ABSTRACT
Hepatitis C virus (HCV) nonstructural protein 5A (NS5A) is essential for HCV genome replication and virion production and is involved in the regulation of multiple host signaling pathways. As a proline-rich protein, NS5A is capable of interacting with various host proteins containing Src homology 3 (SH3) domains. Previous studies have suggested that vinexin, a member of the sorbin homology (SoHo) adaptor family, might be a potential binding partner of NS5A by yeast two-hybrid screening. However, firm evidence for this interaction is lacking, and the significance of vinexin in the HCV life cycle remains unclear. In this study, we demonstrated that endogenously and exogenously expressed vinexin β coimmunoprecipitated with NS5A derived from different HCV genotypes. Two residues, tryptophan (W307) and tyrosine (Y325), in the third SH3 domain of vinexin β and conserved Pro-X-X-Pro-X-Arg motifs at the C terminus of NS5A were indispensable for the vinexin-NS5A interaction. Furthermore, downregulation of endogenous vinexin β significantly suppressed NS5A hyperphosphorylation and decreased HCV replication, which could be rescued by expressing a vinexin β short hairpin RNA-resistant mutant. We also found that vinexin β modulated the hyperphosphorylation of NS5A in a casein kinase 1α-dependent on manner. Taken together, our findings suggest that vinexin β modulates NS5A phosphorylation via its interaction with NS5A, thereby regulating HCV replication, implicating vinexin β in the viral life cycle.
IMPORTANCE Hepatitis C virus (HCV) nonstructural protein NS5A is a phosphoprotein, and its phosphorylation states are usually modulated by host kinases and other viral nonstructural elements. Additionally, cellular factors containing Src homology 3 (SH3) domains have been reported to interact with proline-rich regions of NS5A. However, it is unclear whether there are any relationships between NS5A phosphorylation and the NS5A-SH3 interaction, and little is known about the significance of this interaction in the HCV life cycle. In this work, we demonstrate that vinexin β modulates NS5A hyperphosphorylation through the NS5A-vinexin β interaction. Hyperphosphorylated NS5A induced by vinexin β is casein kinase 1α dependent and is also crucial for HCV propagation. Overall, our findings not only elucidate the relationships between NS5A phosphorylation and the NS5A-SH3 interaction but also shed new mechanistic insight on Flaviviridae NS5A (NS5) phosphorylation. We believe that our results may afford the potential to offer an antiviral therapeutic strategy.
INTRODUCTION
Hepatitis C virus (HCV) infection is a global health disease and is a major cause of chronic liver disease leading to hepatic fibrosis, liver cirrhosis, and hepatic carcinoma. No protective vaccine is available. Some directly acting antiviral agents combining pegylated interferon and ribavirin show therapeutic promise for chronic hepatitis C. However, the mechanisms of drug action, the issues of interferon-free therapy, drug resistance, and broad treatment of all HCV genotypes remain to be addressed (1).
HCV is a single-stranded positive-sense enveloped RNA virus belonging to the Hepacivirus genus in the Flaviviridae family. The RNA genome of HCV consists of about 9,600 nucleotides flanked by 5′ and 3′ untranslated regions (UTR) encoding four structural proteins (core, E1, E2, and P7) and six nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) (2). NS3 to NS5B are sufficient to support viral RNA replication in cultured cells (3). Recently, multifunctional roles for these NS proteins, including NS3, NS4B, and NS5A, also have been shown to regulate HCV particle production (4–6).
HCV NS5A is a proline-rich phosphoprotein with multiple functions in viral replication, pathogenesis, and the innate immunity response (7). On the one hand, phosphorylation of NS5A (NS5) is conserved in members of the Flaviviridae family, including hepaciviruses, pestiviruses, and flaviviruses (8). A hallmark of HCV NS5A is that it exists as two distinct phosphorylated variants termed hypophosphorylated (p56) and hyperphosphorylated (p58). They can be separated by SDS-PAGE on the basis of their sizes; the molecular mass of the former is 56 kDa, and that of the latter is 58 kDa (9). Previous studies have shown that phosphorylation of NS5A is crucial to parts of the HCV life cycle, such as viral genome replication complex formation and infectious particle production (10–12). Although the details of the mechanism regulating NS5A phosphorylation are still not clear, various host factors (HFs) involved in NS5A phosphorylation, such as casein kinase 1α (CK1α) and CK2α, have been identified (6, 13, 14). On the other hand, NS5A has also been implicated in the modulation of host defenses, apoptosis, the cell cycle, and stress-responsive pathways through its interaction with a wide variety of HFs such as PKR, Bin1, P53, and Grb2 (15). Moreover, the proline-rich motifs Pro-X-X-Pro-X-Arg (PxxPxR) of NS5A are thought to interact with proteins containing Src homology 3 (SH3) domains such as Fyn, Hck, and Lck (16). However, the functional consequences of these interactions within two conserved domains during the HCV life cycle have not been clearly elucidated.
The sorbin homology (SoHo) family is a family of adaptors with three members, vinexin, c-Cbl-associated protein (CAP)/ponsin, and Arg-binding protein 2 (Argbp2). All of these members contain one SoHo domain followed by three SH3 domains and have effects on cell adhesion and cytoskeletal organization (17). As one of the SoHo family members, vinexin was first identified via its interaction with vinculin, an abundant cytoskeletal protein localized predominantly at cell-substrate adhesion sites (such as focal adhesions) and cell-cell junctions. Vinexin has several alternative transcripts, including α, β, and γ (17). Vinexin β mainly contains three SH3 domains, and vinexin α contains an additional N-terminal region harboring a SoHo domain. Vinexin β is ubiquitously expressed in mouse liver tissue. In contrast, expression of vinexin α cannot be detected in liver tissue but is high in skeletal muscle (18). Vinexin γ was isolated from mouse fetal gonads and has rarely been reported (19). Vinexin β not only regulates cytoskeletal arrangement, cell adhesion, and spreading but is also involved in cellular signal transduction. For instance, phosphorylated vinexin β, catalyzed by extracellular signal-regulated kinase, inhibits cell spreading and migration (20). Moreover, vinexin β binds to mSos, a Ras-guanine nucleotide exchange factor, and regulates the activation of c-jun N-terminal kinase (JNK) induced by epidermal growth factor (EGF) (21). However, no studies have implicated vinexin β in viral life cycles.
In the present study, we discovered that HCV NS5A specifically interacts with vinexin β in mammalian cells. Truncation and point mutation analysis of NS5A showed that conserved PxxPxR motifs at the NS5A C terminus and conserved SH3 functional residues in the third SH3 domain of vinexin β are critical for their interaction. Furthermore, silencing of endogenous vinexin β suppresses HCV replication and simultaneously decreases NS5A hyperphosphorylation. This phenotype can be rescued by the expression of a short hairpin RNA (shRNA)-resistant mutant form of vinexin β. We further demonstrated that vinexin β modulates NS5A hyperphosphorylation in a CK1α-dependent manner. Moreover, vinexin β plays a role in the regulation of the CK1α-NS5A catalytic reaction in vivo.
Our study provides strong evidence that, through the interaction between the SH3 domain and PxxPxR motifs, vinexin β is capable of modulating NS5A phosphorylation and regulating HCV replication. Furthermore, our work also offers insight into a previously unappreciated interrelationship of the NS5A-SH3 domain-related interaction, phosphorylation of NS5A, and HCV propagation.
MATERIALS AND METHODS
Cell culture.
Human hepatoma Huh7 cells, derivative Huh7-Lunet cells (kindly provided by Ralf Bartenschlager, Heidelberg University, Heidelberg, Germany), and Huh7.5.1cells (kindly provided by Francis V. Chisari, Scripps Research Institute) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, and 100 U/ml streptomycin (Beyotime) at 37°C in a humidified atmosphere with 5% CO2. The 1b Con1 subgenomic HCV replicon pFKI389neo/NS3-3′ (also generously provided by Ralf Bartenschlager) was derived from Huh7-Lunet cells and maintained in the same medium as Huh7-Lunet cells but supplemented with 0.5 mg/ml G418 (Amersco) (22). Human embryonic kidney (HEK) 293T cells and other cell lines were cultured in minimal essential medium with the same reagents as Huh7 cells.
Amplification of HCV stocks and HCVcc infection.
Infectious HCV genotype 2a strain JFH1 (kindly provided by Takaji Wakita, National Institute of Infectious Diseases, Japan) was prepared as described previously (23). Briefly, viral stocks were diluted in DMEM and used to inoculate naive 40% confluent Huh7.5.1 cells at a multiplicity of infection (MOI) of 0.01 in a T75 flask. Infected cells were trypsinized and spliced into two T75 flasks before reaching confluence at 72 h postinfection (hpi). The supernatants from infected cells was then harvested at 7 or 8 days postinfection, aliquoted, and stored at −80°C. The viral titer was measured in focus-forming units (FFU) per milliliter of supernatant and determined by the average number of core-positive foci detected at the highest dilutions (24). Stable vinexin β knockdown Huh7.5.1 cells were infected with HCV JFH1 (MOI = 0.02). At 72 hpi, all of the infected cells with corresponding supernatants were collected and analyzed by quantitative real-time PCR (qRT-PCR) or Western blot (WB) assay.
Plasmids and reagents.
NS5A from HCV of different genotypes (1b Con1, 1b Whu, and 2a JFH1) was cloned into mammalian expression vector pRK-7 (Addgene) with a 5′ Flag tag. Total RNA from Huh7.5.1 cells infected with HCV JFH1 and the HCV Con1 replicon was used as the template. Additionally, plasmid pHCV-WHU-1 was also used as the template for molecular cloning (25). The wild-type cDNAs of cell host factors (HFs), including Argbp2, CAP, vinexin β, CK1α, VAP-A, CK2α, and others, were amplified from cDNAs from Huh7.5.1 or HEK 293T cells by reverse transcription-PCR amplification of total cellular mRNA with SuperScript III reverse transcriptase (Invitrogen). The cDNAs were cloned into mammalian expression vector pEGFP-C1 (Clontech), pmCherry-C1 (Clontech), or pRK-5 (Addgene) with a 5′ hemagglutinin (HA) tag. NS5A from HCV clone JFH1 was also subcloned into mammalian expression vector pcDNA3.1 (Life Technologies) without a tag. Truncation mutant proteins were constructed, and site-directed mutagenesis was done by the overlap extension PCR method with high-fidelity KOD plus Neo DNA polymerase (TOYOBO). Mouse monoclonal antibodies (MAbs) against Flag (M2; Sigma-Aldrich), HA (Proteintech), green fluorescent protein (GFP; Proteintech), glutathione S-transferase (GST; GenScript), actin (Proteintech), HCV core (C7-50; Santa Cruz Biotechnology), HCV 1b NS5A (256-A; Virogen), and HCV 2a NS5A (7B5 and 2F6; BioFront) were purchased from the companies indicated. Rabbit polyclonal antibodies against Flag (GenScript), HA (ABclonal), and GFP (Proteintech) were also purchased from commercial sources. HCV genotype 1b NS5A rabbit antiserum was raised against a recombinant NS5A domain I (genotype 1b strain Whu) protein with the N-terminal helix deleted and expressed with a histidine tag from expression vector pET28a (Novagen) in Escherichia coli. A New Zealand rabbit was purchased from the Experimental Animal Center, Wuhan Institute of Virology of the Chinese Academy of Sciences. This work was approved by Institutional Animal Care and Use Committee. Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Beyotime.
Co-IP and WB assay.
Plasmids were transfected into HEK 293T cells in 10-cm dishes or six-well plates with calcium phosphate. At 36 h posttransfection (hpt), the transfected cells were lysed in 1 ml of immunoprecipitation (IP) buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10 mg/ml aprotinin (Amresco), 10 mg/ml leupeptin (Amresco), and 0.5 mM phenylmethylsulfonyl fluoride (Beyotime) for 25 min on the ice. The lysates were then centrifuged at 16,000 × g for 15 min at 4°C, and the supernatants were collected. An equal volume of supernatants was incubated with 1.2 μg of the indicated antibody or control IgG (Sigma-Aldrich) and 30 μl of a 50% slurry of protein G-Sepharose (GE Healthcare Life Sciences) at 4°C for 5 h. The Sepharose beads were then harvested and washed four times with 1 ml of lysis buffer containing 500 mM NaCl. In the endogenous co-IP assay, the 1b Con1 subgenomic HCV replicon with naive 95% confluent and Huh7.5.1 cells infected with HCV JFH1 (MOI = 0.1, 96 hpi) were directly lysed and subjected to the procedure described above. All of the WB samples were treated by boiling in 2× SDS loading buffer, separated by SDS-PAGE, transferred to polyvinylidene difluoride (Millipore), and blocked with 5% fat-free milk in 50 mM Tris-HCl (pH 7.5)–0.15 M NaCl–0.05% Tween 20. Blots were detected with the primary antibodies indicated, followed by an HRP-conjugated secondary antibody. Finally, imaging was performed with ECL chemiluminescence reagent (Millipore) on X-ray films (Fujifilm). Quantification of blot band intensities was performed by Quantity One software or Image Lab software (Bio-Rad).
In vitro GST pulldown.
For an in vitro binding assay, GST-fused vinexin β or GST alone was expressed in BL21 cells and purified with glutathione Sepharose 4B (GE Healthcare Life Sciences) according to the supplier's instructions. Huh7 cells transfected with the pRK-7 plasmid encoding Flag-NS5A were washed twice with phosphate-buffered saline (PBS) and lysed in IP lysis buffer. The lysate was split into three aliquots and separately incubated with 10 μg of GST-vinexin β, 10 μg of GST, or glutathione Sepharose 4B at 4°C for 12 h. After being washed with IP lysis buffer four times, proteins were extracted from the Sepharose beads by boiling in 2× SDS loading buffer, resolved by SDS-PAGE, and detected by WB assay with the antibodies indicated.
RNA extraction and quantitative reverse transcription-PCR.
Total RNA from cultured cells and HCV RNA from supernatants were extracted with TriPure (Roche) and TRIzol LS (Invitrogen) according to the respective manufacturers' instructions. For intracellular RNA quantity determination, 300 ng of total RNA was used to synthesize cDNA with Moloney murine leukemia virus reverse transcriptase (Promega) and N6 random primer (Bioneer). qRT-PCR experiments were carried out with the CFX96 multicolor real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) with appropriate SYBR green dye (Invitrogen). Relative intracellular HCV RNA levels were examined by determining the quantity of HCV RNA normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNA levels. Relative cellular specific gene RNA levels were also normalized to the GAPDH RNA level. HCV RNA levels in culture supernatants were determined relative to a standard curve composed of serial dilutions of template DNA containing the JFH1 5′ UTR cDNA. The qRT-PCR primers used are as follows: FP (JFH1), TCTGCGGAACCGGTGAGTA; RP (JFH1), TCAGGCAGTACCACAAGGC; FP (vinexin β), CCCACCCAGGAGCCTAGAC; RP (vinexin β), CAAACCAGCCATCGTCACACT; FP (GAPDH), CCACTCCTCCACCTTTGAC; RP (GAPDH), ACCCTGTTGCTGTAGCCA; FP (CK1α), CATCTATTTGGCGATCAACATCA; RP (CK1α), GCCTGGCCTTCTGAGATTCTA.
Immunofluorescence staining and confocal microscopy.
Huh7-Lunet or Huh7.5.1 cells were cultured on a confocal 20-mm glass bottom dish (NEST). After transfection, corresponding cells were washed twice with PBS, fixed with 3.7% paraformaldehyde–PBS for 30 min, and permeabilized with 0.5% Triton X-100–PBS. After samples were blocked with 3% bovine serum albumin–PBS for 1 h, primary antibodies were diluted in blocking buffer and incubated with cells overnight at 4°C. After being washed six times with PBS, the cells were incubated with rhodamine-conjugated or fluorescein isothiocyanate (FITC)-conjugated goat secondary antibodies (Pierce) for 1 h and then treated with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime) for 5 min, washed six times with warm PBS, and then observed with a confocal microscope (Olympus FluoView FV1000). For direct fluorescence observation, Huh7-Lunet cells transfected with a fluorescent fusion protein plasmid were fixed with 3.7% paraformaldehyde for 20 min. At 36 hpt, transfected cells were washed three times and subjected to observation with a confocal microscope. Colocalization efficiency was evaluated quantitatively with ImageJ software (developed by the National Institutes of Health). We calculated the Pearson correlation coefficient to evaluate the pixel-to-pixel covariance of signal levels in two different images and calculated the correlation between the two channels. The values ranged from −1 to 1. Values ranging from 0.4 to 0.699 were indicative of moderate correlations; those between 0.7 and 0.99 were indicative of high correlations (26). All of the image data represent 8 to 10 representative cells in three independent experiments.
Generation of stable knockdown cells with a retroviral shRNA system.
To establish a stable knockdown cell line, shRNA that interferes with the target gene was subcloned into the pSUPER.retro.puro plasmid (oLigoengine) according to the supplier's instructions. These experiments were performed as previously described (27). HEK 293T cells were cotransfected with pSUPER.retro.puro constructs along with packaging plasmids pGag-pol and pVSV-G by the calcium phosphate method. The shRNA retroviral stocks were harvested from the cell supernatants at 72 hpt, filtered with a 0.45-μm filter, and then used to transduce Huh7.5.1, Huh7, and HEK 293T cells with 4 μg/ml Polybrene. At 72 hpt, all of the stable knockdown cells were cultured in puromycin (Amersco) selection medium for at least 3 days (the puromycin concentration was 4 μg/ml for Huh7.5.1 and Huh7 cells and 1 μg/ml for HEK 293T cells). The effects of interference on the surviving colonies were confirmed by qRT-PCR or WB analysis. The vinexin β targeting sequences used for RNA interference (RNAi) were as follows: 1 sh Vinβ, CCCAGAAATTCGGAACGTT; 2 sh Vinβ, GGGTGACATTGTCTACATC; 3 sh Vinβ, GGTGAACGAGAACTGGTAC. The CK1α targeting sequences used for RNAi were as follows: 1 sh CK1α, CAGAATTTGCGATGTACTT; 2 sh CK1α, GGACAATGTTAAAGCAGAA.
As negative controls, short hairpin Renilla luciferase (sh Rluc) with the sequence GTAGCGCGGTGTATTATAC and sh nontarget (sh NT) with the sequence TTCTCCGAACGTGTCACGT were used. In addition, the sequence AGGCCTTGTGGTACTGCCT targeting the HCV 5′ UTR was used as a positive control named sh HCV.
Construction of rescue mutation lentiviral vector to maintain expression of vinexin β in vinexin β knockdown cells.
For experiments involving rescue of vinexin β interference, stable vinexin β knockdown Huh7.5.1 cells (3 sh Vinβ) were transiently transduced with a recombinant lentiviral vector expressing exogenous wobble mutant protein vinexin β (RM3) to resist the cellular interference machinery (3 sh Vinβ-RM3). The rescue mutant form of vinexin β (RM3) was designed so that GGT GAA CGA GAA CTG GTA C was changed to GGC GAG CGC GAG CTA GTC C, which can eliminate the effect of RNAi but does not change the amino acid sequence of the vinexin β protein. Preparation of a recombinant lentiviral vector was based on the lentiviral packaging system consisting of the pHAGE-CMV-MCS-IZsGreen, pMD2.G, and psPAX2 plasmids (a gift kindly supplied by Huang Zan at Wuhan University) (28). The procedure used for the recombinant lentiviral vector was the same as that used for the shRNA retroviral vector described above. As a control, the empty recombinant lentiviral vector was also harvested and used to transiently transduce 3 sh vinexin β Huh7.5.1 cells and sh NT Huh7.5.1 cells (3 sh Vinβ and sh NT), respectively. Equal numbers (3 × 105) of three cell lines were then seeded into a six-well plate. Twenty-four hours later, 3 sh Vinβ and sh NT cells were inoculated with recombinant lentiviral vector RM3 or the empty vector (mock treatment) in the presence of 8 μg/ml Polybrene (Sigma) at an MOI of 3. At 24 hpi, cells were infected with HCV JFH1 (MOI = 0.02) and harvested at 72 h after HCV infection. The samples were subjected to qRT-PCR and WB analyses.
Flow cytometry analyses.
Insofar as the cells were transduced with a retroviral vector, the possibility had to be excluded that overexpression or knockdown of vinexin β did not alter cell proliferation and the cell cycle. Thus, these cells at 80% confluence in the wells of six-well plates were collected after being washed twice with PBS and digested with 0.25% trypsin (Gibco, Life Technologies). After centrifugation (300 × g, 10 min), the cells were fixed with precooled 70% ethanol overnight at −20°C. The next day, the fixed cells were treated with 30 μg/ml propidium iodide (PI; Sigma) and 50 μg of RNase/ml for 60 min at 37°C. Their nuclear DNA content was measured by flow cytometry with a fluorescence-activated cell sorter (EPICS XL/XL-MCL; Beckman Coulter), and data analysis was performed by ModFitLT software (BD Biosciences) to determine the percentage of cells in each phase of the cell cycle.
Statistical analyses.
Statistical graphs were created with GraphPad Prism software, and statistical analyses were performed with two-tailed Student t tests. The graphs represent the mean values ± the standard deviations (SD) of at least three independent experiments. P < 0.05 was considered statistically significant, and P < 0.01 and P < 0.001 were considered highly significant.
RESULTS
HCV NS5A interacts with vinexin β.
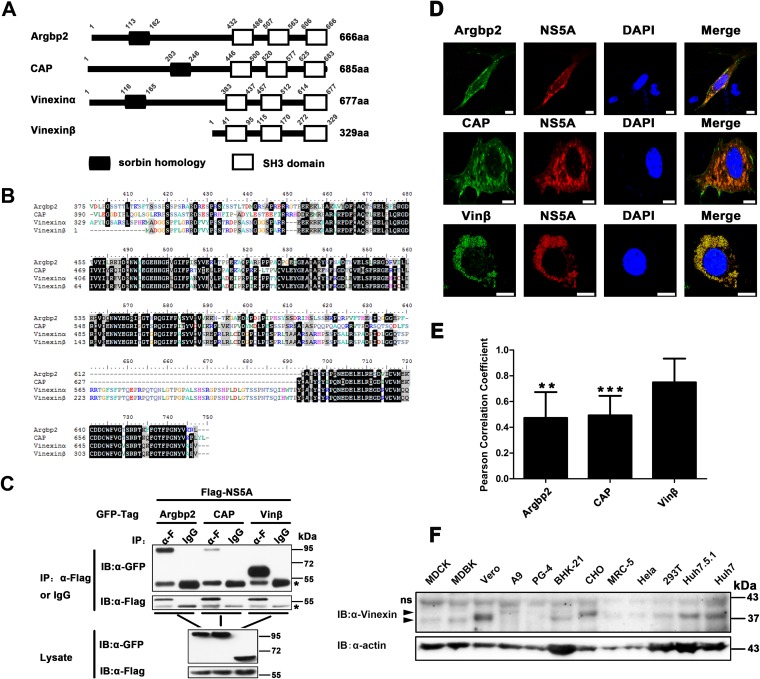
NS5A is a proline-rich protein with two polyproline motifs (29). Since the polyproline motifs of NS5A can interact with proteins containing SH3 domains (30), we sought to identify a new cellular factor(s) associated with NS5A through the SH3 domain. For this purpose, a novel SoHo adaptor family with three SH3 domains at the C terminus captured our attention (Fig. 1A). A previous study showed that the SoHo family is potentially associated with NS5A in a preliminary yeast two-hybrid screening (31). Sequences within the SH3 regions of three members of the SoHo family (Argbp2, CAP, vinexin) are highly homologous (Fig. 1B), so we cloned the genes encoding human Argbp2, CAP, and vinexin β and performed co-IP assays when they were coexpressed with NS5A. The results showed that Argbp2, CAP, and vinexin β could all be specifically pulled down by NS5A (Fig. 1C), and NS5A showed a much stronger interaction with vinexin β than with Argbp2 and CAP. Confocal microscopy analysis showed that Argbp2, CAP, and vinexin β also colocalized with NS5A in the perinuclear region of Huh7-Lunet cells (Fig. 1D). Furthermore, we calculated the Pearson correlation coefficient to quantify the degree of their colocalization; the Pearson correlation coefficients of Argbp2-NS5A, CAP-NS5A, and vinexin β-NS5A colocalization were 0.473, 0.492, and 0.75, respectively (Fig. 1E), indicating that colocalization of vinexin β-NS5A had a stronger interaction than that of Argbp2-NS5A or CAP-NS5A. Taken together, our results demonstrated that Argbp2, CAP, and vinexin β all interact with NS5A and that vinexin β has a much stronger interaction with NS5A (Fig. 1C, D, and E) (18, 32, 33). In the meantime, immunoblot analysis indicated that endogenous vinexin β is ubiquitously expressed as an about 37-kDa protein in many different cell lines, including human 293T, Huh7.5.1, and Huh7 cells (Fig. 1F). As a result, we selected vinexin β for further assay.
FIG 1.
SoHo family proteins interact with HCV NS5A in mammalian cells. (A) Schematic representation of the domain structure of the SoHo family (Homo sapiens). The slanted numbers indicate the sites of SoHo and SH3 domains in the proteins. (B) Alignment of the C-terminal amino acid sequences of H. sapiens Argbp2, CAP, vinexin α, and vinexin β. Multiple sequence alignments were generated with the Clustal X software. (C) pRK-Flag-NS5A (JFH1) was cotransfected with pEGFP-Argbp2, pEGFP-CAP, or pEGFP-vinexin β in HEK 293T cells. Cell lysates were immunoprecipitated and subjected to WB (immunoblot [IB]) analysis as indicated. The IgG heavy chain is indicated by asterisks. (D) pRK-Flag-NS5A was cotransfected with pEGFP-Argbp2, pEGFP-CAP, or pEGFP-vinexin β into Huh7-Lunet cells; immunofluorescence assays were performed with a mouse anti-Flag MAb (α-F) and a rhodamine-conjugated goat anti-mouse secondary antibody; and nuclei were stained with DAPI. Scale bars, 10 μm. (E) Quantification of colocalization was performed by determining the Pearson correlation coefficient with ImageJ software (described in Materials and Methods). Values represent means ± SDs of 8 to 10 representative cells in three independent experiments. Vinexin β served as a control. ns, not significant; **, P < 0.01; ***, P < 0.001. (F) WB analysis of endogenous vinexin β expression in the cell lines indicated. Total cell lysates from the cells indicated were subjected to WB analysis with a mouse polyclonal antibody against the full-length vinexin protein. Arrowheads indicate target bands with corresponding names. ns, nonspecific.
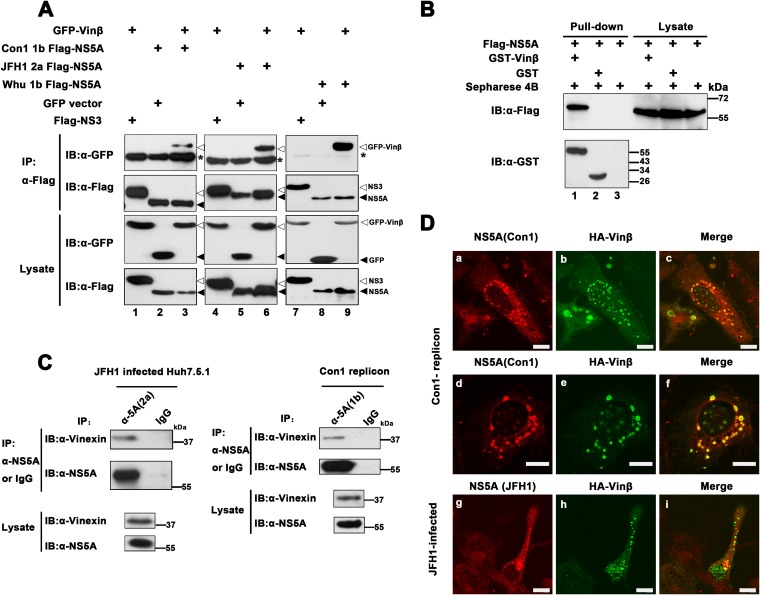
To verify that the interaction of vinexin β with NS5A is strain specific, we coexpressed vinexin β and NS5A derived from either HCV genotype 1b (Con1 and Whu) (25) or 2a (JFH1) in HEK 293T cells. As shown in Fig. 2A, GFP-vinexin β, but not GFP alone, could be coimmunoprecipitated with different subtypes of NS5A (top, lanes 3, 6, and 9), suggesting that the vinexin β-NS5A interplay was not genotype specific. In addition, vinexin β failed to be coimmunoprecipitated with HCV NS3 (Fig. 2A, top, lanes 1, 4, and 7). To further examine whether the interaction can occur in vitro, we performed a GST pulldown assay with purified GST-vinexin β and GST alone mixed with Huh-7 lysates containing exogenously expressed Flag-NS5A. The results indicated that NS5A was specifically pulled down with GST-vinexin β but not GST or Sepharose 4B alone (Fig. 2B, top, lane 1), implying that vinexin β associated with NS5A in vitro. Next, we evaluated whether the interaction occurs in HCV replication cells. Lysates of HCV JFH-infected cells and HCV Con1 replicon cells were subjected to IP analysis with a mouse MAb (34) and a rabbit polyclonal antibody against NS5A (1b), respectively. The results demonstrated that endogenous vinexin β specifically interacted with NS5A of 1b and 2a in a more authentic system (Fig. 2C). Moreover, we also observed that vinexin β and NS5A colocalized in HCV replicon and HCV-infected cells (Fig. 2D). Therefore, we suggest that vinexin β interacts with NS5A in HCV-infected cells.
FIG 2.
Vinexin β interacts with NS5A. (A) Flag-tagged NS5A derived from genotype 1b strain Con1 or Whu or genotype 2a strain JFH1 (pRK-Flag-NS5A) was coexpressed with GFP-tagged vinexin β (pEGFP-vinexin β) in HEK 293T cells. At 36 hpt, transfected cells were immunoprecipitated with a mouse MAb against Flag and subjected to WB (immunoblot [IB]) analysis with a rabbit polyclonal antibody to Flag or a mouse MAb to the Flag or GFP tag. The IgG heavy chain is denoted by asterisks, and triangles indicate proteins. (B) Huh7 cell lysate containing exogenously expressed Flag-NS5A was incubated with GST, GST-vinexin β, or Sepharose 4B alone and then subjected to a pulldown assay. Equal volumes of the proteins bound to the beads and the original lysates (10% of the input) were examined by WB analysis with a rabbit anti-Flag polyclonal antibody or a mouse anti-GST MAb, respectively. (C) HCV JFH1-infected Huh7.5.1 cells were harvested at 96 hpi, and lysates were immunoprecipitated with a mouse anti-NS5A (2a) MAb or control mouse IgG. Con1 replicon cells at 95% confluence were harvested, and cell lysates were immunoprecipitated with anti-NS5A rabbit antiserum (1b) or control rabbit antiserum. The immune complexes were subjected to WB analysis with a mouse anti-vinexin polyclonal antibody and a mouse anti-NS5A MAb (1b or 2a). (D) Vinexin β colocalization with HCV NS5A via immunofluorescence assay. Con1 replicon cells were transfected with pRK-HA-vinexin β (HA-Vinβ). They were fixed at 36 hpt and stained with a rabbit anti-NS5A primary polyclonal antibody (described in Materials and Methods) and a mouse primary MAb against HA, followed by the rhodamine-conjugated goat anti-rabbit secondary antibody and an FITC-conjugated goat anti-mouse secondary antibody for observation via confocal microscopy. Red indicates NS5A; green indicates HA-vinexin β (a to c and d to f). In the bottom row, Huh7.5.1 cells were infected with HCV JFH1 (MOI = 0.2). At 24 hpi, cells were transfected with pRK-HA-vinexin β (HA-Vinβ). At 72 hpt, cells were fixed and incubated with a rabbit anti-HA primary polyclonal antibody and a mouse MAb to 2a JFH1 NS5A (described in Materials and Methods), followed by a rhodamine-conjugated goat anti-mouse secondary antibody and an FITC-conjugated goat anti-rabbit secondary antibody. Red indicates NS5A; green indicates HA-vinexin β (g, h, i) for observation via confocal microscopy. All of the experiments were repeated three times. Scale bars, 10 μm.
Conserved amino acids in the third SH3 domain of vinexin β and PxxPxR motifs at the C terminus of NS5A are critical for the interaction of vinexin β with NS5A.
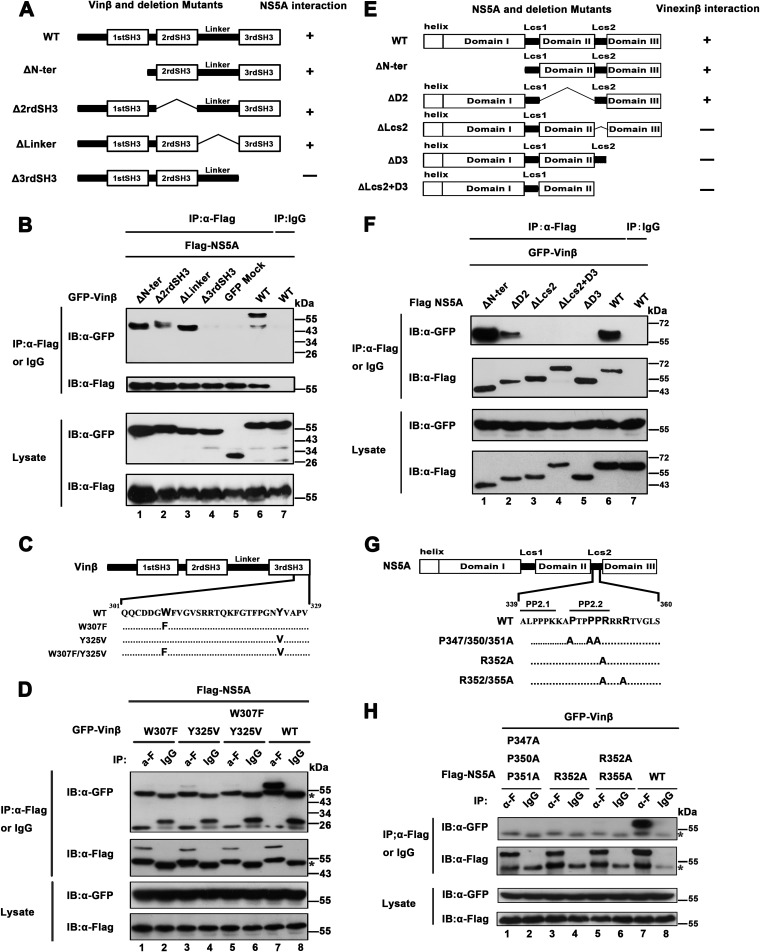
Wild-type vinexin β is composed of 329 amino acids (aa) and contains three SH3 domains covering aa 41 to 95, 115 to 170, and 272 to 329, respectively (35). To determine the regions involved in the interaction of vinexin β with NS5A, we created various truncation mutant variants of vinexin β encoding ΔN-ter (aa 95 to 329), Δ2rdSH3 (aa 1 to 116 and 171 to 329), ΔLinker (aa 1 to 172 and 252 to 329), and Δ3rdSH3 (aa 1 to 262) (Fig. 3A). We then coexpressed these mutant vinexin β proteins with NS5A (JFH1) and preformed co-IP assays. As shown in Fig. 3A and B, the ΔN-ter, Δ2rd SH3, and ΔLinker mutant proteins could still interact with NS5A (Fig. 3B, top, lanes 1 to 3), but the Δ3rd SH3 mutant protein, which lacked the third SH3 domain of vinexin β, failed to associate with NS5A (Fig. 3B, top, lane 4), indicating that the third SH3 domain of vinexin β is critical for the interaction of vinexin β with NS5A. A previous study showed that two conserved amino acid residues, tryptophan (W307) and tyrosine (Y325), located in the third SH3 domain are critical for the function of the SH3 domain (30), so we constructed variants of vinexin β with a tryptophan-to-phenylalanine (W307F) mutation, a tyrosine-to-valine mutation (Y325V), and two mutations (W307F/Y325V) in the third SH3 domain (Fig. 3C) and performed co-IP assays. As shown in Fig. 3D, the W307F and W307F/Y325V mutant proteins failed to interact with NS5A (top, lanes 1 and 5) and the Y325V-NS5A association was also greatly reduced (top, lane 3), suggesting that two conserved sites (W307 and Y325) in the third SH3 domain are essential for the interaction of vinexin β with NS5A.
FIG 3.
Mapping of regions of vinexin β and NS5A involved in the interaction between vinexin β and NS5A. (A) On the left is a schematic diagram of full-length vinexin β and deletion mutant forms thereof, and on the right are the results of vinexin β and mutant protein interactions with NS5A. (B) pEGFP-vinexin β (GFP-Vinβ) or mutant proteins (ΔN-ter, Δ2rdSH3, ΔLinker, and Δ3rdSH3) were used with pRK-Flag-NS5A (JFH1) to cotransfect HEK 293T cells. At 36 hpt, transfected cells were immunoprecipitated with anti-Flag antibody or mouse IgG and subjected to WB (immunoblot [IB]) analysis with a rabbit polyclonal antibody against GFP and a mouse MAb to Flag to detect the Flag and GFP tags, respectively. (C) Schematic diagram of vinexin β and variants thereof with point mutations in the third SH3 domain. (D) HEK 293T cells were cotransfected with pEGFP-vinexin β or point mutant forms thereof and pRK-Flag-NS5A (JFH1). At 36 hpt, transfected cells were immunoprecipitated with anti-Flag antibody or mouse IgG and subjected to WB analysis with mouse MAbs against GFP and Flag to detect the Flag and GFP tags, respectively. (E) The left panel is a schematic diagram of full-length NS5A and deletion mutant forms thereof, and the right panel shows the results of their interactions with vinexin β. (F) pRK-Flag-NS5A (JFH1) or mutant forms thereof (ΔN-ter,ΔD2,ΔLcs2, ΔD3, and ΔLcs2+D3) and pEGFP-vinexin β were used to cotransfect HEK 293T cells. IP and WB analysis were performed as described for panel B. (G) Schematic diagram of full-length NS5A and point mutant forms thereof with changes in the Lcs2 region. (H) HEK 293T cells were cotransfected with pEGFP-vinexin β and pRK-Flag NS5A (JFH1) or point mutant variant plasmids. IP and WB analysis were performed as described for panel D. The data shown are representative of three independent experiments.
Next, we attempted to map the regions in NS5A that are involved in the interaction with vinexin β. NS5A consists of an N-terminal amphipathic helix whose function is to anchor to cytoplasmic membranes and three domains (I, II, and III) separated by two low-complexity sequences (Lcs) (15, 36). A schematic representation of the HCV NS5A (JFH1) and serial truncation mutant proteins is shown in Fig. 3E (top), and these mutant proteins were used for co-IP assays. The results showed that NS5A mutant proteins lacking either low-complexity domain connectors II-Δ Lcs2 (full-length NS5A with aa 339 to 352 deleted) or domain III (ΔD3 from aa 1 to aa 352) failed to interact with vinexin β. Moreover, an NS5A mutant protein with Lcs2 and domain III both deleted (ΔLcs2+D3 from aa 1 to aa 338) naturally did not interact with vinexin β (Fig. 3F, top, lanes 3 to 5). Taken together, our results argue that both Lcs2 and domain III located at the C terminus of NS5A are responsible for the interaction of NS5A with vinexin β.
Previous reports have documented that NS5A contains two proline-rich motifs; one is termed PP1 and covers aa 35 to 39 in domain I, and the other is termed PP2 and covers aa 339 to 352 of the Lcs2 fragment (37). PP2 contains two closely spaced polyproline motifs (PP2.1 and PP2.2) (Fig. 3G) that allow interaction with proteins containing SH3 domains, including Src family tyrosine kinases (38–40). Because the C terminus contains PP2 motifs and mediates the interaction of NS5A with vinexin β, the proline residues at sites 347, 350, and 351 and the arginine residue at site 352 within the PP2.2 motif are highly conserved throughout HCV genotypes (37) and the arginine residue in polyproline motifs could develop a salt bridge with the SH3 domain (16), we constructed a series of point mutations within the PP2 motifs as indicated in Fig. 3G and performed co-IP assay. The results indicated that the substitution of conserved residues 347, 350, 351, and 352 in the PP2.2 motifs led to abrogation of binding to vinexin β (Fig. 3H, top, lanes 1, 3, and 5), suggesting that conserved PxxPxR motifs in the Lcs2 region were indispensable for the binding of NS5A with vinexin β.
Knockdown of vinexin β decreases HCV propagation and NS5A hyperphosphorylation.
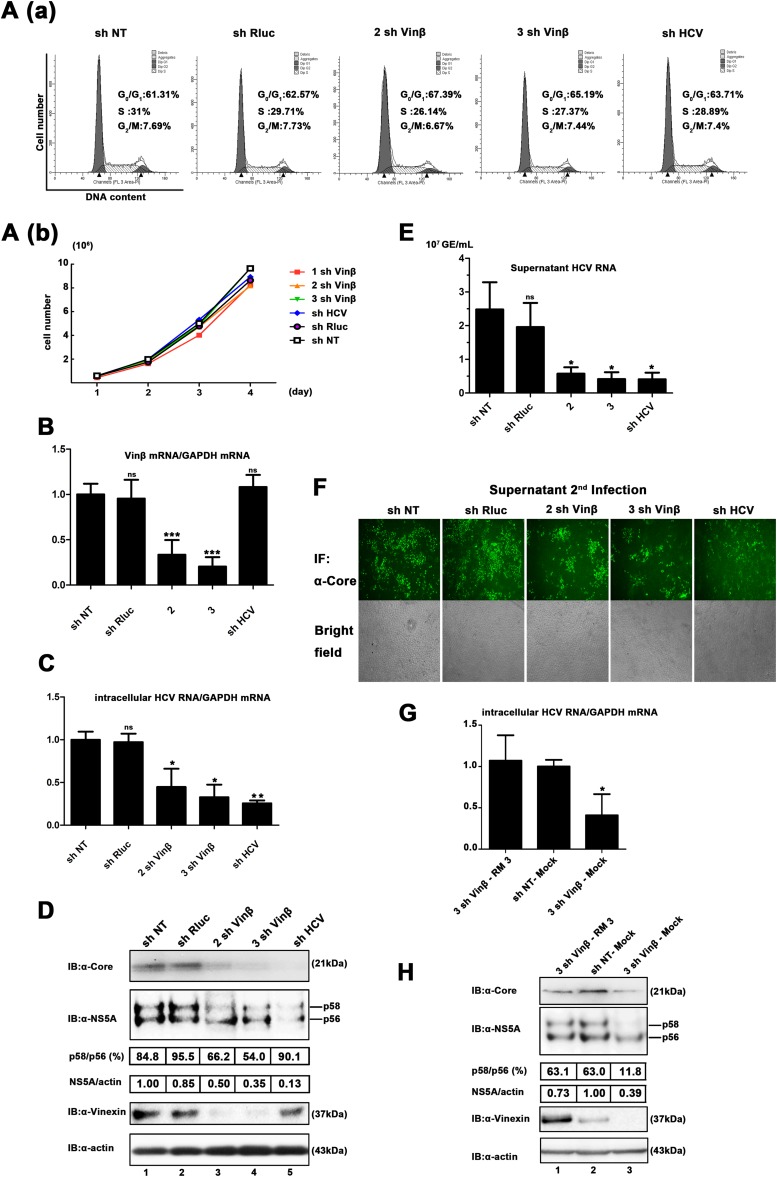
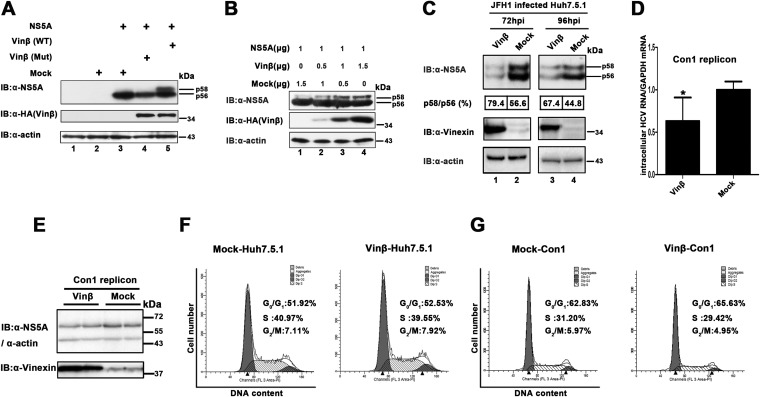
RNAi was introduced to further examine the effect of vinexin β on HCV replication. Three stable vinexin β knockdown Huh7.5.1 cell lines (1 sh Vinβ, 2 sh Vinβ, and 3 sh Vinβ) were established on the basis of a retrovirus-mediated shRNA system. The sh HCV-targeted conserved 5′ UTR of HCV was used as a positive control, sh NT was used as a negative control, and sh Rluc (target Renilla luciferase) was also used, ensuring a consist negative control. We first confirmed that downregulation of vinexin β expression had no significant effect on the cell cycle and growth pattern (Fig. 4A). Vinexin β levels in sh vinexin β-transduced cells (2 sh Vinβ and 3 sh Vinβ) were significantly lower than those in sh NT-, sh HCV-, and sh Rluc-transduced cells (Fig. 4B). Next, shRNA-transduced cells were infected with HCV JFH1 at a low MOI of 0.02, ensuring multiple cycles of infection, which could have a comprehensive effect on the viral life cycle (41–43). At 72 hpi, we found that silencing of vinexin β (2 sh Vinβ and 3 sh Vinβ) reduced the intracellular HCV RNA level to 44.7 and 32.7% of that in control (sh NT) cells, respectively (Fig. 4C). Similarly, WB analysis confirmed that the decreased vinexin β protein level also suppressed the core and NS5A protein expression of HCV (Fig. 4D, lanes 3 and 4). The total relative NS5A protein level decreased to 50 and 36% of that of sh NT, which was consistent with the qRT-PCR results. In addition, we surprisingly observed that the p58/56 ratio, which reflects the level of hyperphosphorylation of NS5A, in sh vinexin β-treated cells was 54% of that in sh NT-treated cells, implying that vinexin β protein expression is also critical for the hyperphosphorylation status of NS5A. When cells are infected at a low MOI, extracellular HCV RNA levels should coincide with those of intracellular RNA in multiple cycles of infection. As shown in Fig. 4E, the genome copy numbers in the supernatants with 2 sh Vinβ and 3 sh Vinβ were 0.58 × 107 ± 0.09 × 107 and 0.41 × 107 ± 0.11 × 107 genome equivalents (GE)/ml, but the genome copy number in the supernatants with the sh NT control was 2.5 × 107 ± 0.4 × 107 GE/ml, implying that downregulation of vinexin β also caused the overall reduction in virion release. Consistently, significant changes in the extracellular HCV RNA were also made in the second round of infection (Fig. 4F). Taken together, these results revealed that knockdown of endogenous vinexin β significantly inhibits HCV propagation in Huh7.5.1 cells with the simultaneous attenuation of NS5A hyperphosphorylation.
FIG 4.
Knockdown of vinexin β inhibits HCV propagation and NS5A hyperphosphorylation. (A) Knockdown of vinexin β does not significantly affect cell proliferation or the cell cycle. (a) Flow cytometry analysis of the knockdown of vinexin β cells. Huh7.5.1 cells transduced with the retroviral vector indicated for knockdown of vinexin β were harvested and mixed with PI for flow cytometry analysis to determine the proportions of the population in the different phases of the cell cycle (details are described in Materials and Methods). The graphs were generated and analyzed with ModFit LT version 2.0. Bold indicates the percentage of cells in each phase of the cell cycle. The data are from one of three independent experiments. (b) Growth curves of vinexin β knockdown cells. Huh7.5.1 cells transduced with the shRNA retroviral vectors indicated were seeded in equal numbers and counted with a Scepter 2.0 Cell Counter (Merck Millipore) at the time points indicated (1st to 4th days). (B) Knockdown of vinexin mRNA was determined by qRT-PCR. Vinexin mRNA levels were normalized to the GAPDH mRNA level. The value of sh NT was used as a control (set at 100%). Values represent means ± SDs from three independent experiments. (C) Huh7.5.1 cells expressing the shRNAs indicated were infected with HCV JFH1 at an MOI of 0.02. At 72 hpi, intracellular HCV RNA levels were measured by relative qRT-PCR. The value of sh NT was used as a control (set at 100%). Values are means ± SDs of three independent experiments. (D) Cells infected with HCV as described for panel C were lysed, and WB (immunoblot [IB]) analysis was performed with the antibodies indicated to determine the corresponding protein levels. The positions of two differently phosphorylated NS5A variants (p56 and p58) are indicated. The relative intensities of HCV NS5A (total, p58, and p56)/cellular actin were quantified by densitometric scanning of the film with Quantity One or Image Lab software (Bio-Rad). The NS5A and actin band intensities were normalized to the intensity of p56 of NS5A (set as 100) in sh NT-treated cells. To analyze the NS5A p58/p56 ratio, the band intensity was normalized to that of p56 of NS5A in sh NT-treated cells (set as 100). Three independent experiments were performed. (E) Extracellular HCV RNA isolated from the culture supernatant was quantified by absolute qRT-PCR, and the HCV 5′ UTR was used for quantification of the template. (F) Huh7.5.1 cells were infected with 10-fold-diluted culture supernatants from the experiment described in panel E. At 72 hpi, all of the cells were stained with a mouse anti-HCV core MAb and then subjected to a fluorescence assay; in the meantime, bright-field images of all of the samples were also obtained. All of the images represent more than 10 random fields. Three independent experiments were performed. (G) Huh7.5.1 cells with stable expression of vinexin β shRNA (3 sh Vinβ) were transiently transduced with vinexin β rescue mutant protein (RM3) resistant to vinexin β RNAi. Twenty-four hours later, transduced cells were infected with JFH1 at an MOI of 0.02. At 72 hpi, intracellular HCV RNA levels were measured by relative qRT-PCR analysis. Values represent means ± SDs of three independent experiments. (H) Huh7.5.1 cells with stable expression of vinexin β shRNA were transduced and infected as described for panel E, and cells were lysed for WB analysis with the antibodies indicated. The positions of two differently phosphorylated NS5A variants (p56 and p58) are indicated on the right. Quantitative analysis of bands was done as described for panel D. Three independent experiments were performed. Error bars indicate standard deviations. Asterisks indicate significant differences (***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, nonsignificant).
Furthermore, to rule out the off-target effects of RNAi, we transiently expressed the wobble sh vinexin β-resistant mutant protein vinexin β (RM3) in vinexin β knockdown cells (3 sh Vinβ) and then infected them with HCV JFH1 (MOI = 0.02). The results indicated that exogenous expression of an sh vinexin β-resistant mutant protein (RM3), but not the empty vector (mock treatment), increased the intracellular HCV RNA level (Fig. 4G). As expected, the core and NS5A protein levels of HCV could also be restored in 3 sh Vinβ-RM3, in contrast to 3 sh Vinβ-mock (Fig. 4H, lane 1 versus lane 3). Similarly, the reduction of NS5A hyperphosphorylation caused by the knockdown of vinexin β was also recovered by exogenous expression of sh vinexin β-resistant vinexin β mutant protein (Fig. 4H). Therefore, we have demonstrated that vinexin β expression may affect viral replication via regulation of NS5A hyperphosphorylation.
Vinexin β can increase levels of hyperphosphorylated NS5A in vivo.
Previous studies have reported that phosphorylation of NS5A and HCV replication are closely linked (7, 15, 44). To further explore whether vinexin β influences the phosphorylation of NS5A, thereby regulating HCV replication, NS5A and vinexin β were coexpressed in HEK 293T cells. We observed that there were two close bands probed with anti-NS5A antibody in cell lysates while performing a WB assay (Fig. 5A, lane 5). The lower band, for the hypophosphorylated variant of NS5A, is commonly termed p56, and the upper band, for the hyperphosphorylated variant of NS5A, is p58 (Fig. 5A) (9). In contrast, when NS5A expressed alone or coexpressed with a vinexin β mutant protein (W307F/Y325V) that cannot interact with the NS5A protein, NS5A could only keep the hypophosphorylated variant (p56) (Fig. 5A, lanes 4 and 3). This suggests that vinexin β promotes the hyperphosphorylation of NS5A in mammalian cells. Furthermore, we found that vinexin β expression has a dose-dependent effect on the hyperphosphorylation of NS5A (from 0 to 1.5 μg) in mammalian cells (Fig. 5B, lanes 1 to 4). Next, we examined whether the expression of vinexin β would affect NS5A phosphorylation in HCV-infected cells. We stably expressed vinexin β and inoculated Huh7.5.1 cells with HCV JFH1 (MOI = 0.02). As shown in Fig. 5C, the p58/p56 ratio also increased slightly when vinexin β was overexpressed compared to that in control-transfected (mock-treated) cells at 72 and 96 hpi, respectively. Overall, our data demonstrated that vinexin β can enhance the hyperphosphorylation of NS5A in mammalian cells. Of note, the total NS5A level was also decreased with the exogenous expression of vinexin β in HCV JFH1-infected cells. We think that the balance of two NS5A phosphorylation statuses, the intracellular signaling events of vinexin β overexpression such as JNK activation and the more unknown complicated relationship between HCV and host cells could be the reasons for this interesting phenotype (9, 14, 15, 45–47). Therefore, we observed that overexpression of vinexin β inhibited HCV infection from intracellular RNA, protein, and extracellular virion levels (data not shown), and viral genome replication was moderately (∼37%) decreased when vinexin β was stably overexpressed in the 1b replicon (Fig. 5D and E). Additionally, the exogenous expression of vinexin β had no impact on the cell division cycle (Fig. 5F and G). Since both knockdown of endogenous vinexin β (which decreased the hyperphosphorylated NS5A level) and exogenous expression of vinexin β (which overenhanced the hyperphosphorylated NS5A level) weakened HCV propagation, we believe that fluctuation of the NS5A phosphorylation proportion regulated by vinexin β may have a negative effect on HCV replication.
FIG 5.
Vinexin β increases levels of hyperphosphorylated NS5A. (A) HEK 293T cells were transfected with the plasmids indicated. At 36 hpt, cells were lysed and WB (immunoblot [IB]) analysis was performed with anti-NS5A (2a), anti-HA, and anti-actin antibodies. The positions of two differently phosphorylated NS5A variants (p56 and p58) are indicated on the right. (B) HEK 293T cells were transfected with gradually increased pRK-HA-vinexin β and a constant quantity of pcDNA3.1-NS5A (JFH1). WB analysis was performed as described for panel A. (C) Stable vinexin β (Vinβ) and empty vector (mock treatment) expression. Huh7.5.1 cells transiently transduced with a lentiviral vector (described in Materials and Methods) were infected with HCV JFH1 (MOI = 0.02). At 72 and 96 hpi, all of the cells were lysed and WB analysis was performed with the antibodies indicated. The positions of two differently phosphorylated NS5A variants (p56 and p58) are indicated on the right. The p58/p56 band intensity ratio was quantified with Quantity One software (details are described in the legend to Fig. 4). The results are representative of three independent experiments. (D and E) Analysis of HCV replication in vinexin β-overexpressing HCV replicon cells. HCV replicon (pFKI389neo/NS3-3′) cells were stably transduced with the lentiviral vector indicated (vinexin β and mock treatment) and reseeded into the wells of a six-well plate (3.5 × 105/well). Upon reaching 95% confluence, all of the cells were harvested and subjected to relative qRT-PCR (D) and WB analysis (E) as described above. (F and G) The cells (Huh7.5.1 and Con1 replicon) stably transduced with the lentiviral vector indicated for overexpression of vinexin β were harvested and mixed with PI for flow cytometry analysis to determine the proportion of the population in each phase of the cell cycle (details are described in Materials and Methods). The graphs were generated and analyzed with ModFit LT version 2.0. Bold indicates the percentage of cells in each phase of the cell cycle. The data are from one of three independent experiments.
Vinexin β interacting with CK1α regulates NS5A phosphorylation.
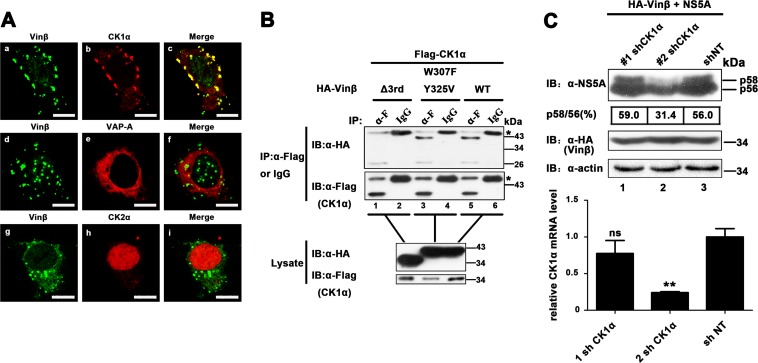
We further explored how vinexin β modulates NS5A hyperphosphorylation. Since vinexin β has no significant intrinsic enzyme activity (48), we speculate that another HF(s) may be involved in the regulation of NS5A phosphorylation via interaction with vinexin β. In addition, various cellular factors involved in viral replication have been identified through interaction with NS5A (44). Thus, we coexpressed GFP-tagged vinexin β and red fluorescent protein (RFP)-fused binding partners of NS5A such as annexin A2, CK1α, CK2α, CyPA, FBXL2, FKBP8, hB-ind1, Hsp72, Pin1, PSTPIP2, VAP-A, and VAP-B in Huh7-Lunet cells (6, 13, 49–57). The results showed that host kinase RFP-CK1α was obviously colocalized with GFP-vinexin β, but RFP-VAP-A and RFP-CK2α were not (6, 58) (Fig. 6A), while the other factors were also not significantly colocalized with vinexin β (data not shown). To further confirm the interaction of CK1α with vinexin β, CK1α was coexpressed with vinexin β or mutant forms thereof in Huh7-Lunet cells. The co-IP result showed that both wild-type vinexin β and the dual point mutant protein (W307F/Y325V) could be pulled down with CK1α (Fig. 6B, top, lanes 3 and 5), but the Δ3rd SH3 vinexin β mutant protein failed to coimmunoprecipitate with CK1α (Fig. 6B, top, lane 1), indicating that essential residues involved in the association of vinexin β with NS5A are not required for the interaction of vinexin β with CK1α. To further confirm the effect of CK1α on the hyperphosphorylation of NS5A, we established HEK 293T cell lines stably expressing lentivirus-delivered shRNAs against CK1α (1 sh CK1α and 2 sh CK1α) to knock down the expression of CK1α (Fig. 6C, bottom). When vinexin β and NS5A were coexpressed, the level of NS5A hyperphosphorylation induced by vinexin β was significantly lower in 2 sh CK1α cells than in cells treated with the sh NT control (Fig. 6C, top, lane 2 versus lane 3), suggesting that vinexin β modulated NS5A hyperphosphorylation in a CK1α-dependent manner.
FIG 6.
Vinexin β regulates NS5A phosphorylation in a CK1α-dependent manner (A) Huh7-Lunet cells were cotransfected with pEGFP-vinexin β and various plasmids encoding red fluorescent protein-fused HFs. At 24 hpt, cells were fixed for immunofluorescence assay. Scale bars, 10 μm. (B) HEK 293T cells were cotransfected with pRK-Flag-CK1α and pRK-HA-vinexin β (W307F/Y325V) or Δ3rd SH3. At 36 hpt, cell lysates were immunoprecipitated with a mouse anti-Flag MAb (α-F) and bound proteins and original lysates were then detected by WB (immunoblot [IB]) analysis. Asterisks indicate the IgG heavy chain. (C) HEK 293T cells stably expressing shRNA of CK1a were cotransfected with pRK-HA-vinexin β and pcDNA3.1-NS5A (JFH1). At 36 hpt, cells were lysed and WB analysis were performed with a mouse anti-NS5A MAb, a mouse anti-HA MAb, or a mouse anti-actin MAb (top). The p58/p56 band intensity ratio was quantified with Quantity One software (details are described in the legend to Fig. 4). Knockdown of CK1α mRNA was determined by relative qRT-PCR (bottom). The level of CK1α mRNA was normalized to that of GAPDH mRNA by using the value for the transduction of the empty vector control (mock treatment) and sh NT, which was set at 100%. Values represent means ± SDs of three independent experiments. Asterisks indicate significant differences (***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, nonsignificant).
Vinexin β is involved in CK1α-NS5A enzymatic reaction in vivo.
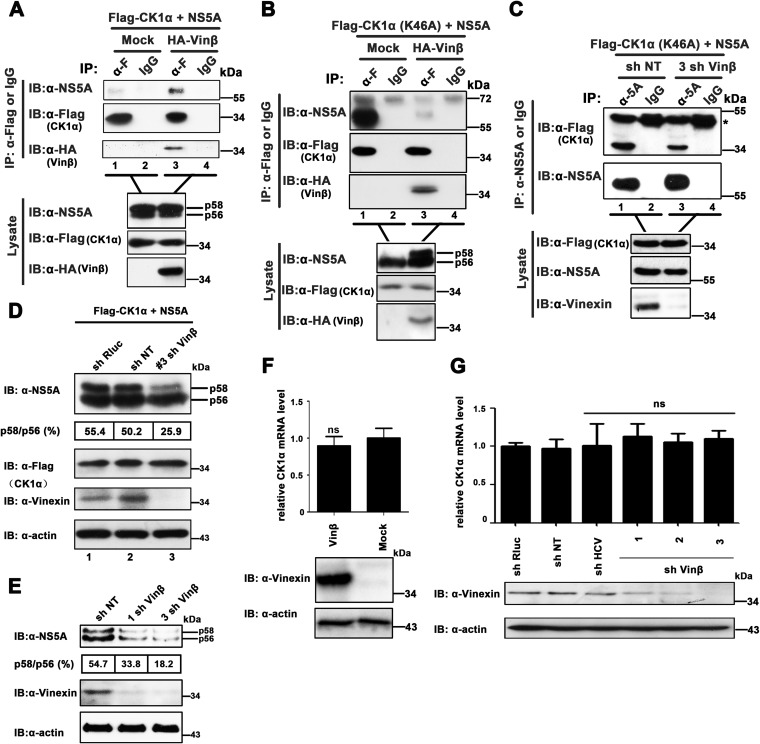
Previous studies have demonstrated that NS5A hyperphosphorylation induced by CK1α regulates the interaction of NS5A with lipid droplets (LDs) and core protein. Moreover, it has been confirmed that the recruitment of hyperphosphorylated NS5A to LDs and the interaction of NS5A with core protein are essential for HCV assembly (10, 11, 13, 14, 59). To further evaluate the contribution of vinexin β to the hyperphosphorylation of NS5A catalyzed by CK1α, we performed co-IP assays to determine the effect of vinexin β on the CK1α-NS5A association. As shown in Fig. 7A, the ability of CK1α to coimmunoprecipitate with NS5A was significantly enhanced in the presence of overexpressed vinexin β (top, lane 3 versus lane 1), while the CK1α and NS5A expression levels in the presence of overexpressed vinexin β were approximately equivalent to those in the absence of overexpressed vinexin β, suggesting that vinexin β promoted the CK1α interaction with NS5A. Since both vinexin β and NS5A were simultaneously coimmunoprecipitated with CK1α (Fig. 7A, lane 3), it was likely that these three proteins could form a complex in vivo. Of note, NS5A was actually hyperphosphorylated by CK1α in mammalian cells irrespective of the presence or absence of exogenous vinexin β (Fig. 7A, top lysate blot). Therefore, we performed a similar co-IP assay with CK1α (K46A), a kinase-inactive mutant form of CK1α, to replace wild-type CK1α (60). To our surprise, we found that hyperphosphorylated NS5A induced by vinexin β decreased the interaction of NS5A with CK1α (K46A) (Fig. 7B, top, lane 1 versus lane 3). It was likely that hyperphosphorylated NS5A catalyzed by CK1α would detach with its direct kinase. On the basis of the results in Fig. 7A and B together, we concluded the following. (i) CK1α and NS5A in a transiently formed enzymatic complex would separate from each other when the catalytic reaction was completed in the absence of vinexin β. In contrast, CK1α and NS5A were still maintained in a complex in the presence of vinexin β (Fig. 1F and 7A and B). (ii) CK1α, vinexin β, and NS5A can form a complex in vivo (Fig. 7A and B), and exogenous expression of vinexin β would connect endogenous CK1α with NS5A for hyperphosphorylation of NS5A (Fig. 7B). To further determine whether complex formation is involved in vinexin β regulation of NS5A hyperphosphorylation and HCV replication, we coexpressed NS5A with CK1α (K46A) in vinexin β knockdown cells. As shown in Fig. 7C, silencing of vinexin β weakened the interaction of CK1α with NS5A (top, lane 1 versus lane 3). In addition, to further explore the critical role of vinexin β in NS5A hyperphosphorylation catalyzed by CK1α, we also coexpressed exogenous NS5A and wild-type CK1α in stably sh NT-, sh Rluc-, and sh vinexin β-transduced Huh7 cells, respectively. We found that the ratio of the hyperphosphorylated NS5A variants (p58/p56) was significantly lower in 3 sh vinexin β cells (25.9%) than in sh NT or sh Rluc cells (50.2 and 55.4%, respectively) (Fig. 7D, top, lane 3 versus lanes 1 and 2). The results indicate that reduction of endogenous vinexin β expression inhibited NS5A hyperphosphorylation mediated by CK1α in vivo. It has been validated that tight control of NS5A phosphorylation is important for the HCV life cycle, and breaking the balance between the hyperphosphorylated variant (p58) and the hypophosphorylated variant (p56) of NS5A is disastrous for HCV propagation (6, 9, 15, 61). Moreover, CK1α has recently been reported to be implicated in NS5A hyperphosphorylation-dependent regulation of HCV virion production (14). On the basis of these studies, we speculated that the modulation of NS5A hyperphosphorylation by vinexin β dependent on CK1α was the reason why HCV failed to reproduce in vinexin β knockdown cells. Therefore, we reconfirmed that the p58/p56 ratio and the total NS5A level were obviously suppressed with the silencing of vinexin β (33.8 and 13.2% in 1 sh Vinβ and 3 sh Vinβ, respectively) compared with the control (54.7% in sh NT) (Fig. 7E) during HCV infection.
FIG 7.
Vinexin β expression regulates the interaction of CK1α with NS5A. (A and B) HEK 293T cells were transfected with pRK-HA-vinexin β (HA-Vinβ), pcDNA3.1-NS5A (JFH1), and pRK-Flag-CK1α or pRK-Flag-CK1α (K46A). At 36 hpt, cell lysates were immunoprecipitated with a mouse anti-Flag MAb (α-F) or control mouse IgG. The immunoprecipitates and the lysates were analyzed with the antibodies indicated. (C) Effect of vinexin β knockdown on the NS5A-CK1α interaction. Huh7 cells stably transduced with sh NT and 3 sh Vin β were cotransfected with pcDNA3.1-NS5A (JFH1) and pRK-Flag-CK1α (K46A) expression plasmids. At 36 hpt, cell lysates were immunoprecipitated with a mouse anti-NS5A MAb or control mouse IgG. The immunoprecipitates and the lysate were analyzed with the antibodies indicated. The IgG heavy chain is denoted by the asterisk. (D) Huh7 cells were transduced with retroviral vector sh NT, sh Rluc, or 3 sh Vinβ and then cotransfected with the pRK-Flag-CK1α and pcDNA3.1-NS5A (JFH1) expression plasmids, respectively. At 36 hpt, cell lysates were separated by 8% SDS-PAGE and immunoblotted (IB) with the antibodies indicated. Quantification of the NS5A p58/p56 ratio was performed by Quantity One software (Bio-Rad). The positions of two differently phosphorylated NS5A variants (p56 and p58) are indicated on the right. (E) Huh7.5.1 cells stably expressing the shRNAs indicated were infected with JFH1 at an MOI of 0.02. At 72 hpi, total cell lysates were resolved by 8% SDS-PAGE and immunoblotted with the antibodies indicated. Quantification of the NS5A p58/p56 ratio was performed by Quantity One software (Bio-Rad). (F and G) Huh7.5.1 cell lines with vinexin β overexpression (F) and knockdown (G) were transduced with the lentiviral vector indicated, and CK1α mRNA expression was determined by relative qRT-PCR. The levels of CK1α mRNA relative to that of GAPDH mRNA were correspondingly normalized by using the values for transduction of the control empty vector or sh NT. Values are means ± SDs of three independent experiments. ns, not significant. Immunoblotting with the antibodies indicated was used to confirm knockdown efficiency.
In addition, considering the role of vinexin β in transcriptional regulation (62), we examined whether overexpression or knockdown of vinexin β would influence the CK1α mRNA level. The results showed that there was no significant difference in the CK1α mRNA level under the conditions of overexpression and downregulation of vinexin β by qRT-PCR assay (Fig. 7F and G, top). Collectively, our data demonstrated that vinexin β regulated NS5A hyperphosphorylation and HCV propagation via its interaction with NS5A and CK1α.
DISCUSSION
In this study, we identified a novel adaptor vinexin β is capable of regulating the hyperphosphorylation of HCV NS5A via its interaction with NS5A in mammalian cells (Fig. 4 and 5). In addition, we observed that two amino acids, W307 and Y325, in vinexin β were critical for the interaction of vinexin β with NS5A (Fig. 3D and 5A). Furthermore, we demonstrated that the PxxPxR motifs located in the Lcs2 region of the C terminus of NS5A were essential for the interaction of NS5A with vinexin β (3F and H). Our results offer a new perspective to understanding the role of host proteins containing SH3 domains interacting with NS5A in viral replication.
HCV NS5A is a proline-rich phosphoprotein (38) that plays a critical role in the control of cellular pathways and participates in various stages of the HCV life cycle through its interaction with many HFs (44, 63). On the one hand, many HFs containing SH3 domains have been reported to interact with NS5A derived from different HCV genotypes through the highly conserved PxxPxR motifs within the SH3 domain. For instance, NS5A binds to tyrosine kinases, including Fyn, Lyn, Hck, and Lck, and regulates their kinase activities, thereby subverting and altering response events of cells to facilitate HCV replication (16); NS5A blocks the cellular apoptosis pathway by binding Bin1 (39) and mixed-lineage kinase 3 (40). Furthermore, the EGF-induced mitogen-activated protein kinase (MAPK) pathway is also perturbed by the interaction of NS5A with Grb2 (38). On the other hand, phosphorylated NS5A has also been shown to play a critical role in the HCV life cycle, especially in maintenance of the balance between RNA replication and viral assembly (9). Thus, investigating the detailed molecular mechanisms by which cellular factors, including some kinases that interact with NS5A and modulate the phosphorylation of NS5A, would be instrumental in the development of antiviral therapy (64, 65). Previous studies have identified various host kinases involved in the regulation of the phosphorylation status of NS5A either directly or indirectly, such as CK1α, CK2α, Plk1, lipid kinase phosphatidylinositol-4 kinase III alpha, glycogen synthase kinase 3, and MAPK (45, 59, 61, 63, 66). Additionally various studies have demonstrated that hyperphosphorylation of NS5A was also modulated by HCV NS proteins (NS3, NS4A, and NS4B) (65, 67). In our study, we have demonstrated that, in addition to the host kinases and viral NS proteins, as an adaptor, vinexin β, can also modulate the phosphorylated variants of NS5A through its interaction with NS5A in vivo (Fig. 2, 4D and H, and 5A to C).
Vinexin β was initially identified as a cytoskeletal protein vinculin binding partner and was a shorter transcriptional variant of vinexin composed of only three SH3 domains (18, 21), acting as a scaffold protein recruiting various signaling and cytoskeletal factors to modulate cell transduction and cytoskeletal rearrangement (48). For instance, vinexin β plays a key role in maintaining EGF receptor phosphorylation on the plasma membrane through the regulation of E3 ubiquitin ligase c-Cbl (68). Another study also indicated that vinexin β regulates the proteasome-dependent degradation of WAVE2 in a protein kinase A-dependent manner (69). To investigate the mechanism of NS5A phosphorylation induced by vinexin β, we examined whether vinexin β might recruit another HF(s) to regulate NS5A phosphorylation. We found that vinexin β specifically interacted with host kinase CK1α through the third SH3 domain of vinexin β without relying upon the two amino acids (W307 and Y325) required for the interaction of vinexin β with NS5A (Fig. 6A and B). However, another casein kinase, CK2α or VAP-A, that was thought to be involved in HCV replication complex formation was not associated with vinexin β in our study (Fig. 6A). Although it has been reported that CK1α is responsible for NS5A hyperphosphorylation (13), it is still possible that another sophisticated regulation process occurs in vivo that mimics the activation of interferon regulatory factor 3 mediated by the adaptor stimulator of interferon genes by recruiting kinase TANK-binding kinase 1 in virus-triggered innate immunity signaling (70, 71). Therefore, we provided further evidence that involvement of vinexin β enhanced the interaction between NS5A and CK1α and that they could form an NS5A-vinexin β-CK1α triple complex in mammalian cells (Fig. 7A). Hence, downregulation of vinexin β expression decreased CK1α-NS5A binding and suppressed NS5A hyperphosphorylation catalyzed by CK1α in human hepatoma cells (Fig. 7C and D). Furthermore, decreased expression of CK1α also inhibited the hyperphosphorylation of NS5A even in the presence of vinexin β expression (Fig. 6C). Taken together, our results suggest that vinexin β serves as a bridge spatially connecting NS5A with CK1α and facilitates a transient kinase-substrate reaction in vivo. Therefore, it is reasonable to conclude that hyperphosphorylated NS5A would detach from its direct kinase, CK1α, when the enzymatic reaction is complete in the absence of vinexin β and still associate with CK1α in the presence of endogenous or exogenous vinexin β, which is ubiquitously expressed in many cell lines, such as in 293T, Huh7, and Huh7.5.1 (Fig. 1F and 7A and B).
It has been demonstrated that there is an inverse relationship between NS5A phosphorylation and the interaction of NS5A with human VAP-A, indicating that hypophosphorylation and hyperphosphorylation of NS5A may play a crucial role in the whole HCV life cycle, one not simply limited to the HCV genome replication process (9, 14, 15). Therefore, it is commonly believed that any events that disrupt the balance of NS5A phosphorylation are harmful to HCV propagation (6, 14, 61). For instance, cell culture-adaptive mutations known to decrease the levels of hyperphosphorylated NS5A prevented the efficient infection of a full-length RNA of HCV in the chimpanzee model (72, 73). We also observed that when endogenous vinexin β expression was downregulated, the interplay between CK1α and NS5A was weakened; thus, the level of NS5A hyperphosphorylated mediated by CK1α was also significantly suppressed (Fig. 7C and D). In contrast, when vinexin β was overexpressed, the state of NS5A hyperphosphorylation was enhanced (Fig. 4H and 5A to C). However, regardless of the knockdown or overexpression of vinexin β, the NS5A phosphorylation (p58/p56) ratio changed and HCV replication was reduced (Fig. 4, 5A to C, and 7E), further suggesting that the normal proportion of hypophosphorylated to hyperphosphorylated NS5A is critical for HCV replication. Of course, we cannot rule out the possibility that the knockdown or overexpression of vinexin β may influence HCV replication in other ways. For instance, previous studies have demonstrated that JNK activation negatively regulates HCV replication. In the meantime, vinexin β could regulate the activation of JNK induced by EGF (21, 46, 47). In addition, it was reported recently that NS5A hyperphosphorylation mediated by CK1α facilitated the interaction of NS5A with the core protein and LDs, which is essential for HCV infectious virus production (14). Therefore, the detailed mechanism of NS5A hyperphosphorylation regulated by vinexin β for viral infectious particle assembly or release and the interplay between NS5A and core protein or LDs needs to be further investigated.
Besides CK1α, it is possible that other kinases can also hyperphosphorylate NS5A by interacting with vinexin β. By using a hyperphosphorylation inhibitor, at least two host kinases (CK1α and Plk1) have been identified that hyperphosphorylate NS5A in vitro and in vivo (59, 61). We also found that vinexin β colocalized with Plk1. Moreover, expression of vinexin β also enhanced the interaction of NS5A with Plk1 (data not shown). Therefore, the critical role of vinexin β and host kinases recruited by vinexin β in cellular events and the viral life cycle needs to be further explored. Additionally, we found that the change in NS5A hyperphosphorylation in the overexpression system was more obvious than that in HCV-infected cells (Fig. 5A and C). Besides much lower transfection efficiency in Huh7.5.1 cells than in HEK 293T cells, it is possible that, in the context of HCV infection, the fluctuation of NS5A phosphorylation is regulated by various HFs and viral elements described previously. Therefore, the physiological contribution of vinexin β to NS5A phosphorylation in the context of HCV infection also remains to be further determined.
In more than 80% of the cases, HCV infection establishes a chronic infection resulting in fibrosis, cirrhosis, and ultimately hepatocellular carcinoma (74). Acquisition of migration properties and reduction of adherence ability are the crucial features of cancer cell invasion and metastasis. Reduced adhesiveness was also observed in HCV-infected cells (75). Furthermore, exogenous expression of NS3 or NS5A significantly inhibited cell adhesion to fibronectin (31). Fibronectin is one of the extracellular matrix proteins associated with the actin cytoskeleton by forming a focal adhesion complex and is an important module in cell migration. Initially, we demonstrated that SoHo family members could interact with NS5A in vivo (Fig. 1C and D). Previous studies showed that the SoHo family is involved in cytoskeleton rearrangement and cell motility, which may explain why HCV infection or NS5A expression leads to weak cell adhesion and promotes cell invasion.
In summary, we have demonstrated that the scaffolding protein vinexin β is a binding partner of NS5A and modulates the phosphorylation state of HCV NS5A in a CK1α-dependent manner. Moreover, vinexin β is also involved in the HCV life cycle via regulation of CK1α-NS5A enzymatic reactions. These findings further deepen our understanding of an unexpected links among the SH3 domain-PxxPxR motif interaction, NS5A phosphorylation, and HCV propagation, which would be beneficial to research and the development of a novel therapeutic target for HCV treatment.
ACKNOWLEDGMENTS
This work was supported by the National Natural Sciences Foundation of China (no. 31370185), the National Basic Research Program of China (no. 2011CB504800), and the National Infrastructure of Natural Resources for Science and Technology Program (no. 2011-572) to C. Zheng.
We thank Guo Jia (George Mason University), Chen Mingzhou (Wuhan University), and John Taylor (Lasell College) for their helpful comments and critical readings of the manuscript.
We are also grateful to Ralf Bartenschlager, Francis V. Chisari, Takaji Wakita, and Huang Zan for generously providing reagents.
REFERENCES
- 1.Paul D, Madan V, Bartenschlager R. 2014. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tellinghuisen TL, Evans MJ, von Hahn T, You S, Rice CM. 2007. Studying hepatitis C virus: making the best of a bad virus. J Virol 81:8853–8867. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 4.Han Q, Manna D, Belton K, Cole R, Konan KV. 2013. Modulation of hepatitis C virus genome encapsidation by nonstructural protein 4B. J Virol 87:7409–7422. doi: 10.1128/JVI.03523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol 82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklan EH, Charuworn P, Pang PS, Glenn JS. 2009. Mechanisms of HCV survival in the host. Nat Rev Gastroenterol Hepatol 6:217–227. doi: 10.1038/nrgastro.2009.32. [DOI] [PubMed] [Google Scholar]
- 8.Reed KE, Gorbalenya AE, Rice CM. 1998. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol 72:6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A 101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 11.Camus G, Herker E, Modi AA, Haas JT, Ramage HR, Farese RV Jr, Ott M. 2013. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem 288:9915–9923. doi: 10.1074/jbc.M112.434910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray CL, Jones CT, Rice CM. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol 6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quintavalle M, Sambucini S, Di Pietro C, De Francesco R, Neddermann P. 2006. The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J Virol 80:11305–11312. doi: 10.1128/JVI.01465-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masaki T, Matsunaga S, Takahashi H, Nakashima K, Kimura Y, Ito M, Matsuda M, Murayama A, Kato T, Hirano H, Endo Y, Lemon SM, Wakita T, Sawasaki T, Suzuki T. 2014. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-alpha in infectious virus production. J Virol 88:7541–7555. doi: 10.1128/JVI.03170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Staschke K, De Francesco R, Tan SL. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9. doi: 10.1016/j.virol.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald A, Mazaleyrat S, McCormick C, Street A, Burgoyne NJ, Jackson RM, Cazeaux V, Shelton H, Saksela K, Harris M. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J Gen Virol 86:1035–1044. doi: 10.1099/vir.0.80734-0. [DOI] [PubMed] [Google Scholar]
- 17.Roignot J, Soubeyran P. 2009. ArgBP2 and the SoHo family of adapter proteins in oncogenic diseases. Cell Adh Migr 3:167–170. doi: 10.4161/cam.3.2.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, Yamada KM, Aota S. 1999. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol 144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama M, Mizusaki H, Shimono A, Mukai T, Okumura K, Abe K, Shimada K, Morohashi K. 2005. A novel isoform of vinexin, vinexin gamma, regulates Sox9 gene expression through activation of MAPK cascade in mouse fetal gonad. Genes Cells 10:421–434. doi: 10.1111/j.1365-2443.2005.00844.x. [DOI] [PubMed] [Google Scholar]
- 20.Mizutani K, Ito H, Iwamoto I, Morishita R, Deguchi T, Nozawa Y, Asano T, Nagata KI. 2007. Essential roles of ERK-mediated phosphorylation of vinexin in cell spreading, migration and anchorage-independent growth. Oncogene 26:7122–7131. doi: 10.1038/sj.onc.1210512. [DOI] [PubMed] [Google Scholar]
- 21.Akamatsu M, Aota S, Suwa A, Ueda K, Amachi T, Yamada KM, Akiyama SK, Kioka N. 1999. Vinexin forms a signaling complex with Sos and modulates epidermal growth factor-induced c-Jun N-terminal kinase/stress-activated protein kinase activities. J Biol Chem 274:35933–35937. doi: 10.1074/jbc.274.50.35933. [DOI] [PubMed] [Google Scholar]
- 22.Pei R, Chen H, Lu L, Zhu W, Beckebaum S, Cicinnati V, Lu M, Chen X. 2011. Hepatitis C virus infection induces the expression of amphiregulin, a factor related to the activation of cellular survival pathways and required for efficient viral assembly. J Gen Virol 92:2237–2248. doi: 10.1099/vir.0.032581-0. [DOI] [PubMed] [Google Scholar]
- 23.Yan R, Zhao Z, He Y, Wu L, Cai D, Hong W, Wu Y, Cao Z, Zheng C, Li W. 2011. A new natural alpha-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides 32:11–19. doi: 10.1016/j.peptides.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Yan R, Xu G, Li W, Zheng C. 2009. Construction of the Vero cell culture system that can produce infectious HCV particles. Mol Biol Rep 36:111–120. doi: 10.1007/s11033-007-9158-3. [DOI] [PubMed] [Google Scholar]
- 26.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu G, Xin X, Zheng C. 2013. GPS2 is required for the association of NS5A with VAP-A and hepatitis C virus replication. PLoS One 8:e78195. doi: 10.1371/journal.pone.0078195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei B, Feng N, Zhou F, Lu C, Su J, Hua L. 2010. Construction and identification of recombinant lentiviral vector containing HIV-1 Tat gene and its expression in 293T cells. J Biomed Res 24:58–63. doi: 10.1016/S1674-8301(10)60009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macdonald A, Crowder K, Street A, McCormick C, Harris M. 2004. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J Gen Virol 85:721–729. doi: 10.1099/vir.0.19691-0. [DOI] [PubMed] [Google Scholar]
- 30.Lim WA, Richards FM, Fox RO. 1994. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 31.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, Andre P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol Syst Biol 4:230. doi: 10.1038/msb.2008.66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Golemis EA, Kruh GD. 1997. ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J Biol Chem 272:17542–17550. doi: 10.1074/jbc.272.28.17542. [DOI] [PubMed] [Google Scholar]
- 33.Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. 1998. A novel, multifunctional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol 18:872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fridell RA, Valera L, Qiu D, Kirk MJ, Wang C, Gao M. 2013. Intragenic complementation of hepatitis C virus NS5A RNA replication-defective alleles. J Virol 87:2320–2329. doi: 10.1128/JVI.02861-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suwa A, Mitsushima M, Ito T, Akamatsu M, Ueda K, Amachi T, Kioka N. 2002. Vinexin beta regulates the anchorage dependence of ERK2 activation stimulated by epidermal growth factor. J Biol Chem 277:13053–13058. doi: 10.1074/jbc.M108644200. [DOI] [PubMed] [Google Scholar]
- 36.Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes M, Gretton S, Shelton H, Brown DD, McCormick CJ, Angus AG, Patel AH, Griffin S, Harris M. 2009. A conserved proline between domains II and III of hepatitis C virus NS5A influences both RNA replication and virus assembly. J Virol 83:10788–10796. doi: 10.1128/JVI.02406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan SL, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs BL, Mayer BJ, Katze MG. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci U S A 96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanda SK, Herion D, Liang TJ. 2006. The SH3 binding motif of HCV NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology 130:794–809. doi: 10.1053/j.gastro.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 40.Amako Y, Igloi Z, Mankouri J, Kazlauskas A, Saksela K, Dallas M, Peers C, Harris M. 2013. Hepatitis C virus NS5A inhibits mixed lineage kinase 3 to block apoptosis. J Biol Chem 288:24753–24763. doi: 10.1074/jbc.M113.491985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo M, Pei R, Yang Q, Cao H, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Lu M, Chen X. 2015. Phosphatidylserine-specific phospholipase A1 is involved in hepatitis C virus assembly through NS2 complex formation. J Virol 89:2367–2377. doi: 10.1128/JVI.02982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Jiang H, Qu L, Yao W, Cai H, Chen L, Peng T. 2014. Hepatocyte nuclear factor 4alpha and downstream secreted phospholipase A2 GXIIB regulate production of infectious hepatitis C virus. J Virol 88:612–627. doi: 10.1128/JVI.02068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96(Pt 4):727–738. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 45.Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. 2013. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog 9:e1003359. doi: 10.1371/journal.ppat.1003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Ishida H, Yamane D, Yi M, Swinney DC, Foung S, Lemon SM. 2013. Contrasting roles of mitogen-activated protein kinases in cellular entry and replication of hepatitis C virus: MKNK1 facilitates cell entry. J Virol 87:4214–4224. doi: 10.1128/JVI.00954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rau SJ, Hildt E, Himmelsbach K, Thimme R, Wakita T, Blum HE, Fischer R. 2013. CD40 inhibits replication of hepatitis C virus in primary human hepatocytes by c-Jun N terminal kinase activation independent from the interferon pathway. Hepatology 57:23–36. doi: 10.1002/hep.25966. [DOI] [PubMed] [Google Scholar]
- 48.Kioka N, Ueda K, Amachi T. 2002. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct 27:1–7. doi: 10.1247/csf.27.1. [DOI] [PubMed] [Google Scholar]
- 49.Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. 2012. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J Virol 86:4139–4150. doi: 10.1128/JVI.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog 5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C, Gale M Jr, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell 18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J 25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taguwa S, Okamoto T, Abe T, Mori Y, Suzuki T, Moriishi K, Matsuura Y. 2008. Human butyrate-induced transcript 1 interacts with hepatitis C virus NS5A and regulates viral replication. J Virol 82:2631–2641. doi: 10.1128/JVI.02153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim YS, Shin KS, Oh SH, Kang SM, Won SJ, Hwang SB. 2012. Nonstructural 5A protein of hepatitis C virus regulates heat shock protein 72 for its own propagation. J Viral Hepat 19:353–363. doi: 10.1111/j.1365-2893.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- 55.Lim YS, Tran HT, Park SJ, Yim SA, Hwang SB. 2011. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J Virol 85:8777–8788. doi: 10.1128/JVI.02533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM. 2012. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol 86:1739–1749. doi: 10.1128/JVI.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T, Moriishi K, Matsuura Y. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol 79:13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 59.Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, Neddermann P. 2007. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem 282:5536–5544. doi: 10.1074/jbc.M610486200. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Sun X, Wu J, Xu BE, Gu C, Wang H, Wang X, Tan F, Peng X, Qiang B, Yuan J, Luo Y. 2008. Casein kinase 1alpha interacts with RIP1 and regulates NF-kappaB activation. Biochemistry 47:441–448. doi: 10.1021/bi7010515. [DOI] [PubMed] [Google Scholar]
- 61.Chen YC, Su WC, Huang JY, Chao TC, Jeng KS, Machida K, Lai MM. 2010. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J Virol 84:7983–7993. doi: 10.1128/JVI.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bour G, Plassat JL, Bauer A, Lalevee S, Rochette-Egly C. 2005. Vinexin beta interacts with the non-phosphorylated AF-1 domain of retinoid receptor gamma (RARgamma) and represses RARgamma-mediated transcription. J Biol Chem 280:17027–17037. doi: 10.1074/jbc.M501344200. [DOI] [PubMed] [Google Scholar]
- 63.Macdonald A, Harris M. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol 85:2485–2502. doi: 10.1099/vir.0.80204-0. [DOI] [PubMed] [Google Scholar]
- 64.Lemay KL, Treadaway J, Angulo I, Tellinghuisen TL. 2013. A hepatitis C virus NS5A phosphorylation site that regulates RNA replication. J Virol 87:1255–1260. doi: 10.1128/JVI.02154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross-Thriepland D, Harris M. 2014. Insights into the complexity and functionality of hepatitis C virus NS5A phosphorylation. J Virol 88:1421–1432. doi: 10.1128/JVI.03017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Street A, Macdonald A, McCormick C, Harris M. 2005. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol 79:5006–5016. doi: 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koch JO, Bartenschlager R. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol 73:7138–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitsushima M, Ueda K, Kioka N. 2006. Vinexin beta regulates the phosphorylation of epidermal growth factor receptor on the cell surface. Genes Cells 11:971–982. doi: 10.1111/j.1365-2443.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 69.Mitsushima M, Sezaki T, Akahane R, Ueda K, Suetsugu S, Takenawa T, Kioka N. 2006. Protein kinase A-dependent increase in WAVE2 expression induced by the focal adhesion protein vinexin. Genes Cells 11:281–292. doi: 10.1111/j.1365-2443.2006.00932.x. [DOI] [PubMed] [Google Scholar]
- 70.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 71.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A 101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 73.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A 99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas DL. 2013. Global control of hepatitis C: where challenge meets opportunity. Nat Med 19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alisi A, Arciello M, Petrini S, Conti B, Missale G, Balsano C. 2012. Focal adhesion kinase (FAK) mediates the induction of pro-oncogenic and fibrogenic phenotypes in hepatitis C virus (HCV)-infected cells. PLoS One 7:e44147. doi: 10.1371/journal.pone.0044147. [DOI] [PMC free article] [PubMed] [Google Scholar]