Abstract
CD8+ T cells are the main effector lymphocytes in the control of hepatitis B virus (HBV) infection. However, limitations of model systems, such as low infection rates, restrict mechanistic studies of HBV-specific CD8+ T cells. Here, we established a novel immunological cell culture model based on HBV-infected HepG2hNTCP cells that endogenously processed viral antigens and presented them to HBV-specific CD8+ T cells. This induced cytolytic and noncytolytic CD8+ T-cell effector functions and reduction of viral loads.
TEXT
Hepatitis B virus (HBV) is a noncytopathic, hepatotropic virus causing acute and chronic necroinflammatory liver disease, which may progress to hepatocellular carcinoma (HCC). It is generally assumed that the pathogenesis of HBV infection is determined by virus-host interactions mediated by the immune system, specifically by virus-specific CD8+ T cells (1–5). Analysis of T-cell antiviral efficacy has been constrained by the limitations of available model systems. Furthermore, hepatoma cells used for in vitro studies were not susceptible to HBV infection except for low-permissive HepaRG cells (6). Recently, human sodium taurocholate-cotransporting polypeptide (hNTCP) was identified as the entry receptor of HBV. Nonpermissive HepG2 cells become susceptible to the virus after hNTCP transduction (7, 8), allowing high infection rates. Here, we used HLA-A*02+ HepG2hNTCP cells to establish a novel immunological cell culture model for HBV infection. In coculture assays, we could analyze antiviral effector functions of HLA-A*02-restricted HBV-specific CD8+ T cells and determine immunological mechanisms of HBV control.
Seven HLA-A*02+ patients with chronic HBV infection presenting at the outpatient hepatology clinic of the University Hospital Freiburg were included in the study after written informed consent was obtained from the patients and approval was given by the ethics committee of the University of Freiburg, Germany. All investigations were conducted according to the principles expressed in the Declaration of Helsinki.
HBV-infected HepG2hNTCP cells induce effector functions in virus-specific CD8+ T cells.
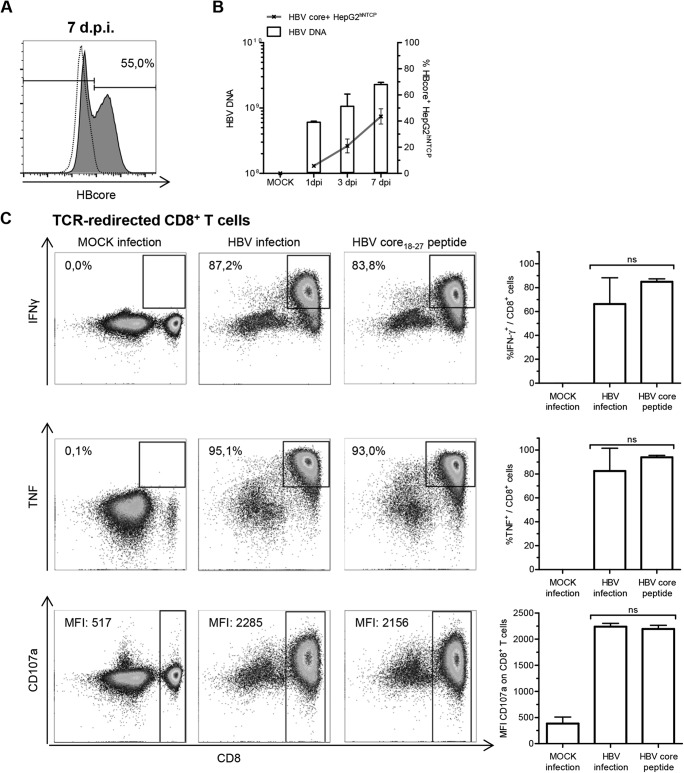
HepG2 cells stably transduced with hNTCP were infected with HBV, genotype D, purified from cell culture supernatant as previously described (7). The efficiency of infection, i.e., HBV protein content on a single-cell basis, was analyzed by flow cytometry using an anti-HBV core antibody (Fig. 1A). Until day 7 postinfection, the frequency of HBV-infected HepG2hNTCP cells and viral loads detected by quantitative PCR (qPCR) (9) increased (Fig. 1B). Subsequently, HBV-infected HepG2hNTCP cells were analyzed for their capacity to induce effector functions in HLA-matched CD8+ T cells from healthy donors retrovirally transduced with a HBV core18–27-specific T-cell receptor (TCR) (10). Indeed, coculture of these TCR-redirected T cells with HBV-infected HepG2hNTCP cells overnight led to a strong production of gamma interferon (IFN-γ) and tumor necrosis factor (TNF) and induced CD107a surface expression/degranulation (Fig. 1C), comparable to peptide stimulation. In sum, these results indicate that viral antigens were efficiently synthesized, endogenously processed, and presented on major histocompatibility complex (MHC) class I molecules, leading to the induction of effector functions in HBV core18–27-specific CD8+ T cells.
FIG 1.
Induction of CD8+ T-cell responses through HBV-infected HepG2hNTCP cells. (A) HBV-infected HepG2hNTCP cells were analyzed for endogenous expression of HBV core antigen (clone 13A9; Thermo Fisher) by flow cytometry 7 days postinfection (dpi). The frequency of HBV core+ HepG2hNTCP cells is indicated in the representative plot. The dotted line indicates mock-infected HepG2hNTCP cells; the solid line indicates HBV-infected HepG2hNTCP cells. (B) Cytoplasmic viral loads on bulk HepG2hNTCP cells were assessed by qPCR at the indicated time points. Frequencies of HBV core+ HepG2hNTCP cells were obtained by flow cytometry (n = 3). (C) TCR-redirected CD8+ T cells specific for HBV-core18–27/HLA-A*0201 were incubated with mock-infected or HBV-infected HepG2hNTCP cells (7 dpi) or with 15 μg/ml HBV core18–27 peptide (FLPSDFFPSV) overnight. Induction of IFN-γ (clone 25723.11; BD Biosciences) and TNF (clone Mab11; BD Biosciences) production and degranulation (CD107a, clone H4A3; BD Biosciences) was analyzed by flow cytometry. (Left) Representative dot plots. Frequencies were calculated as percentages of responding CD8+ T cells in bulk CD8+ T cells. Median fluorescence intensities (MFIs) are those of CD107a on CD8+ T cells. (Right) Statistical graphs of 6 independent assays. One-way analysis of variance (ANOVA) with a subsequent Tukey post hoc test was performed for statistical analysis. Means and standard deviations (SD) are depicted. ns, not significant.
HBV core18–27-specific CD8+ T cells significantly reduce viral loads in HBV-infected HepG2hNTCP cells.
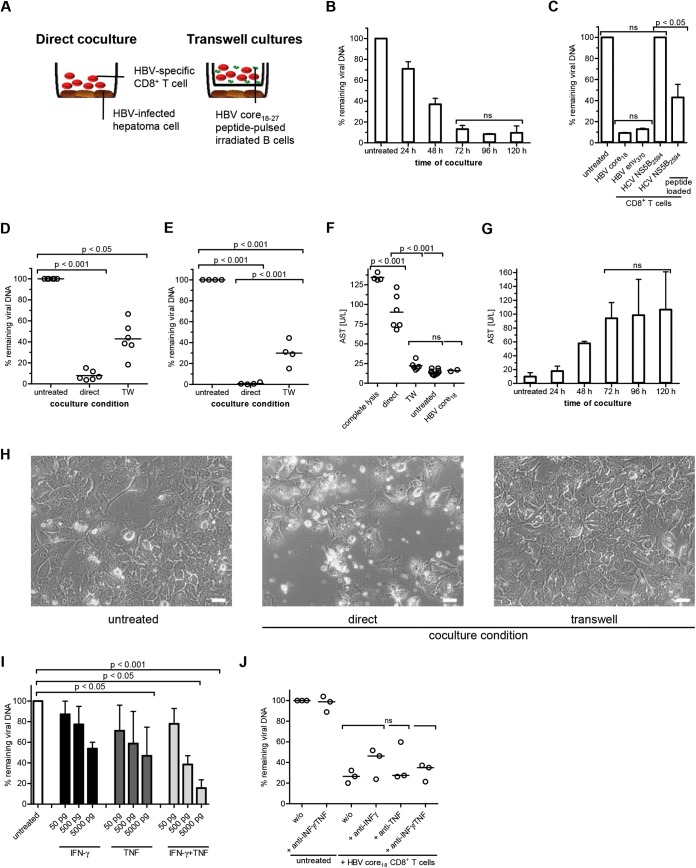
Next, we wanted to quantify the antiviral efficacy of TCR-redirected CD8+ T cells. CD8+ T cells were directly cocultured with HBV-infected HepG2hNTCP cells at an effector-to-target cell (E:T) ratio of 1:1 (Fig. 2A). Viral loads decreased to minimal levels after 72 to 96 h (Fig. 2B). Cocultures with CD8+ T cells specific for two HBV epitopes (HBV core18–27 and HBV env370–379 [10]) confirmed that different viral antigens were presented by HepG2hNTCP cells and led to reduction of viral loads. The absence of antiviral efficacy after incubation of HBV-infected HepG2hNTCP with a HCV NS5B2594–2602-specific CD8+ T-cell clone (11) revealed the specificity of this effect (Fig. 2C). It is well known from cell culture and animal models that viral control requires cytolytic (12, 13) and noncytolytic CD8+ T-cell effector mechanisms (14–16). The antiviral efficacy of both effector functions was assessed by cocultivating HBV core18–27-specific CD8+ T cells in direct contact with their target cells or separated by a semipermeable membrane using Corning Transwell plates (Fig. 2A and D). TCR-redirected CD8+ T cells were stimulated with an equal number of irradiated EBV-transformed B cells pulsed with HBV core18–27 peptide in Transwell cocultures. Importantly, cytoplasmic viral titers were significantly reduced under both coculture conditions. However, HBV DNA was more profoundly diminished in HepG2hNTCP cells from direct cocultures (>90%) than in cells from cocultures made with Transwell plates (>50%) (Fig. 2D). Interestingly, antiviral efficacy of CD8+ T cells is comparably induced in response to HepG2.117 hepatoma cells (17) which were stably transduced with the HBV genome, genotype D (Fig. 2E). These data demonstrate that CD8+ T-cell-mediated HBV control in hepatoma cells is not altered by HBV infection. Moreover, antiviral efficacy of HBV-specific CD8+ T cells is dominated by cell contact-dependent mechanisms in both cell culture systems. The analysis of transaminase levels further revealed that cell killing is required for efficient eradication of HBV, since strong increases in aspartate aminotransferase (AST) levels were observed in direct infectious cocultures (Fig. 2F). Of note, AST was derived only from HepG2hNTCP cells. Transaminase concentrations increased over time, peaking between 72 h and 120 h inversely to viral load (Fig. 2B and G). Histological signs of cytotoxicity, e.g., a destroyed hepatoma cell monolayer and the appearance of cell debris, could also be monitored microscopically (Fig. 2H). Importantly, HBV-infected HepG2hNTCP cells were susceptible to cytokine-mediated antiviral effects (Fig. 2I). To elucidate the importance of the antiviral cytokines IFN-γ and TNF, we performed neutralization assays in direct cocultures. Blocking cytokine activity in this setup had no significant impact on the antiviral efficacy of HBV-specific CD8+ T-cell effector functions (Fig. 2J). In summary, this HepG2hNTCP-based infectious coculture model requires cell killing to eradicate HBV from infected cells, which is in line with data obtained from studies in acutely HBV-infected patients and chimpanzees that reported associations between vigorous HBV-specific CD8+ T-cell responses, elevated ALT levels, and viral clearance (13, 18, 19). However, we could also show significant reductions in viral titers by noncytolytic effector mechanisms (Fig. 2D and E), supporting the hypotheses of several in vitro and in vivo studies (14–16).
FIG 2.
Antiviral efficacy of TCR-redirected CD8+ T cells. HBV-infected HepG2hNTCP cells (7 dpi) were cocultured with TCR-redirected CD8+ T cells specific for HBV core18–27 for 96 h unless otherwise indicated. Cytoplasmic viral loads were determined by qPCR. The amount of HBV DNA remaining was calculated relative to that in untreated samples (without CD8+ T cells). All coculture assays were performed using 250,000 HepG2hNTCP cells. The E:T ratio was 1:1. (A) Schematic experimental setup. (B) HBV DNA was determined at the indicated time points (n = 2). (C) Coculture of CD8+ T cells of indicated specificity with HBV-infected HepG2hNTCP cells. CD8+ T cells from healthy donors were transduced with HBV-specific TCRs (HBV core18–27 and HBV env370–379) (10). HCV-specific CD8+ T-cell clones were generated from patients chronically infected with HCV as described before (11). HCV NS5B2594–2602-specific CD8+ T-cell clones were tested against HBV-infected HepG2hNTCP cells labeled with the corresponding HCV peptide prior to coculture in order to control functionality (n = 2). (D and E) Coculture of TCR-redirected CD8+ T cells specific for HBV core18–27 under direct and indirect (Transwell [TW]) coculture conditions (n = 6) using HBV-infected HepG2hNTCP cells (D) or tetracycline-regulated HBV-producing HepG2.117 cells (E) (n = 4). (F) Comparison of AST levels obtained from 6 infectious coculture assays versus complete lysis using lysis buffer. (G) AST levels corresponding to the viral loads in panel B were determined after coculture with HBV core18–27-specific TCR-redirected CD8+ T cells at the indicated time points. (H) Bright-field microscopic analyses of HBV-infected HepG2hNTCP cells and TCR-redirected CD8+ T cells specific for HBV core18–27 after 96 h coculture. Magnification, ×20; bars, 50 μm. (I) Antiviral efficacy of exogenous recombinant cytokines IFN-γ and TNF (R&D Systems) (n = 3). (J) Cytokine activity of IFN-γ and TNF was blocked by adding neutralizing monoclonal antibodies (MAb) against cytokines (10 μg/ml; anti-human IFN-γ MAb, clone 25723 [R&D Systems]; anti-human TNF MAb, clone 28401 [R&D Systems]) in combination with blocking MAb against cytokine receptors (10 μg/ml; anti-human IFN-γR1/CD119 MAb, clone GIR208 [R&D Systems]; anti-human TNFR1/TNFRSF1A, clone 18605 [R&D Systems]) (n = 3). One-way ANOVA with a subsequent Tukey post hoc test was performed for statistical analysis. ns, not significant. Individual values and means with SD are depicted.
Patient-derived CD8+ T cells reduce viral loads after peptide-specific expansion.
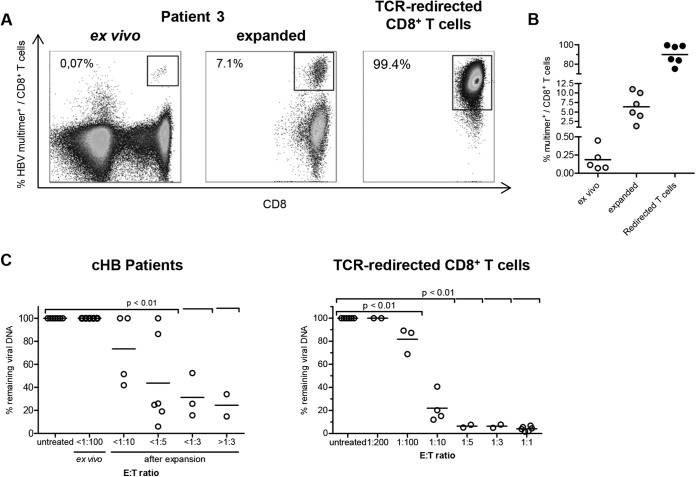
Engineered TCR-redirected CD8+ T cells might respond differently than CD8+ T cells from chronically HBV-infected patients, which are detectable only at very low frequencies ex vivo (Fig. 3A and B; Table 1). In a final set of experiments, we therefore analyzed the antiviral capacity of patient-derived CD8+ T cells directly ex vivo and after peptide-specific expansion in coculture assays with HBV-infected HepG2hNTCP cells. These results revealed that detection of antiviral efficacy requires an E:T ratio of at least 1:100. We could not obtain these critical numbers of virus-specific CD8+ T cells from chronically HBV-infected patients by ex vivo isolation, but we were able to do so after peptide-specific expansion (Fig. 3C). Indeed, we observed an E:T ratio-dependent reduction of viral loads by expanded HBV-specific CD8+ T cells and TCR-redirected CD8+ T cells, respectively (Fig. 3C).
FIG 3.
Antiviral efficacy of HBV-specific CD8+ T cells from patients chronically infected with HBV. (A) Frequencies of TCR-redirected CD8+ T cells specific for HBV core18–27 and of patient-derived HLA-A*02-restricted HBV-specific CD8+ T cells were determined by multimer staining. Stainings of patient samples were performed directly ex vivo and after 14 days of peptide expansion. Frequencies of multimer-positive CD8+ T cells in bulk CD8+ T cells are indicated in representative dot plots. (B) Statistical graph of HBV multimer-positive CD8+ T cells of total CD8+ T cells ex vivo, expanded CD8+ T cells, and HBV core18–27-specific TCR-redirected CD8+ T cells. (C) (Left) CD8+ T cells of 7 patients with chronic HBV infection (cHB) were magnetically isolated by CD8 MicroBeads (Miltenyi Biotec) and subsequently cocultivated with 100,000 HBV-infected HepG2hNTCP cells (7 dpi) at the indicated E:T ratios for 96 h. (Right) E:T ratios of cocultures with patient-derived CD8+ T cells were recapitulated by titration of TCR-redirected CD8+ T cells specific for HBV core18–27. One-way ANOVA with a subsequent Tukey post hoc test was performed for statistical analysis. Individual values and means are depicted.
TABLE 1.
Characteristics of patients included in the studya
| Patient ID | Sex | Age (yr) | HBV genotype | HBV epitope | % multimer positive ex vivo | HBeAg | Viral load (IU/ml) | ALT (U/liter) |
|---|---|---|---|---|---|---|---|---|
| Pat-1 | F | 63 | D | Env183 | 0.45 | Pos | 19,729 | 1,601 |
| Env335 | 0.20 | |||||||
| Pat-2 | M | 57 | A | Core18 | 0.22 | ND | 1,711 | 40 |
| Pat-3 | F | 46 | NS | Core18 | 0.07 | Neg | <34 | 8 |
| Pat-4 | F | 26 | D | Core18 | 0.08 | Neg | 1,822 | 18 |
| Pat-5 | F | 25 | NS | Core18 | 0.11 | Neg | 117 | 20 |
| Pat-6 | M | 68 | A | Pol455 | Pos | <34 | 24 | |
| Pat-7 | M | 25 | NS | Pol455 | Pos | <34 | 24 |
HLA-A*02+ patients with chronic HBV infection presenting at the outpatient hepatology clinic of the University Hospital Freiburg. NS, HBV genotype could not be sequenced; ND, not determined.
In conclusion, we were able to establish an immunological cell culture model of HBV infection based on HepG2hNTCP cells and thereby provide a new tool to study cytolytic and noncytolytic antiviral efficacies of HBV-specific CD8+ T cells.
ACKNOWLEDGMENTS
Recombinant human IL-2 was kindly provided by the NIH AIDS reagent program.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) research consortium FOR1202/UR72/2-5 (TP2 [R.T.], TP5 [S.U.], and TP6 [M.N.]), by the Bundesministerium für Bildung und Forschung (BMBF), and in part by the Excellence Initiative of the German Federal Ministry of Education and Research (GSC-4, Spemann Graduate School) (R.T.).
We have no conflicts of interest to declare.
REFERENCES
- 1.Bertoletti A, Ferrari C. 2012. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut 61:1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 2.Park SH, Rehermann B. 2014. Immune responses to HCV and other hepatitis viruses. Immunity 40:13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Protzer U, Maini MK, Knolle PA. 2012. Living in the liver: hepatic infections. Nat Rev Immunol 12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 4.Schuch A, Hoh A, Thimme R. 2014. The role of natural killer cells and CD8(+) T cells in hepatitis B virus infection. Front Immunol 5:258. doi: 10.3389/fimmu.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieland SF, Chisari FV. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol 79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A 99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS. 2005. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods 126:207–213. doi: 10.1016/j.jviromet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Gehring AJ, Xue SA, Ho ZZ, Teoh D, Ruedl C, Chia A, Koh S, Lim SG, Maini MK, Stauss H, Bertoletti A. 2011. Engineering virus-specific T cells that target HBV infected hepatocytes and hepatocellular carcinoma cell lines. J Hepatol 55:103–110. doi: 10.1016/j.jhep.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 11.Jo J, Aichele U, Kersting N, Klein R, Aichele P, Bisse E, Sewell AK, Blum HE, Bartenschlager R, Lohmann V, Thimme R. 2009. Analysis of CD8+ T-cell-mediated inhibition of hepatitis C virus replication using a novel immunological model. Gastroenterology 136:1391–1401. doi: 10.1053/j.gastro.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 12.Ando K, Guidotti LG, Wirth S, Ishikawa T, Missale G, Moriyama T, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. 1994. Class I-restricted cytotoxic T lymphocytes are directly cytopathic for their target cells in vivo. J Immunol 152:3245–3253. [PubMed] [Google Scholar]
- 13.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. 2003. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol 77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25–36. doi: 10.1016/S1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 15.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 16.Phillips S, Chokshi S, Riva A, Evans A, Williams R, Naoumov NV. 2010. CD8(+) T cell control of hepatitis B virus replication: direct comparison between cytolytic and noncytolytic functions. J Immunol 184:287–295. doi: 10.4049/jimmunol.0902761. [DOI] [PubMed] [Google Scholar]
- 17.Sun D, Nassal M. 2006. Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J Hepatol 45:636–645. doi: 10.1016/j.jhep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Boettler T, Panther E, Bengsch B, Nazarova N, Spangenberg HC, Blum HE, Thimme R. 2006. Expression of the interleukin-7 receptor alpha chain (CD127) on virus-specific CD8+ T cells identifies functionally and phenotypically defined memory T cells during acute resolving hepatitis B virus infection. J Virol 80:3532–3540. doi: 10.1128/JVI.80.7.3532-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maini MK, Boni C, Ogg GS, King AS, Reignat S, Lee CK, Larrubia JR, Webster GJ, McMichael AJ, Ferrari C, Williams R, Vergani D, Bertoletti A. 1999. Direct ex vivo analysis of hepatitis B virus-specific CD8(+) T cells associated with the control of infection. Gastroenterology 117:1386–1396. doi: 10.1016/S0016-5085(99)70289-1. [DOI] [PubMed] [Google Scholar]