Abstract
Although RNA viruses exhibit a high frequency of host jumps, major differences exist among the different virus families. Astroviruses infect a wide range of hosts, affecting both public health systems and economic production chains. Here we delineate the ecological and adaptive processes that drive the cross-species transmission of astroviruses. We observe that distinct transmission zones determine the prevailing astrovirus host and virus diversity, which in turn suggests that no single host group (e.g., bats) can be the natural reservoir, as illustrated through our phylogenetic analysis.
SCALABLE HEIGHTS: FROM EVOLUTION TO ECOSYSTEM
The majority of RNA virus species have a narrow host range, producing progeny that are able to reproduce in a specific environment (1). Though successful cross-species transmission events are rare, replication infidelity and opportunities for ecological exposure that are enhanced through anthropogenic change may lead to sustained infection of incidental hosts. This is often observed for enteric viruses, such as astroviruses, calciviruses, picornaviruses, and rotaviruses, which lack an envelope and show exceptional durability in both the harsh gastrointestinal tract (intrahost survival) and the environment (environmental durability) (2). Astroviruses are a positive-sense, single-stranded RNA virus primarily transmitted via the fecal-oral route by direct ingestion or by fomite or food. There are two groups of astroviruses that have been detected in a variety of mammals (mamastroviruses) and birds (avastroviruses) and which represent the early evolutionary divergence in this family (Fig. 1).
FIG 1.
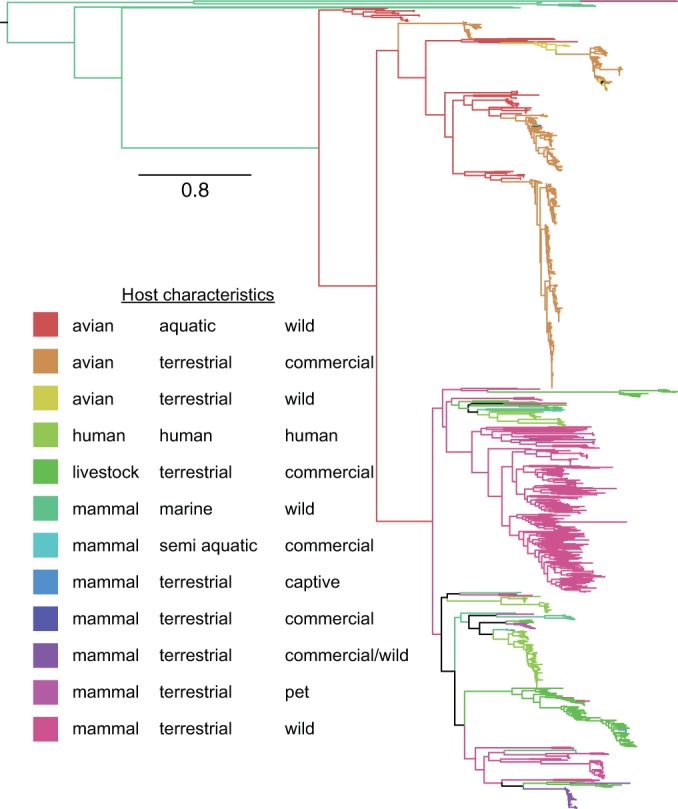
Phylogenetic relationship of the RNA-dependent RNA polymerase (RdRp) genes of 1,259 astroviruses. Tree branches are colored based on host ecology to highlight cross-species transmission between and among ecological niches (key), determined by using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The scale bar indicates nucleotide substitutions per site. The tree was generated based on an alignment of 540 nucleotides of the most conserved region of the RdRp region, which is routinely used for the PCR diagnostics of RNA viruses. This final data set was curated from a preliminary analysis of all publicly available sequence data (∼4,300 sequences deposited in NCBI GenBank) and by picking the most conserved region with the highest numbers of sequences available. The capsid gene had greater representation than the selected RdRp region but was not considered because the genes had greater family-level variation and, hence, could not be unambiguously aligned.
Several ecological and evolutionary processes have led to the large diversity of astroviruses that infect a myriad of hosts (3); astroviruses have been detected from over 80 avian and mammalian host species to date (Fig. 1 and 2). Phylogenetic analysis of the RNA-dependent RNA polymerase (RdRp) region suggests that the long-term evolution of astroviruses is determined by cross-species transmission events that occur among distinct ecological scenarios. The majority of astrovirus lineages naturally infect only one or a few closely related host species; however, more than one lineage can consistently transmit in a single host, as observed in humans (Fig. 1). Virus recombination has also been reported to drive the increasing genomic diversity of this virus family (3, 4), which may arise from infection of individuals with multiple virus strains belonging to different lineages, although reporting of multiple infections is poor. Systematic astrovirus surveillance in swine in the United States revealed a high proportion of pigs (13.9%) coinfected with viruses from two or more porcine astrovirus lineages (5), signifying the potential for recombination. Furthermore, the continual circulation of strains in a population provides the stock for infections of neighboring conspecifics, agricultural species, and peridomestic animals (dogs, cats, and rats) or to seed the environment. Intensification of agriculture, poor sanitation, and continued encroachment and degradation of the environment will ensure that conditions for exposure to and the spread of astroviruses will persist.
FIG 2.
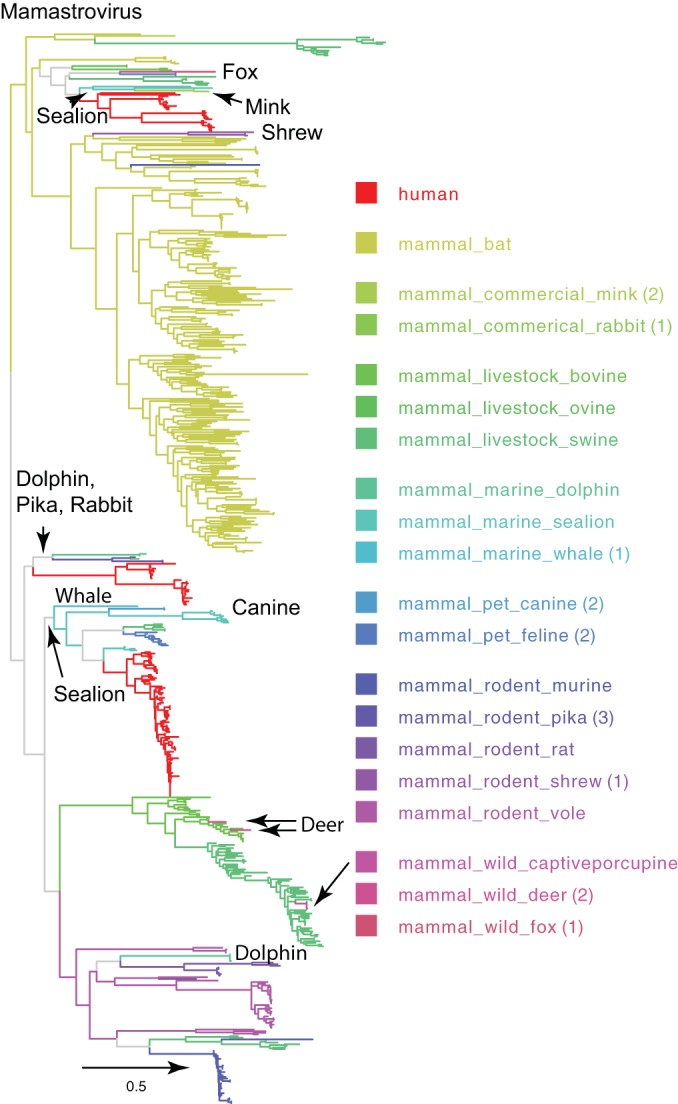
Phylogenetic relationship of mamastroviruses. The mamastrovirus lineage shown in Fig. 1 is enlarged, and tree branches are colored based on host species and group (key) to highlight the relationships between astroviruses isolated from various mammals. Numbers of sequences for severely undersampled hosts (<4 sequences) are shown in parentheses. The scale bar indicates nucleotide substitutions per site.
Here we discuss the different ecological zones in which astroviruses occur and the factors that drive cross-species transmission, and we identify directions to best monitor these viruses.
AGRICULTURAL AND COMMERCIAL TRANSMISSION
Astroviruses have been identified in numerous livestock species, including chickens, turkeys, guinea fowl, ducks, rabbits, sheep, pigs, and cows, and in commercial production of mice, minks, and dogs (3), which are often globally distributed (6). Production of these animals occurs at several different scales, from modern industrial production of isolated single species in midrange facilities down to family farms, where animals often mingle with other reared animals, peridomestics, pets, and wild individuals. Each system facilitates and excludes specific contacts depending on whether the animals are confined or free range, in addition to other biosecurity measures. Large production facilities have mechanical barriers to exposure to other species but present a monoculture with high levels of contact that can allow the rapid spread of astrovirus infections. High-density rearing can increase contact frequency and astrovirus infection, as witnessed in large breeding facilities (7). Astroviruses have been detected in a number of companion animals, including cats and dogs, though it is uncertain if these individuals were infected as juveniles in breeding facilities.
Astrovirus infections have caused great economic losses. Although often asymptomatic or only causing mild disease, young individuals are particularly vulnerable to disease and postinfection complications, such as stunting in broiler chickens (6) and preweaning diarrhea syndrome in minks. Infected individuals may shed virus for long periods of time; for example, minks have tested positive for virus up to 7 weeks after initial detection (8). Asymptomatic shedding has been found in pigs, and individuals can be coinfected with multiple strains at the same time (5); this may stymie intervention methods that are based on quarantine. Though mammalian and avian livestock are commonly reared on the same farm, cross-transmission between the groups has rarely been observed and there is no evidence of recombination between these two groups, although few whole-genome sequences are available for this analysis.
TERRESTRIAL TRANSMISSION
There are a large number of terrestrial species from which astroviruses have been detected, including humans, doves and pigeons, domestic dogs and cats, rabbits, pikas, wild boars, red deer and roe deer, a cheetah, shrews, rodents, and bats, demonstrating the wide host range of astroviruses (3). As with commercial and agricultural rearing, animal density appears to be an important factor for transmission in non-livestock animal infections.
The majority of nonhuman astrovirus detections have been in the two most species-rich mammalian groups, rodents and bats. This may be a function of either the sheer numbers of species where diversifying selection has taken place after introductions or the surveillance intensity of these two groups of animals. The genetic diversity of astroviruses in both small mammals and bats demonstrates that individual species can host a wide range of strains (Fig. 2) (9). However, it is not known whether these were the result of a recent cross-species transmission, persistent infection, or an exceptional diversity of unsampled astroviruses. The primary question with these infected terrestrial animals is the source of primary infection. Two possibilities are that infection originates from familial contacts or from chance contact events. Feces are often inspected by conspecifics, predators, and prey to determine the presence of animals, and this may create the opportunity for inhalation or ingestion of virus. Additionally, certain animals, such as pigs, are coprophagous and will readily consume feces. Bat roosts can have millions of individuals in close contact, with physical communication between neighbors and social grooming. Similarly, outbreaks have been observed in social birds that congregate, such as pigeons and doves.
AQUATIC TRANSMISSION
Astroviruses are transmitted fecal-orally and persist in water, making the aquatic environment ideal for cross-species transmission (2). Astroviruses have been detected in wild ducks, wading birds, and shore birds. When defecation and foraging occur in the same aquatic habitat, susceptible individuals are exposed to shed virus. During bird migration, the commingling and density of birds from different geographic areas at collective resting or breeding sites may allow multiple strains to infect an individual, providing an opportunity for virus recombination. Avastroviruses have been detected in several aquatic bird species, including four species of wild ducks (9). A recent study found evidence of astrovirus infections in several species of sea-dwelling mammals, including a bottlenose dolphin, an orca, a Minke whale, a Stellar's sea lion, and California sea lions (10). A new strain of astrovirus discovered in sea lions is thought to have emerged from a recombination event between a California sea lion astrovirus and a human astrovirus (10), highlighting the potential for reverse zoonosis to a marine environment and successful perpetuation.
Aquatic environments are a rich source of potential infection due to their durability and the fecal route of transmission. Untreated or poorly treated sewage from human domiciles or from agricultural areas can flow into water bodies, such as lakes and oceans. Untreated wastewater can expose incidental hosts to numerous viruses from very different hosts, often with devastating consequences (11). Astroviruses can also bioaccumulate in shellfish (12). The aquatic milieu is also a prime example of where agricultural and terrestrial habitats can contribute astrovirus diversity into water bodies, highlighting the importance of this interface. Multiple clades with astroviruses both from aquatic avian species and poultry suggest previous cross-species transmission events (Fig. 1).
ECOTONES
Ecotones are ecological transitional areas, where opportunities arise for cross-species transmission. For example, on small and medium-sized farms, different species share the same space. This is common with poultry (domestic ducks, chickens, turkeys, and guinea fowl), where coreared and free-ranging birds can interact with each other and with foraging wild birds. This can lead to coinfection with different strains and opportunities for recombination, evidenced by viruses detected in guinea fowl that originated in turkeys or those detected in domestic ducks that are turkey and chicken derived (13, 14). Ecotones also drive zoonoses, as evidenced by seroconversion of farmers and infection of an immunocompromised individual (15, 16). Companion animals and peridomestic animals on farms also have contact with livestock. The presence of asymptomatic hosts allows the virus to be shed across a wider temporal and spatial scale than by symptomatic individuals, whose movement may be limited by the infection. This leads to the question about whether there are trade-offs between these transmission scales and how fitness is affected by the ecology of the host.
FUTURE DIRECTIONS
It is inevitable that many more astroviruses are likely to be discovered, given the enormous advances in virus surveillance and next-generation sequencing technology. Little is known, however, about the actual transmission efficiency of these viruses, particularly those that confer cross-species transmission capability. Efforts are being made to quantify transmission rates in agricultural or production animals (summarized in reference 17), although data from wild hosts are not available.
One striking need in astrovirus research is to determine what host cell receptors it uses for entry. Clathrin-dependent pathways appear to be used in Caco-2 cells (18). Crystal structure analysis of the capsid spike domain (the primary binding site) demonstrates drastic differences in the morphology of avastroviruses and mamastroviruses, suggesting a potential host limitation, yet the presence of human seroconversion against turkey astroviruses reveals their immunogenicity when incidental hosts are exposed (15, 19). The disparity of astrovirus growth rates in different mammalian cell lines indicates these viruses may use multiple receptors or binding sites, making it difficult to discern the susceptible individuals in an environment if there is a mixture of species. Additionally, there is an abundance of RdRp gene sequences available but relatively few structural sequences and only 27 whole-genome sequences, the majority belonging to human astroviruses (whole genomes are available for 10 human, 5 bovine, and 4 swine astroviruses and for 8 astroviruses from miscellaneous hosts) (http://www.ncbi.nlm.nih.gov/genome/?term=astrovirus; accessed 20 April 2015). The lack of genome-level data inhibits determination of the risk for cross-species transmission potential due to prevailing capsid construct diversity.
The defining arenas where transmission can occur are well established and influenced by animal density and food security measures. Determining the breadth of the host range by identifying key receptors and capsid entry dynamics will demonstrate the potential risk for cross-species transmission. Understanding these factors and the ecological drivers that facilitate the evolution, perpetuation, amplification, and dampening of astroviruses across interfaces will assist in mitigating future outbreaks.
ACKNOWLEDGMENTS
We are supported by the National Institutes of Health (R21AI105050) and by the Duke-NUS Signature Research Program funded by the Agency of Science, Technology and Research, Singapore, and the Ministry of Health, Singapore.
REFERENCES
- 1.Holmes EC. 2009. The molecular epidemiology, phylogeography and emergence of RNA viruses, p 131–155. In Holmes EC. (ed), The evolution and emergence of RNA viruses. OUP, Oxford, United Kingdom. [Google Scholar]
- 2.Bishop R, Kirkwood C. 2010. Enteric viruses, p 30–36. In Mahy BWJ, van Regenmortel MHV (ed), Desk encyclopedia of human and medical virology. Academic Press, Oxford, United Kingdom. [Google Scholar]
- 3.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol 11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Grazia S, Medici MC, Pinto P, Moschidou P, Tummolo F, Calderaro A, Bonura F, Banyai K, Giammanco GM, Martella V. 2012. Genetic heterogeneity and recombination in human type 2 astroviruses. J Clin Microbiol 50:3760–3764. doi: 10.1128/JCM.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao CT, Gimenez-Lirola LG, Gerber PF, Jiang YH, Halbur PG, Opriessnig T. 2013. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J Gen Virol 94:570–582. doi: 10.1099/vir.0.048744-0. [DOI] [PubMed] [Google Scholar]
- 6.Koci MD, Schultz-Cherry S. 2002. Avian astroviruses. Avian Pathol 31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds DL, Saif YM, Theil KW. 1987. A survey of enteric viruses of turkey poults. Avian Dis 31:89–98. doi: 10.2307/1590779. [DOI] [PubMed] [Google Scholar]
- 8.Larsen PF, Møller S, Clausen T, Hammer A. 2012. Proceedings of the Xth International Scientific Congress in Fur Animal Production. Wageningen Academic Publishers, Wageningen, The Netherlands. [Google Scholar]
- 9.Chu DK, Poon LL, Guan Y, Peiris JS. 2008. Novel astroviruses in insectivorous bats. J Virol 82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera R, Nollens HH, Venn-Watson S, Gulland FM, Wellehan JF Jr. 2010. Characterization of phylogenetically diverse astroviruses of marine mammals. J Gen Virol 91:166–173. doi: 10.1099/vir.0.015222-0. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland KP, Shaban S, Joyner JL, Porter JW, Lipp EK. 2011. Human pathogen shown to cause disease in the threatened eklhorn coral Acropora palmata. PLoS One 6:e23468. doi: 10.1371/journal.pone.0023468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elamri DE, Aouni M, Parnaudeau S, Le Guyader FS. 2006. Detection of human enteric viruses in shellfish collected in Tunisia. Lett Appl Microbiol 43:399–404. doi: 10.1111/j.1472-765X.2006.01978.x. [DOI] [PubMed] [Google Scholar]
- 13.Bidin M, Bidin Z, Majnaric D, Tisljar M, Lojkic I. 2012. Circulation and phylogenetic relationship of chicken and turkey-origin astroviruses detected in domestic ducks (Anas platyrhynchos domesticus). Avian Pathol 41:555–562. doi: 10.1080/03079457.2012.733340. [DOI] [PubMed] [Google Scholar]
- 14.De Battisti C, Salviato A, Jonassen CM, Toffan A, Capua I, Cattoli G. 2012. Genetic characterization of astroviruses detected in guinea fowl (Numida meleagris) reveals a distinct genotype and suggests cross-species transmission between turkey and guinea fowl. Arch Virol 157:1329–1337. doi: 10.1007/s00705-012-1311-1. [DOI] [PubMed] [Google Scholar]
- 15.Meliopoulos VA, Kayali G, Burnham A, Oshansky CM, Thomas PG, Gray GC, Beck MA, Schultz-Cherry S. 2014. Detection of antibodies against Turkey astrovirus in humans. PLoS One 9:e96934. doi: 10.1371/journal.pone.0096934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz-Cherry S. (ed). 2012. Astrovirus research: essential ideas, everyday impacts, future directions. Springer, New York, NY. [Google Scholar]
- 18.Méndez E, Munoz-Yanez C, Sanchez-San Martin C, Aguirre-Crespo G, Banos-Lara Mdel R, Gutierrez M, Espinosa R, Acevedo Y, Arias CF, Lopez S. 2014. Characterization of human astrovirus cell entry. J Virol 88:2452–2460. doi: 10.1128/JVI.02908-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DuBois RM, Freiden P, Marvin S, Reddivari M, Heath RJ, White SW, Schultz-Cherry S. 2013. Crystal structure of the avian astrovirus capsid spike. J Virol 87:7853–7863. doi: 10.1128/JVI.03139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


