIP1 and induces signaling similarly to TNFR1. CTAR1 primarily induces noncanonical NF-κB, PI3K, and ERK signaling, while CTAR2 induces canonical NF-κB and JNK signaling. Although much is known about many of the factors required for induction of signaling and the resulting gene expression changes induced by LMP1, the technical challenges of working with membrane proteins and the dynamic nature of the assembly of the signaling complex have made some of the mechanisms of LMP1 signaling difficult to discern. For example, whether TRAF binding and raft association are sequential, interdependent, or independent remains unclear.
Our lab has previously utilized bimolecular fluorescence complementation (BiFC) to examine the assembly of the LMP1 protein complex in the membrane of mammalian cells. BiFC between LMP1-LMP1 and LMP1-TRAF combinations was localized to previously observed cellular locations, i.e., perinuclear regions and patches within the membranes of cells. Mutation of the TRAF-binding sites of LMP1 resulted in decreased BiFC between LMP1-TRAF combinations. We have used BiFC as a screening tool to identify new LMP1-binding proteins. LMP1 fused to the N terminus of yellow fluorescent protein (NYFP) was used as a bait protein, and the C terminus of yellow fluorescent protein (CYFP) was contained in an enhanced retroviral mutagen (ERM) that forms a gene trap with cellular proteins. We identified that transmembrane protein 134 (Tmem134) is a membrane protein of unknown function induced by BiFC with LMP1. A portion of Tmem134 fractionated with LMP1 in the lipid rafts of cells, and inhibition of Tmem134 affected LMP1 signaling.
In the current study, we report the results of a genome-wide screen with an ERM vector that is compatible with genes that are trapped where the first base of the exon is the first base of the codon triplet. YFP-positive cells were sorted by fluorescence-activated cell sorting (FACS), and single-cell clones were examined for the targeted open reading frames. A number of new potential LMP1-binding proteins were identified. In particular, several proteins associated with the actin cytoskeleton were identified in our screen, including actinin 1 (Actn1), actinin 4 (Actn4), gelsolin (Gsn), and tropomyosin (Tpm). BiFC between LMP1 and the cytoskeletal proteins was decreased by LMP1 signaling mutants and was restricted to lipid rafts. Overexpression of actinin 1, actinin 4, and gelsolin decreased LMP1-induced gene reporter activity. In addition, regions of the cytoskeletal proteins that were required for BiFC with LMP1 were mapped by the use of deletion mutants. Finally, LMP1 fused to a promiscuous biotin ligase induced the biotinylation of the cytoskeletal proteins, which could be observed by streptavidin pulldown. Together these data suggest that the cytoskeleton underlying lipid rafts is associated with the LMP1 signaling complex and that LMP1-induced raft localization may be induced through this interaction.
MATERIALS AND METHODS
Plasmids.
The enhanced retroviral mutagen (ERM) vector VC1 was generously provided by Z. Ding and Z. Songyang (27, 28). VC1 is an inducible exon trap vector which splices to cellular exons to create CYFP-tagged proteins encoded by genes in which the first base of the exon is the first base of the codon triplet. The various vectors used in the current study were described previously (29, 30) and included wild-type LMP1 expression vectors LMP1-NYFP (with a C-terminal NYFP tag), LMP1-CYFP (with a C-terminal CYFP tag), LMP1-NYFP-TH (where TH represents an inducible retrovirus), and M3-LMP1 (where M3 represents an N-terminal triple myc tag). Previously described mutant LMP1 vectors with C-terminal NYFP tags were also used and included LMP1-A5-NYFP (a CTAR1 mutant with 5 A residues [A5]), LMP1-Y384G-NYFP (a CTAR2 mutant), and LMP1-A5-Y384G-NYFP (a CTAR1/CTAR2 mutant). LMP1-A5-Y384G was subcloned into the myc-tagged vector to create M3-LMP1-A5-Y384G (a CTAR1/CTAR2 mutant with an N-terminal triple myc tag). LMP1-CYFP was cloned into the inducible retrovirus vector RetroX-Tight-Pur (Clontech) to produce LMP1-CYFP-TP for use as a BiFC-positive control. Because LMP1-A5-Y384G-NYFP had some residual reporter activity (30), two additional CTAR2 mutants were constructed by PCR mutagenesis. The CTAR2 sequence was deleted by fusion to the NYFP tag following leucine 378 of LMP1 to create LMP1-378-NYFP. In addition, tyrosine 385 was also mutated to glycine in the mutant with the Y384G CTAR2 mutation to create LMP1-GG-NYFP (with Y384G and Y385G mutations). Both CTAR2 mutations were also introduced into CTAR1 mutants to yield LMP1-A5-378-NYFP and LMP1-A5-GG-NYFP.
CYFP-tagged proteins from the screen were subcloned from cDNA that had been PCR amplified with primers CYFP′ and T7′ from the BiFC-positive cloned cell lines as described previously (29). Actinin 1, actinin 4, and gelsolin were cloned with the CYFP-hemagglutinin (HA) tags from cDNA into pcDNA3 (Invitrogen). Actinin 4 and gelsolin were cloned with gene-specific primers. Actinin 1 was cloned into a shuttle vector using an In-Fusion cloning kit (Clontech), which induces homologous recombination between vector and insert fragments on the basis of 15 to 20 bp of homology on the ends of the fragments. The shuttle vector CYFP-Zip+AscI/T7 was made by full-plasmid PCR with primers to create a unique AscI restriction site and to insert the T7′ sequence in the reverse orientation immediately downstream of the CYFP′ sequence in the CYFP-coding sequence of CYFP-Zip (30). Linearization of the vector with AscI leaves the CYFP′ and T7′ sequences on the ends of the vector fragment that are homologous to the CYFP′ and T7′ sequences of the PCR products that are required for the In-Fusion reaction.
Actinin 1, actinin 4, and gelsolin deletion mutants were constructed by full-plasmid PCR. Outward-facing primers flanking specific domains were designed with unique restriction sites. Following PCR amplification, products were digested using the unique restriction enzyme, gel purified, and ligated. Deletion of two domains at once was similarly accomplished using primers specific for regions flanking both domains. All constructs were tested for expression of proteins of the appropriate molecular mass and sequenced to confirm that the desired deletions with no other changes were obtained. Both actinin 1 and actinin 4 have two N-terminal calponin homology (CH) domains (the CH1/2 domains), internal spectrin repeats, and C-terminal EF-hand domains. Each of these domains was deleted individually to produce delta CH1/2, delta spectrin, and delta EF-hand mutants for both actinin 1 and actinin 4. Deletion of both the CH1/2 and spectrin domains to produce an EF-hand-only mutant and deletion of both the spectrin and EF-hand domains to produce an CH1/2-only mutant were also performed for actinin 1 and actinin 4. The N terminus of gelsolin contains three gelsolin-like repeats and an actin-severing domain, and the C terminus contains three more gelsolin-like repeats and a calcium-sensitive actin-binding domain. Gsn mutants with deletion of the N terminus and C terminus were constructed to create mutants Gsn-dN and Gsn-dC, respectively.
Vectors containing the biotin ligase used for biotin identification (BioID) (31) were obtained from Addgene (vectors 36047 and 35700). The BioID tag was cloned into the LMP1-NYFP and LMP1 mutant constructs to replace the NYFP tag downstream of the linker (GGGGSGGGG). The resulting constructs were subcloned into the inducible retrovirus vector to produce LMP1-BioID-TH and mutants LMP1-A5-BioID-TH, LMP1-GG-BioID-TH, and LMP1-A5-GG-BioID-TH.
Cell culture, transfections, and retrovirus.
Human embryonic kidney (HEK-293T), SiHa, and Rat-1 (rodent fibroblast) cells were maintained in Dulbecco modified Eagle medium (DMEM; Mediatech) supplemented with an antibiotic-antimycotic mixture and 10% (vol/vol) heat-inactivated fetal bovine serum (FBS). For BiFC assays and pulldown experiments, HEK-293T cells were transfected by use of the Transit-LT1 transfection reagent (Mirus) according to the manufacturer's directions. Stably transduced cells were selected with G418 (neomycin [Neo]; 1 mg/ml; Mediatech), hygromycin B (Hygro; 0.5 mg/ml; Mediatech), or puromycin (Puro; 5 μg/ml; Mediatech). Tetracycline-inducible promoters were induced with the less toxic tetracycline analog doxycycline (Dox; Clontech).
Small-scale retrovirus production was accomplished as previously described (29, 30, 32) by transfection of HEK-293T cells with retrovirus expression vectors with plasmids expressing vesicular stomatitis virus G glycoprotein and gag-pol using the Transit-LT1 transfection reagent. At 24 h posttransfection, the medium was changed and the cells were moved to 33°C. At 48 h posttransfection, clarified supernatants were collected and used to infect Rat-1 cells. For production of ERM retrovirus, HEK-293T cells were transfected as described above, except that calcium phosphate transfection in multiple plates was used. Supernatants were collected at 48, 72, and 96 h posttransfection, and the plates were replenished with fresh medium. Supernatants from each day were pooled and filtered using 0.45-μm-pore-size filter flasks to remove cellular debris, and small aliquots and pooled retrovirus were frozen. The titers of aliquots of frozen ERM retrovirus were determined with serial dilutions of virus aliquots on SiHa cells by counting the puromycin-resistant colonies present at 1 week postinfection. ERM retrovirus was concentrated using a Retro-X concentrator (Clontech) according to the manufacturer's directions and resuspended in an appropriate volume of DMEM for bait cell infection. Retrovirus infection was performed in the presence of 8 μg/ml Polybrene.
Western blotting.
Cells were washed with ice-cold phosphate-buffered saline (PBS; Mediatech) and lysed with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1% deoxycholic acid, protease and phosphatase inhibitors [Pierce]). Cell lysates were clarified by centrifugation and quantitated by use of a Bio-Rad DC protein assay system (Bio-Rad). Samples were then boiled in SDS sample buffer, the amounts of protein indicated below were separated using SDS-polyacrylamide gel electrophoresis, and the separated proteins were transferred to nitrocellulose membranes (LI-COR) for Western blotting. LMP1 was detected with a mixture of four rat monoclonal antibodies diluted 1:500 each (Cao 7E10, Cao 8G3, LMP1 IG6, and Cao 7G8; Ascenion GmbH). Actinin 1, actinin 4, gelsolin, TRAF2, and TRAF3 antibodies were purchased from Santa Cruz, and HA-tagged and actin antibodies were purchased from Cell Signaling. myc-tagged and gelsolin antibodies (EMD) and YFP antibodies (catalog number 632381; Clontech) were also used. Primary antibodies were detected with IRDye-labeled secondary antibodies (LI-COR) and by scanning with a LI-COR Odyssey imaging system. Bands were quantitated using LI-COR imaging software.
Reporter assays.
Reporter assays were performed as previously described (30, 32). HEK-293T cells were transfected with test plasmids, NF-κB–luciferase (Stratagene), and control renilla luciferase (Promega)-simian virus 40 (RL-SV40). At 40 h posttransfection, the cells were harvested and dual-luciferase assays were performed using a dual-luciferase reporter assay system (Promega) according to the manufacturer's directions. Triplicate samples were analyzed for each condition, and each experiment was repeated three times on different days.
ERM screen.
The ERM screen, which is based on previously described strategies (27, 28, 33), was performed as previously described (29) with the following modification. Rat-1 Tet-On LMP1-NYFP bait cells were cloned by limiting cell dilution in 96-well plates. Clones were screened for Dox-dependent LMP1 expression by In-Cell Western blotting according to the manufacturer's directions (LI-COR) and flow cytometry as previously described (29). Bait cells were infected with the ERM vector VC1 at a multiplicity of infection of 0.3 and selected with Neo, Hygro, and Puro. In parallel, BiFC-control cells were made by infection of Rat-1 Tet-On LMP1-NYFP cells with LMP1-CYFP-TP, followed by Neo, Hygro, and Puro selection. The concentration and duration of Dox incubation were optimized to determine the minimum concentration required to induce BiFC in control cells. Induced and uninduced BiFC-control cells were used to establish gating for BiFC-positive cells that excluded BiFC-negative cells. VC1-infected cells were induced with 50 ng/ml doxycycline overnight prior to cell sorting. BiFC-positive VC1 cells were sorted by fluorescence-activated cell sorting (FACS) using a FACSAria flow cytometer (BD Biosciences) in the Rosalind Franklin University of Medicine and Science (RFUMS) Flow Cytometry Core Facility. Sorted cells were pooled, replated at a low density, and grown for several days without Dox. Pooled cells were reinduced as described above, cells with high levels of YFP fluorescence were sorted, and some of the cells were plated in limiting dilution in 96-well plates. The remaining cells were grown for a few days at a low density and induced, and total RNA was purified for possible future analyses. Single-cell clones were expanded from 96-well plates, plated in replicates, and tested for BiFC by addition of 500 ng/ml Dox overnight, followed by observation by fluorescence microscopy and flow cytometry. One hundred single-cell clones were analyzed further.
Clone identification.
Clone identification was performed as previously described (29). Total RNA was purified from induced VC1 clones and treated with DNase (Qiagen). First-strand cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen) was performed with anchored dT-primed primer RT-1T (GCTAATACGACTCACTATAGGGATCCTTTTTTTTTTTTTTTTV). Primer RT-1T has T7′ primer sequences (underlined) at its 5′ ends for subsequent PCR and sequencing steps. The cDNA was PCR amplified with primers T7-2′ (GCTAATACGACTCACTATAGGGATC) and CYFP′ (ACTTCAAGATCCGCCACAACATCGAG) and AccuPrime Pfx (Invitrogen). Reaction mixtures with or without reverse transcriptase were used to control for amplification of contaminating genomic DNA. PCR-amplified bands were gel purified and directly sequenced with the CYFP′ primer. The sequence downstream of the ERM splice junction was used to search the GenBank database by BLAST analysis to determine the targeted open reading frame.
BiFC assays.
BiFC assays were performed as previously described (30). Different combinations of BiFC plasmids were transfected into HEK-293T cells and examined for fluorescence by flow cytometry. Cells were cotransfected with pmCherry-N1 (Clontech) to gate on transfected cells. At 48 h posttransfection, the cells were trypsinized, washed, and resuspended in PBS. Fluorescence was determined using an LSRII flow cytometer (BD Biosciences) in the Flow Cytometry Core Facility of RFUMS. The main cell population was gated using the forward scatter-versus-side scatter dot plot. Transfected cells were enriched by gating for mCherry-fluorescent cells. YFP gating was determined by comparing the histograms of cells transfected with mCherry alone with those of cells transfected with the BiFC plasmid. The YFP fluorescence of 1 × 104 mCherry-positive cells was analyzed for each combination of plasmids. Flow cytometry data were analyzed with BD FACSDiva (BD Biosciences) and FlowJo (TreeStar) software. The remaining cells were harvested for Western blotting to confirm the expression of the BiFC plasmids.
Pulldown assays.
Pulldown of LMP1-binding proteins using LMP1 with an N-terminal tandem triple myc tag was performed as previously described (29). HEK-293T cells were transfected with CYFP-tagged clones individually or with M3-LMP1 or M3-LMP1-A5-Y384G. At 48 h posttransfection, the cells were harvested for pulldown. Ten percent of the lysates were set aside for use as directly loaded samples, and the rest of the lysates were subjected to pulldown with a mammalian c-myc tag immunoprecipitation/coimmunoprecipitation kit (Pierce) according to the manufacturer's directions. Directly loaded, pulled down, and unbound proteins were analyzed by Western blotting for myc-tagged (LMP1) and clone proteins (YFP antibody) (Fix It).
Confocal microscopy.
Cloned cells from the ERM screen were plated on coverslips, induced with Dox (at 20 ng/ml overnight), fixed at 18 h after Dox induction with 4% paraformaldehyde in PBS for 10 min at 4°C, and washed with PBS. Coverslips were blocked for 10 min at room temperature in blocking solution (0.2% fish skin gelatin, 0.2% Triton X-100, phosphate-buffered saline) and stained in primary antibody for 1 h in a humidified chamber. The following primary antibodies were used in blocking solution: rat anti-LMP1 (1:100) and lipid raft marker goat anti-flotillin-1 (1:25) (Abcam, Cambridge, MA). Coverslips were then washed with PBS and stained with secondary antibodies for 30 min in a humidified chamber. Donkey anti-goat Alexa Fluor 594 and chicken anti-rat Alexa Fluor 647 fluorescent secondary antibodies were used at a 1:1,000 dilution in blocking solution. The coverslips were washed and mounted with ProLong Gold antifade reagent containing 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Note that paraformaldehyde fixation preserves the YFP fluorescence after BiFC. An Olympus FV10i confocal microscope at the Microscopy Core of the Rosalind Franklin University of Medicine and Science was used for data collection at a ×60 magnification under oil immersion, and analysis was performed using FluoView FV1000 software (Olympus, Melville, NY). z-stacked images represent a 0.5-μm section of the cell in the x, y, and z planes. Uninduced cells, cells stained with secondary antibody alone, and cells stained with a single antibody were used to ensure specificity.
BioID assays.
Inducible cell lines were created by infection of C33A cells with Tet-On retrovirus and infection with LMP1-BioID-TH retrovirus. Inducible cells were incubated overnight in the presence or absence of Dox and biotin (50 μM). On the following day, the cells were lysed in RIPA buffer and quantitated, and equal amounts of protein for each condition were pulled down with streptavidin beads (Sigma) overnight. Bound proteins were eluted by boiling with SDS-polyacrylamide gel loading buffer containing biotin (3 mM) and were analyzed by Western blotting as described above. Control blots including input, unbound, and pulldown fractions were reacted with streptavidin-680 (LI-COR) to control for labeling and pulldown.
RESULTS
Seminal work with yeast two-hybrid screens using the cytoplasmic domain of LMP1 identified the TRAFs and other proteins to be critical cellular proteins that are required for LMP1 signaling. However, membrane proteins are generally not suitable for traditional two-hybrid approaches due to the requirement for nuclear translocation of bait and prey proteins. In addition, screens performed in lower organisms often lack the accessory proteins necessary to detect secondary interactions that may be critical for function. In contrast, our recent studies with full-length LMP1 have utilized BiFC in the membrane of mammalian cells within the physiological context (30). We recently identified Tmem134 to be a novel LMP1-interacting protein during the optimization of the BiFC screen with an ERM (29). In the present study, we report the results of a screen with one of the ERM vectors, VC1.
To identify new proteins that are important for LMP1 signaling, a BiFC screen was performed using our previously described approach (29). The screen was performed in Rat-1 rodent fibroblasts because they tolerate LMP1-NYFP bait expression and can be recovered at a high frequency following cell sorting. Attempts to perform the screen in several human epithelial cell lines were unsuccessful because of bait cell line instability and an inability to recover viable cells following induction and cell sorting. Rat-1 cells were infected with Tet-On and LMP1-NYFP-TH retrovirus to create a bait cell line for BiFC which expresses LMP1-NYFP in a Dox-dependent manner (LMP1-Bait). LMP1-Bait cells were cloned by limiting cell dilution, and single-cell clones were screened by In-Cell Western blotting and flow cytometry to confirm the inducible expression of LMP1-NYFP (data not shown). Previous studies demonstrated that LMP1-NYFP and LMP1-CYFP produce BiFC, and a positive-control cell line for BiFC (LMP1-BiFC) was created by infection of LMP1-Bait cells with LMP1-CYFP-TP. LMP1-BiFC and LMP1-Bait cells were tested for the minimum Dox concentration and duration of incubation required to induce BiFC and to ensure maximum recovery following induction, respectively. A LMP1-Bait cloned cell line was chosen, and a Dox concentration of 50 ng/ml was chosen for the ERM screen.
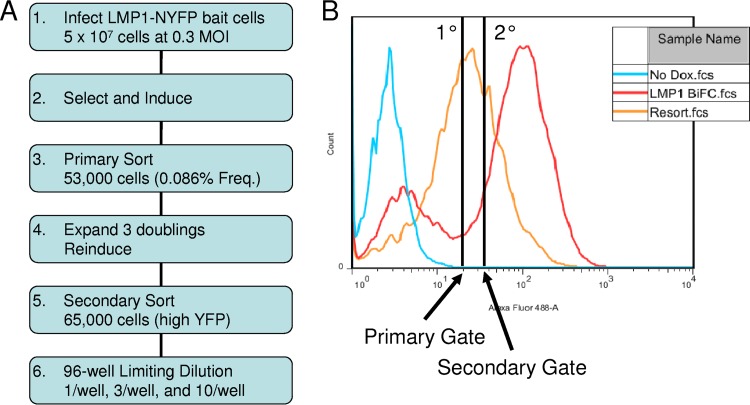
The ERM screen was performed as shown in Fig. 1A. LMP1-Bait cells were infected with VC1 at a multiplicity of infection of 0.3 to ensure no more than one infection per cell. VC1 is the ERM exon trap retrovirus that is in frame with cellular exons in which the first base of the exon is the first base of the codon triplet. Infected cells were selected with Neo, Hygro, and Puro. After two doublings, cells were induced with 50 ng/ml Dox overnight and sorted on the basis of YFP fluorescence. The YFP-positive gate was determined on the basis of the histograms of uninduced and induced LMP1 BiFC-control cells (Fig. 1B, blue and red histograms, respectively). The gate for the primary sort was set to exclude cells with fluorescence similar to that of uninduced cells (Fig. 1B, primary gate). Out of 50 million infected LMP1-Bait cells, about 53,000 YFP-positive events were sorted. Because of the low frequency of positive cells and to avoid false-positive events, all of the sorted cells were replated in the absence of Dox at a low density, grown for three doublings, and reinduced with Dox as described above. The fluorescence of the entire reinduced sorted pool shifted (Fig. 1B, orange histogram). This confirms that the sorted cells reinduce BiFC and suggests that in the screen the ERM successfully integrated into loci where the encoded proteins produce BiFC with LMP1.
FIG 1.
Flowchart of BiFC and secondary sorting. (A) The flowchart depicts the experimental design for BiFC that we used for the VC1 screen. (B) Flow histograms for YFP fluorescence of the control (LMP1-NYFP and LMP1-CYFP) and secondary sorting of the sorted ERM pool. Results for uninduced and induced control cell lines are shown (blue and red histograms, respectively). The induced ERM pool (orange histogram) and gates from primary sorting and secondary sorting are shown. MOI, multiplicity of infection; Freq., frequency; fcs, flow cytometry standard.
Because our previous studies indicated that low-level BiFC may arise from nonphysiological interactions (30), a secondary sort was performed on the reinduced cells to isolate the cells with the highest fluorescence (Fig. 1B, secondary gate). In the secondary sort, about 65,000 YFP-positive events out of nearly 300,000 total cells were sorted. Following the secondary sort, about one-third of the cells with high-level YFP fluorescence were plated in limiting dilution in 96-well plates to isolate single-cell clones. The remaining cells were plated at a low density, grown for 1 week, and induced overnight, and total RNA was isolated. The purified RNA was frozen for possible future analyses by next-generation sequencing or a similar approach to determine the YFP-positive messages in the pool of sorted cells without single-cell cloning.
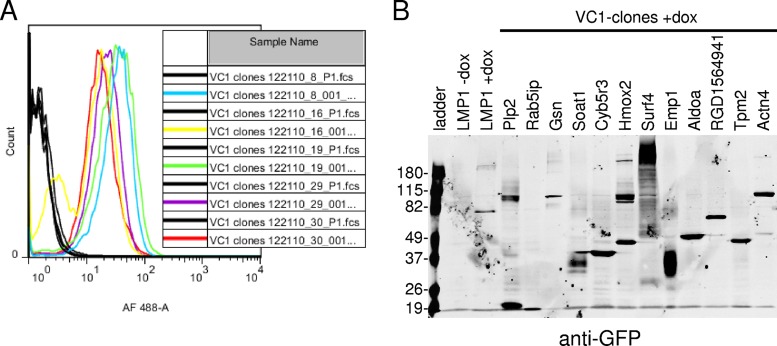
One hundred single-cell clones were chosen for further analyses. Clones were expanded and examined for BiFC induction by fluorescence microscopy and flow cytometry. All clones induced BiFC following Dox induction by both methods. Representative histograms of uninduced and induced cells of several clones are given in Fig. 2A (black and colored histograms, respectively). Clones were also analyzed by Western blotting for expression of CYFP-tagged fusion protein expression with a green fluorescent protein (GFP)-specific monoclonal antibody that specifically recognizes the CYFP tag. A representative Western blot for induced clones is given in Fig. 2B. Results for uninduced and induced cells with LMP1 BiFC are also shown. Proteins of various sizes were observed in different clones.
FIG 2.
VC1 clone BiFC and fusion protein expression. (A) VC1 clones were tested for BiFC induction by flow cytometry. Clones were plated in duplicate and tested for BiFC following overnight induction with Dox at 500 ng/ml. The YFP fluorescence of uninduced cells (black histograms) was compared to that of induced cells (colored histograms). AF, Alexa Fluor. (B) Induced clones were examined for CYFP-fusion proteins by Western blotting with a GFP-specific monoclonal antibody that specifically recognizes the CYFP domain. Uninduced and induced control cells with BiFC (LMP1 without Dox and LMP1 with Dox, respectively) and the ladder are indicated. Control cells tested for BiFC inducibly express LMP1-NYFP and LMP1-CYFP. Numbers on the left are molecular masses (in kilodaltons).
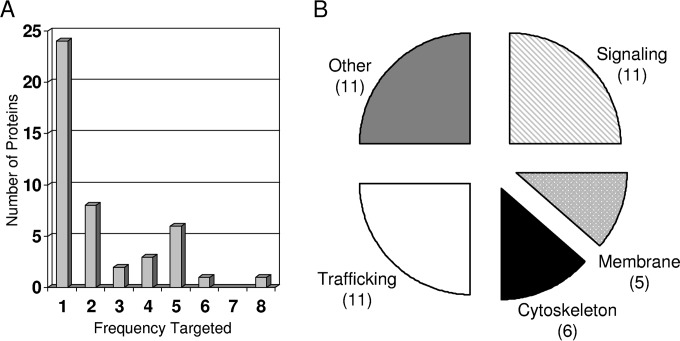
The mRNA containing the CYFP tag was identified through DNA sequencing as described previously (29). cDNA was synthesized with anchored oligo(dT) primers containing the T7′ sequence at the 5′ end and PCR amplified with T7′- and CYFP-specific primers. Bands were gel purified and directly sequenced with the CYFP-specific primer. Cellular proteins were identified by a BLAST search of sequences downstream of the CYFP-coding sequence and splice junction of the ERM vector. The targeted open reading frames of the VC1 clones are given in Table 1 and summarized in Fig. 3. The tagged proteins of 22 of the clones were targeted by only 1 of the clones, while a number of the proteins were targeted by several of the clones from two to eight times (Fig. 3A). On the basis of the fact that cells were selected and expanded at two steps in our experimental design, it is certain that clones targeting the same protein were duplicates of the same initially infected cell or independently targeted the same protein. However, it seems unlikely that the same initial clone would be randomly picked from among our 100 clones multiple times in 65,000 events in the secondary sort, but the possibility cannot be ruled out. The proteins were grouped into several groups on the basis of published or hypothesized functions (Fig. 3B). Many of the proteins that have known functions in trafficking and signaling could be envisioned to affect LMP1 maturation and function. The likely regulation of LMP1 function by a number of other proteins that are associated with metabolism and that have an unknown function is less clear.
TABLE 1.
Proteins encoded by genes identified in BiFC screen
| Protein type and name | Description | GenBank accession no. | No. of clonesa |
|---|---|---|---|
| Signaling proteins | |||
| Emp1 | Epithelial membrane protein 1 | NM_012843.2 | 5 |
| Kidins220 | Kinase D-interacting substrate 220 | NM_053795.1 | 1 |
| C1qtnf2 | C1q and tumor necrosis factor-related protein 2 | NM_001191918.1 | 3 |
| Tm4sf4 | Transmembrane 4 L 6 family member 4 | NM_053785.1 | 1 |
| Lgals1 | Lectin, galactoside-binding, soluble, 1 | NM_019904.1 | 1 |
| Mall | Mal, T cell differentiation protein-like | NM_001014182.1 | 1 |
| Ifitm3 | Interferon-induced transmembrane protein 3 | NM_001136124.1 | 1 |
| Tspan1 | Tetraspanin 1 | NM_001004236.1 | 1 |
| Scn2a1 | Sodium channel, voltage gated, type II, alpha1 | NM_012647.1 | 1 |
| Sh3d21 | SH3 domain containing 21 | NM_001162535.1 | 6 |
| Maml2 | Mastermind-like 2 | XM_008774867.1 | 1 |
| Membrane proteins | |||
| Vezt | Vezatin, adherens junctions transmembrane protein | NM_001006984.1 | 1 |
| Tmem134 | Transmembrane protein 134 | NM_001078647.1 | 1 |
| Plp2 | Proteolipid protein 2 | NM_207601.1 | 2 |
| Fam69a | Family with sequence similarity 69, member A | NM_001170456.1 | 1 |
| Tmem43 | Transmembrane protein 43 | NM_001007745.1 | 2 |
| Cytoskeleton proteins | |||
| Gsn | Gelsolin | NM_001004080.1 | 1 |
| Tpm2 | Tropomyosin 2beta | NM_001024345.1 | 1 |
| Actn1 | Actinin alpha1 | NM_031005.3 | 2 |
| Actn4 | Actinin alpha4 | NM_031675.2 | 5 |
| Actg1 | Actin gamma1 | NM_001127449.1 | 1 |
| Mical2 | Microtubule-associated monooxygenase, calponin, and LIM domain-containing protein 2 | NM_001139508.1 | 2 |
| Trafficking proteins | |||
| Rab5ip | RAB5-interacting protein | NM_001013922.1 | 2 |
| Surf4 | Surfeit 4 | NM_001033868.1 | 5 |
| Bnip1 | BCL2/adenovirus E1B-interacting protein 1 | NM_080897.1 | 3 |
| Ranbp1 | RAN binding protein 1 | NM_001108324.1 | 1 |
| Sept11 | Septin 11 | NM_001107208.2 | 4 |
| Spcs1 | Signal peptidase complex subunit 1 homolog | NM_001131006.1 | 4 |
| Vamp8 | Vesicle-associated membrane protein 8 | NM_031827.1 | 1 |
| Dpm2 | Dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit | NM_019252.1 | 1 |
| Sec62 | SEC62 homolog | NM_001034129.1 | 1 |
| Sft2d1 | SFT2 domain-containing protein 1 | NM_001008302.1 | 1 |
| Syne1 | Spectrin repeat-containing, nuclear envelope protein 1 | XM_006227849.1 | 1 |
| Other proteins | |||
| Slc27a4 | Solute carrier family 27 (fatty acid transporter), member 4 | NM_001100706.1 | 2 |
| RGD1564941 | Ankyrin repeat domain-containing protein 7-like | XM_006227851.2 | 4 |
| Cyb5r3 | Cytochrome b5 reductase 3 | NM_138877.1 | 8 |
| Hmox2 | Heme oxygenase (decycling) 2 | NM_001277073.1 | 5 |
| Aldoa | Aldolase A, fructose-bisphosphate | NM_001177305.1 | 5 |
| Ctps1 | CTP synthase 1 | NM_001134873.1 | 2 |
| Slc38a4 | Solute carrier family 38, member 4 | NM_130748.1 | 5 |
| Soat1 | Sterol O-acyltransferase 1 | NM_031118.1 | 1 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | NM_017008.4 | 1 |
| Fa2h | Fatty acid 2-hydroxylase | NM_001135583.1 | 2 |
| Rpl4 | Ribosomal protein L4 | NM_022510.1 | 1 |
Number of clones with the indicated CYFP-fusion protein.
FIG 3.
Summary of hits from our screen for VC1 BiFC. (A) Number of clones which target the same protein in our screen. The clones are displayed according to the frequency at which different proteins were targeted. About 50% of the identified proteins were targeted by a single clone (24 proteins, frequency 1). The other identified proteins were targeted by multiple clones from 2 to 8 times. (B) The identified proteins were grouped on the basis of published or hypothesized functions. The number of proteins in each functional group is indicated in parentheses.
The current study focused on several proteins associated with the actin cytoskeleton, including actinin 1 (Actn1; also called alpha-actinin), actinin 4 (Actn4), and gelsolin (Gsn). Previous studies have demonstrated both an interaction between LMP1 and the intermediate filament protein vimentin and the regulation of vimentin by LMP1 (34, 35). In addition, although previous studies have indicated that LMP1 signaling induces actin cytoskeleton rearrangement, motility, and invasion (36), the direct regulation of LMP1 signaling by the actin cytoskeleton has not been observed. Finally, there is an extensive literature suggesting that the cytoskeleton is important for the structure and function of the lipid raft domains of the membrane (37).
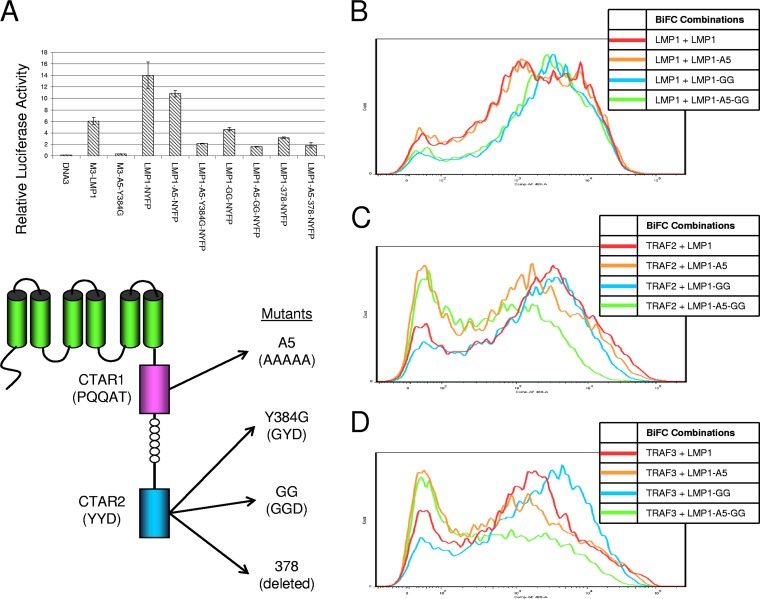
To determine if the signaling domains of LMP1 are important for the potential association with the actin-binding proteins, reporter and BiFC assays were performed. Several LMP1 mutants with different tags were tested by an NF-κB reporter assay (Fig. 4A) to ensure that NYFP-tagged LMP1 signaling activity could be correlated to BiFC. Vector control (pcDNA3) or LMP1 constructs were transfected into HEK-293T cells with the NF-κB luciferase reporter and control renilla luciferase reporter, and dual-luciferase reporter assays were performed at 48 h posttransfection. Triple myc-tagged LMP1 (M3-LMP1) activated the reporter more than 50-fold more than the vector control cells did, and M3-LMP1-A5-Y384G (a CTAR1/2 mutant) activated the reporter slightly, about 3-fold, more than the vector control cells did. Consistent with the findings of our previous studies with LMP1 with the NYFP tag (30) and numerous other studies with LMP1 signaling mutants in NF-κB reporter assays, mutation of CTAR1 (LMP1-A5-NYFP), CTAR2 (LMP1-Y384G-NYFP), and CTAR1 and CTAR2 (LMP1-A5-Y384G-NYFP) decreased the level of reporter induction. Additional CTAR2 mutations were also tested with the A5 CTAR1 mutant, including the A5-Y384G/Y385G CTAR2 double point mutant (LMP1-A5-GG-NYFP) and a CTAR2 deletion mutant which fuses the NYFP domain to amino acid 378 of LMP1 (LMP1-A5-378-NYFP). These additional mutations resulted in a slight decrease in reporter induction. LMP1-A5-GG-NYFP, which had the least reporter activity, was chosen for use as the LMP1 CTAR1 and CTAR2 mutant for BiFC assays.
FIG 4.
Activity of LMP1 mutants. (A) Various LMP1 mutants were tested for their ability to activate an NF-κB reporter. M3 constructs contain an N-terminal triple myc tag. LMP1 constructs for BiFC contain a C-terminal NYFP tag. Mutants with mutations in CTAR1 (A5) and CTAR2 (Y384G, Y384G/Y385G [GG], and amino acid 378) were tested. The structures and sequences are given at the bottom. Relative luciferase activity was determined by a dual-luciferase assay. (B to D) Several LMP1 mutant constructs were analyzed in BiFC assays with LMP1-CYFP (B), CYFP-TRAF2 (C), and CYFP-TRAF3 (D). Representative histograms of YFP fluorescence are displayed for LMP1-NYFP, LMP1-A5-NYFP, LMP1-GG-NYFP, and LMP1-A5-GG-NYFP.
NYFP-tagged wild-type LMP1 and LMP1 CTAR1 and/or CTAR2 mutants were tested in control BiFC assays with LMP1-CYFP, CYFP-TRAF2, and CYFP-TRAF3 (Fig. 4B to D, respectively) as previously described (30). This was repeated to ensure that differences in BiFC were observed at the DNA concentrations used in the transfections in the current study because we have previously shown that nonphysiological BiFC can occur through overexpression of the proteins. Since the self-association of LMP1 is independent of the signaling domains, LMP1-LMP1 BiFC (Fig. 4B) was unaffected by mutation of CTAR1 and/or CTAR2. In contrast, LMP1-TRAF BiFC was greatly decreased by mutation of CTAR1 and/or CTAR2 because TRAF binding requires functional CTAR1 and CTAR2 domains (Fig. 4C and D).
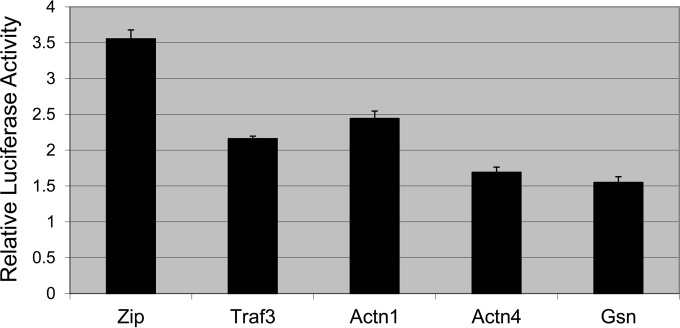
To determine if LMP1 signaling was influenced by Actn1, Actn4, and Gsn, reporter assays were performed (Fig. 5). Reporter assays were performed as described above for Fig. 4A with M3-LMP1, except that the CYFP-tagged proteins indicated in Fig. 5 were also included. M3-LMP1, which contains a triple myc tag, was used to avoid any confounding effects of LMP1-target protein BiFC complexes. LMP1 activated the reporter in the presence of a control plasmid expressing CYFP-Zip, and this activation was inhibited by the addition of CYFP-TRAF3, which can act as a dominant negative construct in LMP1 signaling. Addition of CYFP-Actn1, -Actn4, and -Gsn also decreased LMP1-induced NF-κB reporter activation. Actn1 was slightly less effective than TRAF3 in reducing reporter activity, while Actn4 and Gsn were more effective than TRAF3 and decreased reporter activity to less than 50% of that for the vector control reporter. This suggests that the actin-binding proteins are involved in LMP1 signaling or could be involved in LMP1 trafficking or maturation.
FIG 5.
Regulation of LMP1-induced reporter activity. Relative luciferase activity was determined by a dual-luciferase assay as described in the legend to Fig. 4. M3-LMP1 was transfected with the NF-κB reporter, control renilla luciferase, and effector BiFC plasmids. The BiFC plasmids included a vector control (CYFP-Zip) and CYFP-tagged TRAF3, Actn1, Actn4, and Gsn. The mean relative luciferase activity for triplicate samples for each condition is plotted, and error bars represent the standard deviation for the triplicate samples. Data from a representative experiment are given, and similar results were observed in three independent experiments.
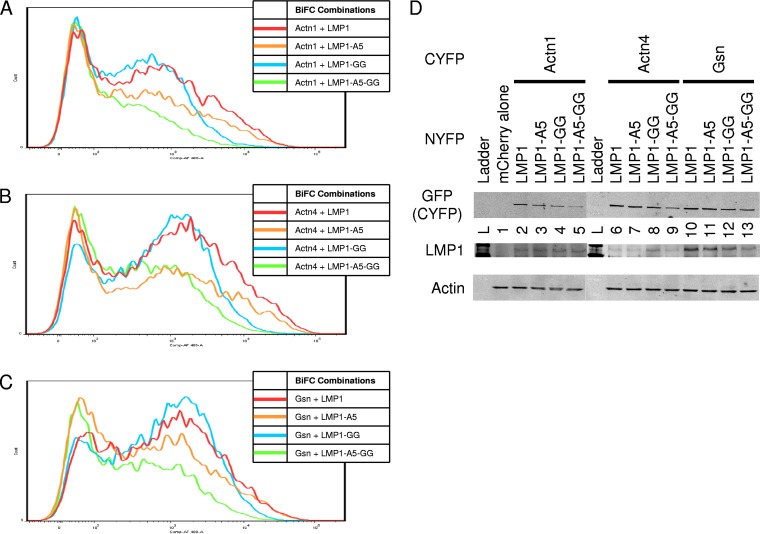
Alteration of reporter activity is suggestive of a functional relationship between LMP1 signaling and the actin-binding proteins. Gene knockdown studies were attempted for Actn1, Actn4, and Gsn using small interfering (siRNA) and short hairpin RNA, but despite the validation of the action of siRNA against GFP-tagged versions of the proteins, significant transient or stable knockdown was not successful. As an alternative approach, we have previously shown that mutation of functional domains of LMP1 results in a decrease in BiFC between proteins, including LMP1-TRAF BiFC (Fig. 4C and D) (30). If LMP1- and actin-binding proteins are functionally related in LMP1 signaling, then BiFC should be sensitive to mutations in CTAR1 and/or CTAR2. To determine if functional LMP1 signaling domains are required for BiFC with actin-binding proteins, BiFC assays were performed (Fig. 6). CYFP-Actn1, -Actn4, and -Gsn were tested with LMP1-NYFP and LMP1 mutants in BiFC assays, and histograms of YFP fluorescence from representative experiments are given. Interestingly, the relative fluorescence of all three proteins with the different LMP1 mutants was similar. BiFC between LMP1 (red histograms) and Actn1, Actn4, and Gsn was confirmed (Fig. 6A to C, respectively). BiFC was greatly diminished with LMP1-A5-GG (green histograms in Fig. 6A to C), which contains mutated CTAR1 and CTAR2, in combination with the actin-binding proteins. BiFC with CTAR2 mutant LMP1-GG-NYFP (blue histograms in Fig. 6A to C) was high and similar to BiFC with wild-type LMP1. BiFC with CTAR1 mutant LMP1-A5-NYFP (orange histograms in Fig. 6A to C) was diminished relative to BiFC with LMP1 or LMP1-GG but slightly higher than BiFC with LMP1-A5-GG. Cells were also harvested and examined using Western blotting for GFP (which recognizes the CYFP tag), LMP1, and the actin loading control to confirm the expression of the different constructs and mutants (Fig. 6D). Although there was some minor variation among the different combinations, all proteins tested for BiFC were expressed. Actn1, Actn4, and Gsn were all of approximately the same molecular mass (100 kDa), and the identity of the different proteins was confirmed by Western blotting with Actn1-, Actn4-, and Gsn-specific antibodies (data not shown). Together the results of the BiFC assays suggest that functional CTAR1 alone is sufficient to induce BiFC and that CTAR2 is not required in the presence of CTAR1 but that the CTAR2 mutation in the presence of the CTAR1 mutation further decreases BiFC. Counterscreening of other proteins found in our BiFC screen with the LMP1 mutants revealed two patterns of BiFC (data not shown). Like LMP1-LMP1 BiFC, BiFC with some proteins was unaffected by the CTAR1 and CTAR2 mutations, including Surfeit 4, Aldoa, BNIP1, and Plp2. Other proteins had similar BiFC patterns that were sensitive to the CTAR1 and CTAR2 mutations, like the actin-binding proteins and TRAFs, including RanBP1 and Cyb5r3. The latter group would be predicted to be important for LMP1 functions specifically required for signaling.
FIG 6.
LMP1 mutant BiFC assays. (A to C) Various LMP1 CTAR1 and CTAR2 mutants from the assay whose results are presented in Fig. 4 were tested for BiFC with CYFP-Actn1 (A), CYFP-Actn4 (B), and CYFP-Gsn (C). Representative histograms of YFP fluorescence are displayed as described in the legend to Fig. 4. (D) Cells were also analyzed by Western blotting for LMP1, GFP (CYFP tag), and actin (loading control). Lanes 2 to 5 (Actn1), 6 to 9 (Actn4), and 10 to 13 (Gsn) correspond to LMP1, LMP1-A5, LMP1-GG, and LMP1-A5-GG, respectively, and lanes containing the ladder (L) and mCherry-transfected cells (lane 1) are indicated.
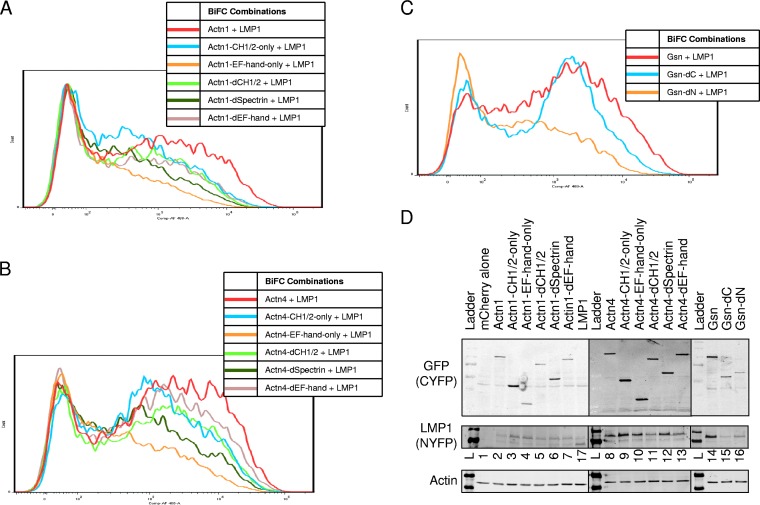
In order to determine the domains of the actin-binding proteins that are important for BiFC with LMP1, deletion mutants were constructed and tested in BiFC assays (Fig. 7). Actn1 and Actn4 are nonmuscle actinins, have 87% amino acid identity, and have similar domain organizations. Actn1 and Actn4 have N-terminal calponin homology (CH1/2) domains, internal spectrin repeats, and C-terminal EF-hand domains. Various deletion mutants were constructed by PCR mutagenesis to delete one or more of these domains in the context of the CYFP-tagged proteins. The mutants of Actn1 and Actn4 were tested for BiFC with LMP1-NYFP (Fig. 7A and B, respectively). As expected on the basis of their high degree of identity, mutants of Actn1 and Actn4 had similar profiles of BiFC with LMP1. Full-length Actn1 and Actn4 (red histograms in Fig. 7) had high levels of BiFC, and deletion of the spectrin repeats (dark green histograms in Fig. 7) greatly decreased the fluorescence. This suggests that the spectrin repeats contribute to BiFC with LMP1. Deletion of the EF-hand domain (beige histograms in Fig. 7) slightly decreased the BiFC between LMP1 and Actn4 (Fig. 4B), and BiFC was lower with Actn1 (Fig. 4A). Mutants containing only the EF-hand domain (orange histograms in Fig. 7) had low levels of BiFC. This suggests that the EF-hand domain does not contribute greatly to BiFC. Deletion of the CH1/2 domain (light green histograms in Fig. 7) slightly decreased the level of BiFC, and expression of the CH1/2 domain alone (blue histograms in Fig. 7) resulted in a relatively high level of BiFC, similar to the results obtained by deletion of the EF-hand domain and deletion of the CH1/2 domain (beige and light green histograms, respectively, in Fig. 7). This indicates that the CH1/2 domain also contributes to BiFC with LMP1. Together the results for the Actn1 and Actn4 mutants demonstrate that both the spectrin repeat and CH1/2 domains but not the EF-hand domain are required for BiFC with LMP1.
FIG 7.
Cloned mutant BiFC assays. Various mutants were tested for BiFC. Actn1 and Actn4 contain N-terminal calponin homology (CH1/2) domains, internal spectrin repeats, and C-terminal EF-hand domains. (A and B) Various deletion mutants were tested by BiFC with LMP1-NYFP Actn1 (A) and Actn4 (B). Representative histograms for LMP1-NYFP plus either full-length proteins, mutants with a deletion of one domain (deletion of the CH1/2 [dCH1/2], spectrin [dSpectrin], or EF-hand [dEF-hand] domain), or proteins with single domains (CH1/2 only or EF-hand only) are shown. (C) Gsn mutants were similarly tested. The N terminus of gelsolin contains three gelsolin-like repeats and an actin-severing domain, and the C terminus contains three more gelsolin-like repeats and a calcium-sensitive actin-binding domain. Histograms for full-length Gsn, a Gsn mutant with a C-terminal deletion, and a Gsn mutant with an N-terminal deletion were tested as described in the legend to panels A and B. (D) Cells were also analyzed by Western blotting for LMP1, GFP (CYFP tag), and actin (loading control). The contents of lanes 2 to 7 (Actn1), 8 to 13 (Actn4), and 14 to 16 (Gsn) are indicated above the gel. Lanes containing the ladder (lane L), mCherry-transfected cells (lane 1), and LMP1-CYFP (lane 17) are also indicated.
Gsn is naturally cleaved into N-terminal and C-terminal domains during various cellular processes. Constructs for BiFC with deletion mutations similar to those described above were constructed. The N terminus of Gsn contains three gelsolin-like repeats and an actin-severing domain, and the C terminus contains three more gelsolin-like repeats and a calcium-sensitive, actin-binding domain. Gelsolin with a deletion of the C terminus (Gsn-dC; blue histogram in Fig. 7C) had a high level of BiFC that was similar to that of the full-length gelsolin (red histogram in Fig. 7C). In contrast, deletion of the N terminus of gelsolin (Gsn-dN; orange histogram in Fig. 7C) resulted in a very low level of BiFC. This indicates that BiFC between LMP1 and gelsolin is through the N terminus and not the C terminus of gelsolin. The expression of all constructs, including Actn1, Actn4, and gelsolin mutants, was confirmed by Western blotting for GFP (which recognizes the CYFP tag), LMP1, and an actin loading control (Fig. 7D). The sizes of the various deletion mutants are consistent with the predicted molecular size for each protein. Lanes 2 to 7 and 8 to 13 in Fig. 7D contain the Actn1 and Actn4 mutants, respectively. The constructs are in the following order for both Actn1 and Actn4: full-length protein, protein with the CH1/2 domain only, protein with the EF-hand domain only, protein with the CH1/2 deletion, protein with the spectrin deletion, and protein with the EF-hand deletion. Lanes 14 to 16 in Fig. 7D contained full-length gelsolin, gelsolin with deletion of the C terminis, and gelsolin with deletion of the N terminus, respectively. Lanes 1 and 17 are BiFC-negative (mCherry alone) and BiFC-positive (LMP1-NYFP/LMP1-CYFP) controls, respectively.
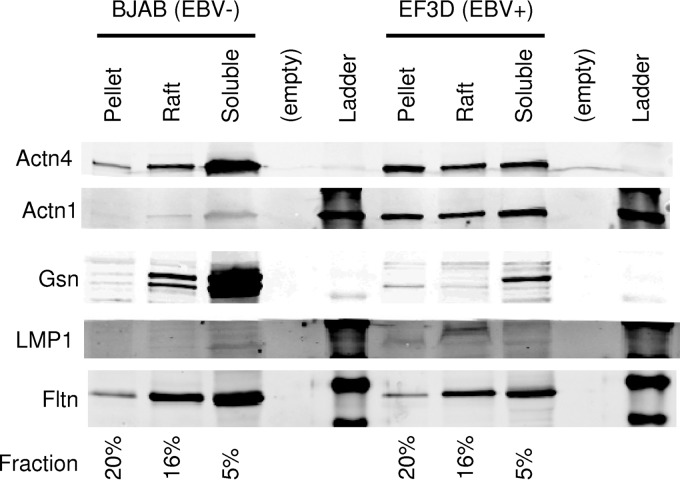
BiFC and reporter assays suggest that a functional relationship exists between the actin-binding proteins and LMP1 signaling. If there is a functional relationship between the proteins, then the proteins should be present together in the lipid raft domains, where LMP1 signaling occurs. LMP1 is known to be constitutively present in the lipid raft domains. To determine if the actin-binding proteins are present within lipid rafts, fractionation of BJAB (EBV-negative) and EF3D (EBV-infected) cells was performed as described elsewhere (38). Cells were fractionated into soluble (Brij 58-soluble), raft (octyl glucoside-soluble), and pellet (octyl glucoside-insoluble) fractions and Western blotting was performed (Fig. 8). Actn4 was present in the soluble, raft, and pellet fractions in both BJAB and EF3D cells. Expression was roughly equal between the two cell lines, and there was a slight increase in the amount of Actn4 in the pellet of EF3D cells and an increase in the amount of soluble Actn4 in BJAB cells. Actn1 was also present in the soluble, raft, and pellet fractions of both BJAB and EF3D cells in relatively equal quantities between the fractions. EF3D cells had increased amounts of Actn1 compared to the amounts in BJAB cells. Gsn was primarily present in the soluble fractions of both EF3D and BJAB cells and was expressed at much higher levels in BJAB cells. However, a small amount of Gsn was present in the lipid rafts of both BJAB and EF3D cells, and little Gsn was present in the pellet fractions. This indicates that a portion of each of the proteins is present in the lipid rafts of EBV-negative and EBV-infected cells.
FIG 8.
Lipid raft fractionation. EBV-negative (BJAB) and EBV-positive (EF3D) cells were fractionated into soluble (1% Brij 58-soluble), raft (60 mM octyl glucoside-soluble), and pellet (10 mM octyl glucoside-insoluble) fractions. The fractions were examined by Western blotting for the presence of Actn1, Actn4, Gsn, LMP1, and flotillin (Fltn). The percentage of each fraction that was loaded on the gel is indicated at the bottom.
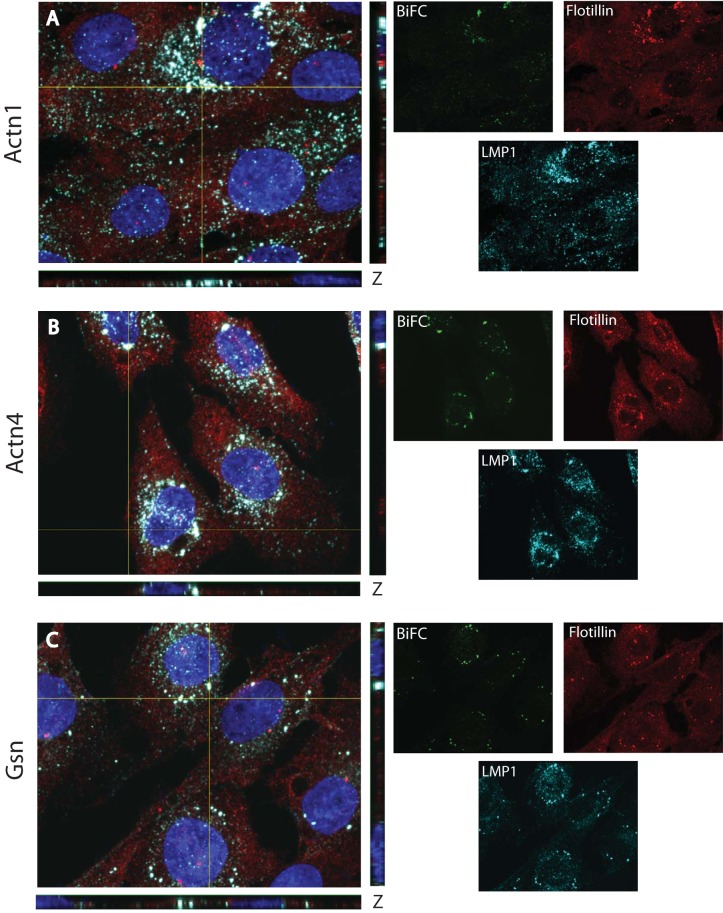
An advantage of the BiFC approach is that cells can be examined for the subcellular localization of fluorescence. To determine the subcellular localization of BiFC between LMP1 and the actin-associated proteins, confocal microscopy was performed. Cloned cell lines in which Actn1, Actn4, and Gsn were identified in the BiFC screen were used. Cells were plated on coverslips and induced overnight with the minimum amount of Dox required to observe fluorescence (20 ng/ml). In previous experiments, this low Dox concentration induced nearly physiological levels of LMP1 expression (29). After overnight induction, cells were fixed with paraformaldehyde to preserve the YFP fluorescence and stained for LMP1 and flotillin (a lipid raft marker) by immunofluorescence, as described previously (29). Stained cells were examined by four-color confocal immunofluorescence for BiFC (green), LMP1 (cyan), flotillin (red), and DAPI counterstain (blue). Representative micrographs are presented in Fig. 9. Single channels are on the right, and 4-channel x, y, and z confocal planes in which signal overlap was observed are on the left. Triple colocalization (white) between LMP1, flotillin, and BiFC was observed for Actn1 (Fig. 9A), Actn4 (Fig. 9B), and Gsn (Fig. 9C) in punctate perinuclear regions. This staining is consistent with the previously described LMP1 localization and LMP1-TRAF BiFC (30, 39, 40). Some LMP1 staining (cyan) was also observed in smaller specks that were not in the overlapping regions with BiFC or flotillin. The nonoverlapping staining suggests that LMP1 is trafficking through the cell but is not associated with lipid rafts and is associated with the actin-binding proteins. In contrast, virtually all BiFC overlapped with the lipid raft marker (flotillin). In other experiments, immunofluorescence staining of endogenous Actn1, Actn4, or Gsn with flotillin and/or LMP1 yielded limited overlap (data not shown). This finding suggests that all LMP1-Actn1, LMP1-Actn4, and LMP1-Gsn BiFCs are raft associated and the potential regulation of LMP1 signaling by the actin cytoskeleton through the actin-binding proteins occurs only in the lipid raft domains of the membrane. Differences in Actn1, Actn4, and Gsn localization were not observed in the presence and absence of LMP1 (data not shown), which suggests that LMP1 is not altering the functions of these proteins but is utilizing their normal function.
FIG 9.
Raft localization of BiFC. Rat-1 clones from the VC1 ERM screen were plated overnight on coverslips and then induced with 20 ng/ml Dox for 18 h. Cells were fixed with formaldehyde to maintain BiFC (green) and stained by immunofluorescence for LMP1 (cyan) and flotillin (a lipid raft marker; red). Cells were counterstained with DAPI (blue). Confocal images were acquired, and representative four-color images in the x, y, and z planes are displayed in the large panels on the left. Single channel colors for each image are displayed on the right. Clones containing CYFP-tagged Actn1 (A), Actn4 (B), and Gsn (C) are displayed. The overlap of BiFC, LMP1, and flotillin is shown in white.
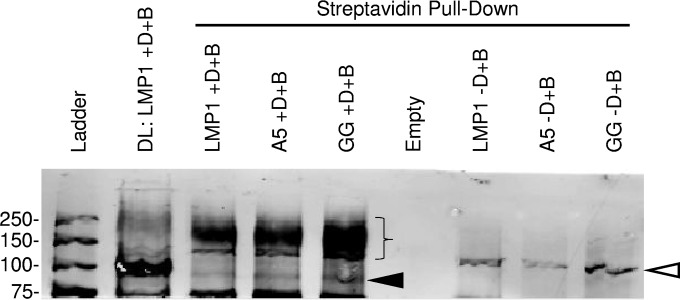
BiFC with LMP1 and the actin-binding proteins in lipid rafts suggests that these proteins are contained within the LMP1 signaling complex. To determine if these proteins were present in a complex, immunoprecipitations were performed. Triple myc-tagged LMP1 was cotransfected with CYFP-tagged actin-binding proteins, and myc-tagged LMP1 was immunoprecipitated with myc-labeled beads. Pulldown of the actin-binding proteins with myc-tagged LMP1 was not observed (data not shown). The interaction between LMP1 and actin-binding proteins may be transient or disrupted during immunoprecipitation. Another recently described technique for identifying proteins in larger complexes directly in eukaryotic cells is BioID (31). BioID utilizes a fusion protein containing a biotin ligase (BirA). Addition of biotin induces the biotinylation of proximal proteins close to the fusion protein. Biotinylated proteins can be isolated with streptavidin beads and identified by Western blotting or proteomic approaches. LMP1-BioID, containing the biotin ligase at the C terminus, was cloned into an inducible retrovirus, and inducible C33A cells were created. In parallel, mutant LMP1 with a mutation(s) in CTAR1 and/or CTAR2 was also made. Inducible cells were induced overnight with biotin in the presence or absence of Dox. Cells were harvested, and biotinylated proteins were pulled down and examined by Western blotting for Actn1 (Fig. 10). Actn1 was clearly visible in the direct loading of LMP1 with Dox and biotin at its normal molecular mass of 100 kDa (Fig. 10, arrowhead). A very faint band was present in the pulldowns of LMP1 and mutant LMP1 cells at the Actn1 native molecular mass. In addition, a diffuse band was also present in the pulldowns between 150 and 250 kDa, which is much stronger than the native Actn1 (Fig. 10, brace). In contrast, the amount of high-molecular-mass Actn1 was much less in the directly loaded lane. Neither Actn1 with the native molecular mass nor Actn1 with the higher molecular mass was observed in pulldowns from uninduced cells in the presence of biotin or in induced cells in the absence of biotin (data not shown). Similar pulldowns for Actn4 were not observed (data not shown). It is not clear if this is due to the lower reactivity of the Actn4 antibody or if it reflects a difference in the association of LMP1 with Actn1 and Actn4. The pulldown of high-molecular-mass Actn1 suggests that Actn1 is modified by posttranslational modification and that this modification is greatly enriched in proximity to LMP1. Together these findings indicate that the LMP1 signaling complex is in contact with Actn1 within lipid rafts and that this results in the posttranslational modification of Actn1.
FIG 10.
Biotinylation of actinin 1 in proximity to LMP1. Dox-inducible C33A cells were analyzed for actinin 1 by Western blotting. LMP1, LMP1-A5 (A5; CTAR1 mutant), and LMP1-GG (GG; CTAR2 mutant) were tagged with the BioID tag and inducibly expressed in the presence of 50 μM biotin (+D+B). Uninduced cells were also incubated with biotin (−D+B). Arrowhead, size of actinin 1 (100 kDa); brace, size of higher-molecular-mass actinin 1 (150 to 250 kDa). The lane containing the ladder and empty lanes are also indicated. A nonspecific band at about 125 kDa was also observed (open arrowhead). Numbers on the left are molecular masses (in kilodaltons).
DISCUSSION
In the current study, we utilized BiFC to perform a genome-wide screen for LMP1-binding proteins. We have identified a number of new proteins that may potentially be important for different aspects of the LMP1 life cycle. Although signaling and lipid raft-related proteins are of primary interest for the development of novel therapies to inhibit LMP1 signaling, many other aspects of cell biology related to LMP1 may also be isolated using this screen. These include proteins that translocate into the endoplasmic reticulum (ER), trafficking proteins (from the ER to the Golgi apparatus and from the Golgi apparatus to lipid rafts), and turnover-related proteins. Indeed, a number of proteins related to these functions were identified in our screen.
As with all screening technologies, there is a concern for both false-positive and false-negative results with our BiFC approach. Because splicing with the ERM vectors occurs at the next cellular exon, any binding that requires protein domains contained in exon 1 of the targeted proteins would not be contained in the CYFP-fusion proteins. However, the ERM vectors preferentially integrate near transcription start sites, which should minimize false-negative results from the N terminus of targeted proteins. The TRAFs in particular should be isolated in our screen, on the basis of the findings of our published studies of LMP1-TRAF BiFC. However, the second exons of TRAF2 and TRAF3 are not compatible with the establishment of functional fusion proteins with the ERM vector used in this study. A larger concern for most screening technologies is false-positive results. About half of the proteins that were identified were membrane proteins, and many of them have an unclear connection to LMP1 signaling. It seems likely that being constrained by the membrane may drive false-positive interactions between LMP1 and other membrane proteins. From our previous studies, our screen is performed with nearly physiological levels of LMP1 expression (29), but to fully demonstrate a functional relationship, the proteins need to be analyzed at physiological levels. Future experiments will determine if any of these proteins has a functional relationship with LMP1. The assay with BioID tag-containing LMP1 was useful for confirming the results for proteins in close proximity to LMP1. Others have used the BioID approach coupled to mass spectrometry analysis to determine protein complexes in cells. Such an approach could also be used with LMP1.
Of particular interest for the current study were the cytoskeletal proteins. Several findings from our study support a functional role for these proteins in LMP1 signaling. There are a number of cytoskeletal structures that underlie the lipid raft domains of the membrane, and disruption of the cytoskeleton results in the loss of the raft structure. Actinin 1 and actinin 4 are nonmuscle actin-associated proteins that were identified in several of our positive clones. BiFC between LMP1 and the actinin proteins and gelsolin was confirmed and limited to lipid raft domains in cells. A portion of the actinins and gelsolin fractionated into lipid rafts independently of LMP1 expression. BiFC was dependent on functional LMP1 signaling domains and was mapped to specific domains of the actinins and gelsolin. Although actinin 1 and actinin 4 are highly homologous, in work by others, actinins 1 and 4 were found to have different roles in cellular survival, motility, and RhoA signaling in astrocytoma cells (41). This suggests that they may have functional roles in the induction of signaling. In our study, we were not able to transiently or stably knock down actinin 1 and actinin 4 protein expression, and the specific role of these proteins in LMP1 signaling could not be determined (data not shown). Gelsolin is a calcium-sensitive, actin-severing protein that is important for actin cytoskeleton dynamics. Tropomyosin 2 is a nonmuscle actin motor protein. Binding of these proteins to LMP1 could be important for targeting LMP1 to lipid rafts or for regulating the motility and invasion of LMP1-expressing cells.
Nondegradative ubiquitination of proteins in the TNFR pathway is critical to activation of signaling. Both lysine-63 (K-63) and methionine-1 (M-1; linear) polyubiquitination are critical for the assembly of the TNFR1 signaling complex. This leads to lysine-48 ubiquitination of the inhibitor of NF-κB signaling, IκBa, and its degradation. Ubiquitination of other proteins is critical for intracellular trafficking. Higher-molecular-mass actinin 1 was pulled down with streptavidin beads from the inducible cells expressing LMP1 with the BioID tag. This modified actinin was only a minor part of the total actinin 1 from the direct load. Since the higher-molecular-mass actinin 1 was not observed in uninduced cells, this suggests that only the actinin 1 that is in close proximity to LMP1 is modified, likely in lipid rafts. Whether this modification is indeed ubiquitination and which type of linkage is associated with actinin 1 are the subjects of ongoing experiments. Actinin 1 could be ubiquitinated at K-63 or M-1 to help potentiate signaling within the rafts, or it could be modified by another linkage that is critical for the targeting of actinin 1 or other proteins to the lipid rafts.
The current BiFC screening approach is limited by the number of single-cell clones that could be analyzed: 100 clones out of 65,000 sorted events. An alternative approach would be to determine the targeted open reading frames from the entire sorted population by next-generation sequencing. cDNA can be synthesized from sorted cells and amplified with YFP-specific primers. The results could then be compared with those obtained by an analogous approach with various LMP1 signaling mutants. Screens run with LMP1 CTAR1 and/or CTAR2 mutants would allow the determination of the proteins that are important for CTAR1 and CTAR2 signaling. Genes that are isolated with the mutants will be either genes that have false-positive results or genes that are important for functions other than LMP1 signaling. Genes that are isolated with signaling-competent LMP1 but not the mutants would be genes critical for signaling.
Together the findings of our study suggest that BiFC is a novel technology for analysis of LMP1 signaling. BiFC is performed with LMP1 in the membrane of mammalian cells. These types of approaches will be critical to determine how the various proteins are recruited to the LMP1 signaling complex. In addition future studies will help to determine the dynamics of this process. Such information will be useful to design specific interventions that can block this process.
ACKNOWLEDGMENTS
This work was supported by the H. M. Bligh Cancer Research Laboratories, American Cancer Society, Illinois Division, Inc. (grant 08-35), and American Cancer Society Illinois Division Research Scholar grant RSG-12-229-01-MPC to D.N.E.
We thank Bob Dickinson of the RFUMS Flow Cytometry Core Facility for assistance with FACS analysis.
REFERENCES
- 1.Kieff ED, Rickinson AB. 2007. Epstein-Barr virus and its replication, p 2603–2654. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol II Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol II Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol 12:431–441. doi: 10.1016/S1044579X0200086X. [DOI] [PubMed] [Google Scholar]
- 4.Young L, Alfieri C, Hennessy K, Evans H, O'Hara C, Anderson KC, Ritz J, Shapiro RS, Rickinson A, Kieff E, Cohen JI. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med 321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 5.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 6.Magrath I. 1990. The pathogenesis of Burkitt's lymphoma. Adv Cancer Res 55:133–270. doi: 10.1016/S0065-230X(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi MK, Tellam JT, Khanna R. 2004. Epstein-Barr virus-associated Hodgkin's lymphoma. Br J Haematol 125:267–281. doi: 10.1111/j.1365-2141.2004.04902.x. [DOI] [PubMed] [Google Scholar]
- 8.Carbone A. 2002. AIDS-related non-Hodgkin's lymphomas: from pathology and molecular pathogenesis to treatment. Hum Pathol 33:392–404. doi: 10.1053/hupa.2002.124723. [DOI] [PubMed] [Google Scholar]
- 9.Diamond C, Taylor TH, Aboumrad T, Anton-Culver H. 2006. Changes in acquired immunodeficiency syndrome-related non-Hodgkin lymphoma in the era of highly active antiretroviral therapy: incidence, presentation, treatment, and survival. Cancer 106:128–135. doi: 10.1002/cncr.21562. [DOI] [PubMed] [Google Scholar]
- 10.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baichwal VR, Sugden B. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461–467. [PubMed] [Google Scholar]
- 12.Everly DN Jr, Mainou BA, Raab-Traub N. 2004. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J Virol 78:13470–13478. doi: 10.1128/JVI.78.24.13470-13478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mainou BA, Everly DN Jr, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 14.Moorthy RK, Thorley-Lawson DA. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J Virol 67:1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 16.Dawson CW, Eliopoulos AG, Blake SM, Barker R, Young LS. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204–217. doi: 10.1006/viro.2000.0344. [DOI] [PubMed] [Google Scholar]
- 17.Eliopoulos AG, Young LS. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 18.Izumi KM, Kieff ED. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci U S A 94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller WE, Mosialos G, Kieff E, Raab-Traub N. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-kappaB activation. J Virol 71:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paine E, Scheinman RI, Baldwin AS Jr, Raab-Traub N. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-kappa B/Rel family proteins. J Virol 69:4572–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts ML, Cooper NR. 1998. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240:93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- 22.Thornburg NJ, Kulwichit W, Edwards RH, Shair KH, Bendt KM, Raab-Traub N. 2006. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene 25:288–297. doi: 10.1038/sj.onc.1209023. [DOI] [PubMed] [Google Scholar]
- 23.Mainou BA, Everly DN Jr, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J Virol 81:9680–9692. doi: 10.1128/JVI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung CP, Meckes DG Jr, Raab-Traub N. 2011. EBV latent membrane protein 1 (LMP1) activates EGFR, STAT3 and ERK through effects on PKCδ. J Virol 85:4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem 278:3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 26.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. 2004. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol 78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Z, Liang J, Lu Y, Yu Q, Songyang Z, Lin SY, Mills GB. 2006. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A 103:15014–15019. doi: 10.1073/pnas.0606917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Songyang Z. 2008. ERM-mediated genetic screens in mammalian cells. Methods Enzymol 446:409–419. doi: 10.1016/S0076-6879(08)01624-8. [DOI] [PubMed] [Google Scholar]
- 29.Talaty P, Emery A, Holthusen K, Everly DN Jr. 2012. Identification of transmembrane protein 134 as a novel LMP1-binding protein by using bimolecular fluorescence complementation and an enhanced retroviral mutagen. J Virol 86:11345–11355. doi: 10.1128/JVI.00523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talaty P, Emery A, Everly DN Jr. 2011. Characterization of the latent membrane protein 1 signaling complex of Epstein-Barr virus in the membrane of mammalian cells with bimolecular fluorescence complementation. Virol J 8:414. doi: 10.1186/1743-422X-8-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everly DN Jr, Mainou BA, Raab-Traub N. 2009. Transcriptional downregulation of p27KIP1 through regulation of E2F function during LMP1-mediated transformation. J Virol 83:12671–12679. doi: 10.1128/JVI.01422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Yang X, Yang D, Songyang Z. 2000. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene 19:5964–5972. doi: 10.1038/sj.onc.1203992. [DOI] [PubMed] [Google Scholar]
- 34.Meckes DG Jr, Menaker NF, Raab-Traub N. 2013. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol 87:1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebowitz D, Kopan R, Fuchs E, Sample J, Kieff E. 1987. An Epstein-Barr virus transforming protein associates with vimentin in lymphocytes. Mol Cell Biol 7:2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson CW, Port RJ, Young LS. 2012. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol 22:144–153. doi: 10.1016/j.semcancer.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Allen JA, Halverson-Tamboli RA, Rasenick MM. 2007. Lipid raft microdomains and neurotransmitter signalling. Nat Rev Neurosci 8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- 38.Xie P, Bishop GA. 2004. Roles of TNF receptor-associated factor 3 in signaling to B lymphocytes by carboxyl-terminal activating regions 1 and 2 of the EBV-encoded oncoprotein latent membrane protein 1. J Immunol 173:5546–5555. doi: 10.4049/jimmunol.173.9.5546. [DOI] [PubMed] [Google Scholar]
- 39.Hennessy K, Fennewald S, Hummel M, Cole T, Kieff E. 1984. A membrane protein encoded by Epstein-Barr virus in latent growth-transforming infection. Proc Natl Acad Sci U S A 81:7207–7211. doi: 10.1073/pnas.81.22.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultheiss U, Puschner S, Kremmer E, Mak TW, Engelmann H, Hammerschmidt W, Kieser A. 2001. TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J 20:5678–5691. doi: 10.1093/emboj/20.20.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quick Q, Skalli O. 2010. Alpha-actinin 1 and alpha-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res 316:1137–1147. doi: 10.1016/j.yexcr.2010.02.011. [DOI] [PubMed] [Google Scholar]