ABSTRACT
Rift Valley fever (RVF) is a mosquito-borne zoonotic disease endemic to Africa and characterized by a high rate of abortion in ruminants and hemorrhagic fever, encephalitis, or blindness in humans. RVF is caused by Rift Valley fever virus (RVFV; family Bunyaviridae, genus Phlebovirus), which has a tripartite negative-stranded RNA genome (consisting of the S, M, and L segments). Further spread of RVF into countries where the disease is not endemic may affect the economy and public health, and vaccination is an effective approach to prevent the spread of RVFV. A live-attenuated MP-12 vaccine is one of the best-characterized RVF vaccines for safety and efficacy and is currently conditionally licensed for use for veterinary purposes in the United States. Meanwhile, as of 2015, no other RVF vaccine has been conditionally or fully licensed for use in the United States. The MP-12 strain is derived from wild-type pathogenic strain ZH548, and its genome encodes 23 mutations in the three genome segments. However, the mechanism of MP-12 attenuation remains unknown. We characterized the attenuation of wild-type pathogenic strain ZH501 carrying a mutation(s) of the MP-12 S, M, or L segment in a mouse model. Our results indicated that MP-12 is attenuated by the mutations in the S, M, and L segments, while the mutations in the M and L segments confer stronger attenuation than those in the S segment. We identified a combination of 3 amino acid changes, Y259H (Gn), R1182G (Gc), and R1029K (L), that was sufficient to attenuate ZH501. However, strain MP-12 with reversion mutations at those 3 sites was still highly attenuated. Our results indicate that MP-12 attenuation is supported by a combination of multiple partial attenuation mutations and a single reversion mutation is less likely to cause a reversion to virulence of the MP-12 vaccine.
IMPORTANCE Rift Valley fever (RVF) is a mosquito-transmitted viral disease that is endemic to Africa and that has the potential to spread into other countries. Vaccination is considered an effective way to prevent the disease, and the only available veterinary RVF vaccine in the United States is a live-attenuated MP-12 vaccine, which is conditionally licensed. Strain MP-12 is different from its parental pathogenic RVFV strain, strain ZH548, because of the presence of 23 mutations. This study determined the role of individual mutations in the attenuation of the MP-12 strain. We found that full attenuation of MP-12 occurs by a combination of multiple mutations. Our findings indicate that a single reversion mutation will less likely cause a major reversion to virulence of the MP-12 vaccine.
INTRODUCTION
Rift Valley fever (RVF) is a mosquito-borne zoonotic disease endemic to sub-Saharan Africa (1, 2). Rift Valley fever virus (RVFV) belongs to the genus Phlebovirus, family Bunyaviridae, and the genome is comprised of tripartite negative-stranded RNA consisting of the S, M, and L segments (3). In areas of endemicity, RVFV is considered to be maintained by transovarial transmission among floodwater species of mosquitoes, such as Aedes vexans (4). An increase in mosquito populations through environmental changes that sustain conditions for favorable breeding of mosquitoes, such as heavy rainfall for long periods or dam construction, triggers hatching of infected mosquitoes and leads to the transmission of RVFV into susceptible animal species (5–7). Sheep, goats, and cattle are the most susceptible animal hosts affected during RVF outbreaks (1). RVF causes lethal hepatitis in newborn lambs and high rates of abortion and fetal malformation in pregnant sheep, cattle, and goats. Humans become infected with RVFV through close contact with aerosols of body fluids from infected animals or from bites of infected mosquitoes (1, 2, 8). Most RVF patients develop a biphasic febrile illness, while in some patients the disease progresses into more severe forms of disease, i.e., hemorrhagic fever, encephalitis, or blindness (9). Since the first reported outbreak of RVF in Kenya in 1930 (10), RVFV has been identified in most countries in sub-Saharan Africa and has further spread into Madagascar, Egypt, Saudi Arabia, and Yemen (11–13). The introduction of RVFV into countries where the disease is not endemic can have a negative impact on the agricultural industry. Therefore, an effective countermeasure to prevent the further spread of RVFV is important (14). RVFV is classified as a category A priority pathogen by the National Institutes of Health (NIH) in the United States and an overlap select agent by the U.S. Department of Health and Human Services (HHS) and the U.S. Department of Agriculture (USDA) (15–17).
Vaccination is considered an effective strategy to prevent the spread of RVFV. In countries where the disease is endemic, the live-attenuated Smithburn vaccine has been used since the 1950s (18, 19). Due to residual virulence, the Smithburn vaccine is not used on pregnant animals or humans, and it cannot be used outside countries where the disease is endemic. In the United States, a formalin-inactivated RVF vaccine (TSI-GSD-200) has been developed from the pathogenic wild-type Entebbe strain (20–22). Later, the live-attenuated MP-12 strain was generated from pathogenic wild-type strain ZH548, which was derived from a febrile RVF patient from the 1977-1978 RVF outbreak in Egypt (23, 24). Master seed and vaccine lots (25) of the MP-12 strain have been generated, and their safety and efficacy have been evaluated in ruminants (26–29) and nonhuman primates (30–32). Currently, the MP-12 vaccine is conditionally licensed for use for veterinary purposes and was also tested for human use in a phase II clinical trial (33). Though MP-12 is highly immunogenic in ruminants, there is a lack of knowledge about the mechanism of MP-12 attenuation. Thus, it has been difficult to demonstrate whether a reversion to virulence, e.g., whether it causes abortion in vaccinated animals, occurs in MP-12. To better understand the potential risk of MP-12 (34, 35) and to control the quality of the master seed and vaccine lots of the MP-12 vaccine, it is important to understand the attenuation mechanism.
The RVFV genome encodes 7 proteins; i.e., the S segment carries the genes for the nucleoprotein (N) and NSs; the M segment carries the genes for the 78-kDa protein, NSm, Gn, and Gc; and the L segment carries the gene for the RNA-dependent RNA polymerase (L) (Fig. 1A). Viral genomic RNA is encapsidated with N proteins, and both N and L proteins are essential for mRNA transcription and genomic RNA replication (36–38). Gn and Gc are envelope glycoproteins (39–41) which interact with the viral ribonucleocapsid at the Golgi apparatus (42, 43) and trigger virion formation. The glycoprotein ectodomain(s) attaches to the cellular receptors, e.g., DC-SIGN (44), of target cells, which leads to exposure of the fusion domain of Gc through a caveola-mediated endocytosis pathway (45). A major virulence factor, NSs, is a nonstructural protein dispensable for viral replication (46, 47) and plays a major role in counteracting the innate immunity of the host, e.g., by (i) shutting off of host cell transcription by interacting with TFIIH p44 (48) and by promoting the degradation of TFIIH p62 (49, 50), (ii) specific inhibition of the beta interferon (IFN-β) promoter (51, 52), and (iii) posttranslational degradation of double-stranded RNA-dependent protein kinase (PKR) (53, 54). Another nonstructural protein, NSm, has a role in delaying apoptosis in mammalian cells (55, 56), while it is involved in the efficient transmission of virus in mosquitoes (57, 58). The 78-kDa protein is a structural protein for virions derived from mosquito C6/36 cells but not for those derived from VeroE6 cells (3, 59).
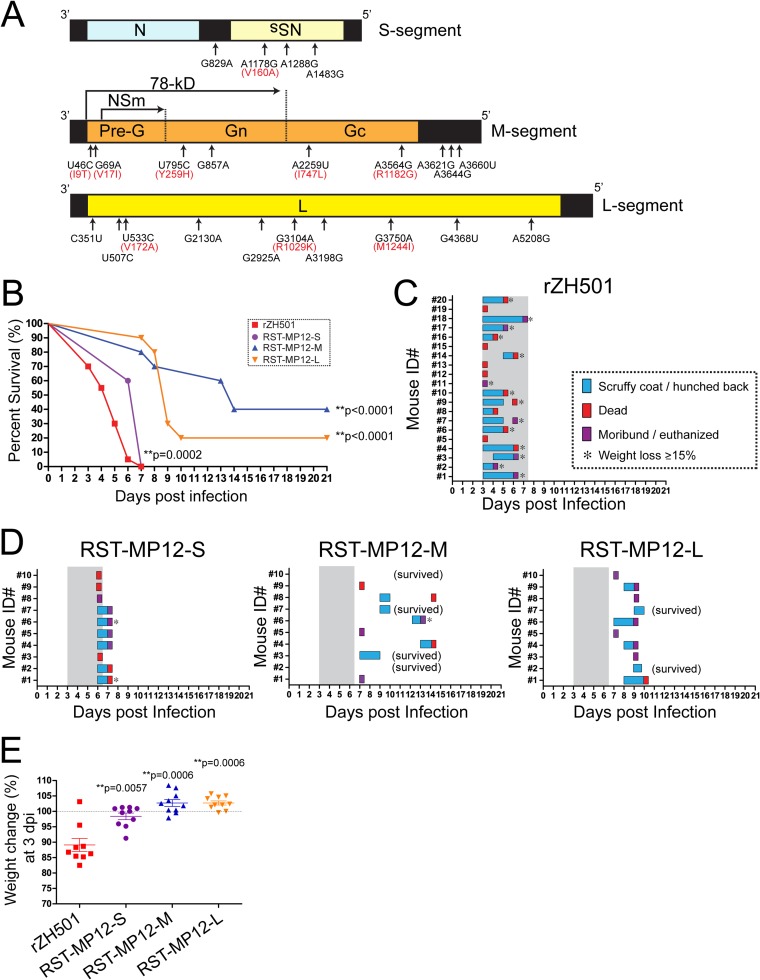
FIG 1.
Challenge of mice with rZH501 with the MP-12 S, M, or L segment. (A) Schematics of the MP-12 S, M, and L segments and 23 mutations. Amino acid substitutions are shown in red. (B) Survival curves for outbred CD1 mice (n = 10 per group) challenged with a 1 × 103-PFU dose (i.p.) of rZH501, RST-MP12-S, RST-MP12-M, or RST-MP12-L. P values obtained by the log-rank test (versus the results for parental strain rZH501) are shown (**, P < 0.01). (C) Clinical signs of disease in mice challenged with parental strain rZH501. Scruffy coat/hunched back or other signs of disease, such as ataxia or partial paralysis in a leg(s) (blue), death (red), or a moribund state (purple), were recorded for individual mice. A loss of body weight exceeding 15% of the initial body weight is indicated by an asterisk. (D) Clinical signs of diseases in mice challenged with RST-MP12-S, RST-MP12-M, or RST-MP12-L. Gray shading, period of disease in parental strain rZH501-infected mice. (E) Change in body weight (in percent) at 3 dpi compared to the body weight at 0 dpi. The bars represent the mean and standard error. P values obtained by the Mann-Whitney test (versus the results for parental strain rZH501) are shown (**, P < 0.01). The dotted line indicates a 100% change in body weight. Mouse ID#, mouse identifier.
RVFV strain MP-12 is derived from a plaque isolate of strain ZH548, and its genome encodes 23 nucleotide mutations (25), i.e., 4 mutations in the S segment, 9 mutations in the M segment, and 10 mutations in the L segment (Fig. 1A). Among those mutations, amino acid substitutions occur in the following 9 locations: V160A (NSs), I9T (78-kDa protein), V17I (78-kDa protein), Y259H (Gn), I747L (Gc), R1182G (Gc), V172A (L), R1029K (L), and M1244I (L). We previously found that parental strain ZH548 has genetic subpopulations whose genomes encode the I9T (78-kDa protein), V17I (78-kDa protein), or I747L (Gc) mutation, as well as 3 other silent mutations which are identical to the mutations in MP-12 (25). It indicates that those 6 mutations in MP-12 have been derived from a genetic subpopulation of strain ZH548. A study using RVFV reassortants between strains MP-12 and ArD38661 (a strain isolated in Senegal in 1983) indicated that each S, M, and L segment of MP-12 is independently attenuated (60). Later, it was claimed that the ArD38661 strain is not sufficiently virulent in mice, and thus, pathogenic wild-type strain ZH548 was used as a backbone RVFV strain for the testing of MP-12 attenuation using reverse genetics. The ZH548 reassortant with the MP-12 S segment was shown to cause 80% mortality in 4- to 5-week-old Swiss strain mice within 7 to 14 days postinfection (dpi) when it was intraperitoneally (i.p.) inoculated at 1 × 104 PFU (61). A similar mortality rate was observed by using a strain ZH548 reassortant with the MP-12 NSs gene, indicating that MP-12 NSs may partly contribute to attenuation (61). The MP-12 NSs gene has a single amino acid change (V160A) and two other silent mutations (25), which may contribute to the weak attenuation of the MP-12 S segment.
We hypothesized that mutations within the M and L segments play a major role in the attenuation of the RVFV MP-12 vaccine strain. In this study, we aimed to characterize the role of individual MP-12 mutations in the attenuation of pathogenic wild-type strain ZH501, which is one of the best-characterized strains in terms of RVF pathology, and to characterize the virulence of MP-12 with reversion mutations.
MATERIALS AND METHODS
Media, cells, and viruses.
VeroE6 cells (ATCC CRL-1586), Vero cells (ATCC CCL-81), or Hepa1-6 cells (ATCC CRL-1830) were maintained in Dulbecco's modified minimum essential medium (DMEM) containing 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). BHK/T7-9 cells that express T7 RNA polymerase (62) were maintained in alpha minimum essential medium containing 10% FBS, penicillin-streptomycin, and 600 μg/ml of hygromycin. Rescue and work with infectious recombinant RVFV strain ZH501 (rZH501) or recombinant strain MP-12 (rMP-12) were performed at The University of Texas Medical Branch at Galveston (UTMB) Robert E. Shope biosafety level 4 (BSL-4) laboratory as described previously (63). Rescued viruses were amplified once in VeroE6 cells after recovery in BHK/T7-9 cells and used for subsequent experiments. Virus titration was performed by plaque assay using VeroE6 cells with either a 0.3% tragacanth gum or a 0.6% Noble agar overlay as described previously (64, 65).
Plasmids.
Six plasmids (S, M, and L genomic RNA expression plasmids and N, Gn/Gc, and L protein expression plasmids) were used for recovery of rMP-12, rZH501, or the reassortants. Plasmids carrying a full-length S, M, or L segment flanked by the T7 promoter and hepatitis delta virus ribozyme sequences were described previously, i.e., for strain ZH501, pProT7-wS(+), pProT7-wM(+), or pProT7-wL(+), and for strain MP-12, pProT7-avS(+), pProT7-avM(+), or pProT7-avL(+) (55, 65, 66). Plasmids carrying open reading frames (ORFs) for the N, M, or L segment were also described previously, i.e., for strain ZH501, pProT7-IRES-wN, pCAGGS-wG, and pProT7-IRES-wL, and for strain MP-12, pProT7-IRES-vN, pCAGGS-vG, and pProT7-IRES-vL (55, 65, 66). In addition, individual MP-12 mutations were introduced separately or in combination into pProT7-wM(+) (U46C, G69A, U795C, G857A, A2259U, A3564G, A3621G, A3644G, or A3660U) or pProT7-wL(+) (C351U, U507C, U533C, G2130A, G2925A, G3104A, A3198G, G3750A, C4368U, or A5208G) by site-directed mutagenesis using Pfu Ultra high-fidelity DNA polymerase (Agilent Technologies). Further, a reversion mutation was introduced separately or in combination into pProT7-vM(+) (C795U or G3564A) or pProT7-vL(+) (C533U, A3104G, or G3198A).
Mouse challenge experiments with recombinant RVFV.
Groups of 10 CD1 mice (5- to 6-week-old female mice; Charles River) were i.p. inoculated with 1 × 103 PFU of rZH501 or the individual mutants. The mice were observed daily for the development of disease, while individual body weights were measured daily for 7 days and subsequently every 3 days until the termination of the experiment at 21 days postchallenge. Moribund mice, including those that had a body weight loss of more than 15% and/or that showed clinical signs, such as viral encephalitis or severe lethargy, were humanely euthanized, while all surviving mice were euthanized at 21 days postchallenge. Tissues were collected and placed in 10% buffered formalin for histopathological analysis. The sera of the survivors were tested for levels of neutralizing antibody and anti-N IgG. Survival curves for infected mice were analyzed by the use of the GraphPad Prism (version 5.03) program (GraphPad Software Inc., La Jolla, CA). All work with infectious rZH501 was performed in the Robert E. Shope BSL-4 laboratory, UTMB.
Antibody titration.
An 80% plaque reduction neutralization test (PRNT80) was performed as described previously using MP-12 and VeroE6 cells (67). An IgG enzyme-linked immunosorbent assay (ELISA) for detection of anti-RVFV N antibody using His-tagged RVFV N proteins derived from recombinant baculovirus was described previously (67). The cutoff value of the IgG ELISA was determined to be 0.245 from the geometric mean + 3 times the standard deviation for normal sera (a 1:100 dilution of 10 normal serum samples per plate).
Droplet digital PCR analysis.
For the measurement of viral RNA (S RNA and N mRNA), mouse liver or spleen tissues were homogenized in TRIzol reagent (Life Technologies), followed by the extraction of total RNA by use of a Direct-zol RNA miniprep kit (Zymo Research) according to the manufacturer's instruction. DNA was digested with the DNase I provided in the kit. The total RNA concentration was measured by use of a Qubit 2.0 fluorometer (Life Technologies), and total RNA was used for first-strand cDNA synthesis by using an iScript cDNA synthesis kit (Bio-Rad), which contains a random hexamer and RNase H. cDNA derived from total RNA (25 or 250 ng), 250 nM TaqMan probe, 5′-HEX-CAG GCT TTG GTC GTC TTG AG-BHQ1-3′ (where HEX is hexachlorofluorescein and BHQ1 is black hole quencher 1), and 900 nM forward primer (5′-GGC TGG CTG GAC ATG-3′) and 900 nM reverse primer (5′-AGT GAC AGG AAG CCA CTC A-3′), which are specific for the RVFV N ORF, were added to the PCR mixture (25 μl), which also contained ddPCR supermix for probes (Bio-Rad). The iScript non-reverse transcription control reagent (Bio-Rad) was used to make a control reaction mixture with which the reverse transcription step was not performed. Oil droplets were generated by a QX100 droplet generator (Bio-Rad), and PCR was performed by initial heating at 95°C for 10 min, 40 cycles of 94°C (30 s) and 60°C (1 min), and a final step at 98°C for 10 min. PCR data were analyzed by a QX100 droplet reader (Bio-Rad) with QuantaSoft (version 1.4) software according to the manufacturer's instructions.
Blood chemistry.
Alanine aminotransferase (ALT), albumin, alkaline phosphatase (ALP), amylase, total calcium, creatinine, globulin, glucose, phosphorus, K+, Na+, total bilirubin, total protein, and blood urea nitrogen levels in mouse heparinized whole blood collected at 3 dpi were measured by use of the VetScan comprehensive diagnostic profile (Abaxis) according to the manufacturer's instruction.
Immunohistochemistry.
Immunohistochemistry of tissue sections was performed using anti-RVFV N antibody as described previously (68).
Statistical analysis.
The log-rank test, the Mann-Whitney test, and Student's unpaired t test were performed using the GraphPad Prism (version 5.03) program (GraphPad Software Inc.).
Ethics statement.
All experiments using recombinant DNA and RVFV were performed upon the approval of the Notification of Use by the Institutional Biosafety Committee at UTMB. Mouse studies were performed in the UTMB Robert E. Shope BSL-4 laboratory, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) in accordance with the Animal Welfare Act, NIH guidelines, and U.S. federal law. The animal protocol was approved by the UTMB Institutional Animal Care and Use Committee (IACUC).
RESULTS
Replacement of the rZH501 S, M, or L segment with the MP-12 S, M, or L segment differentially attenuates rZH501.
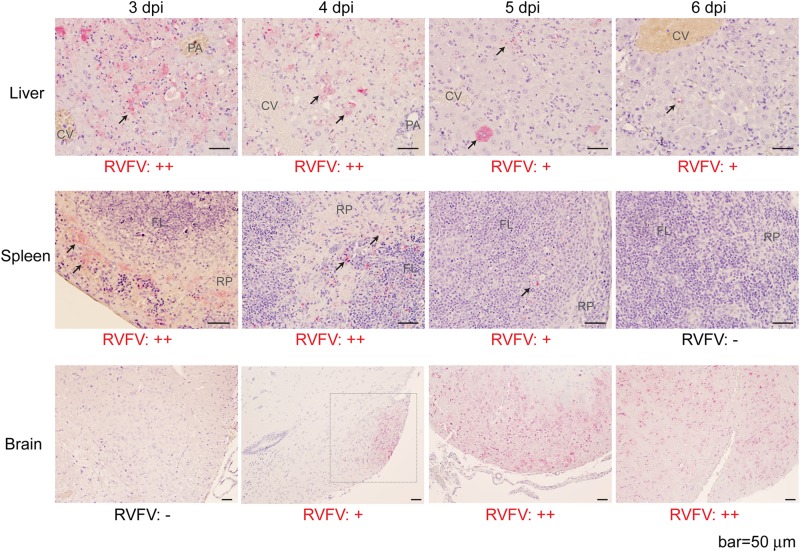
Strain ZH501 is highly virulent in mice, with a 50% lethal dose (LD50) of 0.5 to 3 PFU (i.p.) in outbred CD1 (ICR) mice (24, 69), and the viral distributions and pathological changes that occur during infection have been well characterized (70). Taking advantage of the well-characterized mouse model using strain ZH501, we analyzed the MP-12 mutation(s) in the ZH501 backbone by reverse genetics. To determine if either the S, M, or L segment of MP-12 can attenuate the virulence of ZH501 in mice, outbred CD1 mice were challenged (i.p.) with 1 × 103 PFU of rZH501, rZH501 with the MP-12 S segment (RST-MP12-S), rZH501 with the MP-12 M segment (RST-MP12-M), or rZH501 with the MP-12 L segment (RST-MP12-L). All mice infected with rZH501 succumbed to infection within 7 days postinfection (dpi) (Fig. 1B). All mice that were moribund or died at 3 dpi did not have any detectable clinical signs of disease at 2 dpi, while those that were moribund or died at 4, 5, 6, or 7 dpi showed clinical signs, such as a scruffy coat or a hunched back, before death (Fig. 1C). Interestingly, the survival curves for mice infected with RST-MP12-S, RST-MP12-M, or RST-MP12-L were significantly different from those for mice infected with parental strain rZH501 (log-rank test) (Fig. 1C), and mice infected with RST-MP12-S, RST-MP12-M, or RST-MP12-L showed a delayed onset of clinical signs (Fig. 1D). All mice infected with RST-MP12-S were dead at either 6 dpi (40% of mice) or 7 dpi (60% of mice), while RST-MP12-M or RST-MP12-L infection resulted in mouse death at 7 to 14 dpi (60% death rate), or at 7 to 10 dpi (80% death rate), respectively. All survivors had detectable anti-N IgG at 21 dpi (data not shown). At 3 dpi, the body weights of mice challenged with RST-MP12-S, RST-MP12-M, or RST-MP12-L were significantly higher than those of mice challenged with parental strain rZH501 (Fig. 1E). In histopathological analyses of parental strain rZH501-infected mice, abundant viral N antigens were detected in necrotic hepatocytes in the liver or necrotic red pulp cord or mononuclear cells in follicles, and lymphoid depletion was detected in the spleen at 3 and 4 dpi (Fig. 2). The amount of viral antigen in the liver and spleen became less abundant at 5 and 6 dpi than it was at 3 or 4 dpi. On the other hand, a viral antigen-positive focus was detected at 4 dpi in the hypothalamus of an rZH501-infected mouse, while further spread of the viral antigen occurred in the brain, including the cerebral cortex, hippocampus, brain stem, and cerebellum, in most infected mice at 5 and 6 dpi (Fig. 2). Histopathologically, moribund mice infected with RST-MP12-S, RST-MP12-M, or RST-MP12-L had abundant viral antigens in the central nervous system (CNS) but not in the liver or spleen (data not shown). These results indicate that the MP-12 S, M, and L segments are all attenuated and that the MP-12 M and L segments are more attenuated than the MP-12 S segment in the mouse model. Meanwhile, we previously showed that 90 to 100% of mice infected with MP-12 survive challenges with 1 × 103 or 1 × 105 PFU (i.p.), while up to 10% of mice die or become moribund at 11 dpi or later and have abundant viral antigens in the brain (68). Thus, RST-MP12-S, RST-MP12-M, and RST-MP12-L are more pathogenic than MP-12.
FIG 2.
Immunohistochemistry of liver, spleen, and brain sections of mice infected with parental strain rZH501. (A) Immunohistochemistry of livers, spleens, and brains from mice infected with parental strain rZH501 at 3, 4, 5, or 6 dpi using anti-RVFV N antibody. Arrows, locations of RVFV N antigens; square with dotted lines, viral antigen location in brain section at 4 dpi. CV, central vein; PA, portal area; RP, red pulp cord; FL, follicle. The relative abundance of N antigens is indicated by the symbols −, +, and ++.
Y259H (Gn) and R1182G (Gc) independently contribute to MP-12 M-segment attenuation.
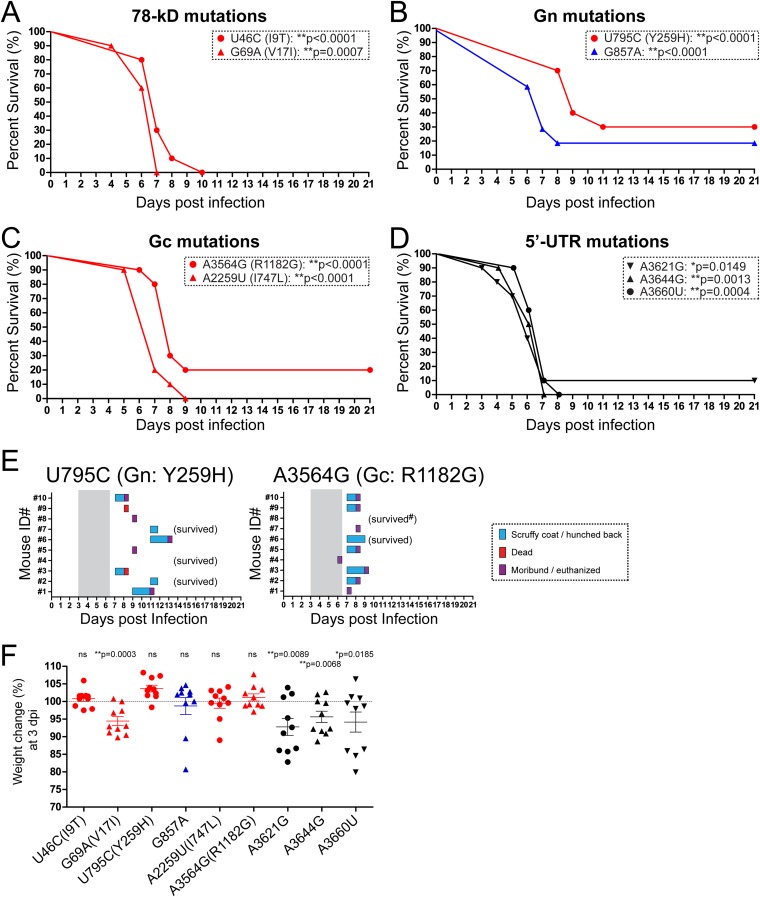
Next, we introduced MP-12 M-segment-specific mutations individually into the rZH501 backbone to further characterize their potential contribution to attenuation. These mutations are located within the 78-kDa protein, Gn, Gc, and 5′ untranslated region (UTR) (Fig. 1A). As described above, mice were challenged with rZH501 or one of the nine individual rZH501 M-segment point mutants, i.e., a mutant with the U46C (I9T, 78-kDa protein), G69A (V17I, 78-kDa protein), U795C [Y259H (Gn)], G857A, A2259U [I747L (Gc)], A3564G [R1182G (Gc)], A3621G, A3644G, or A3660U mutation. The survival curves for all mice infected with those mutants were significantly different from those for mice infected with parental strain rZH501 (log-rank test) (Fig. 3A to D). The rate of death for mice infected with the U795C [Y259H (Gn)] mutant was 70%, while that for mice infected with the A3564G [R1182G (Gc)] mutant was 80% (Fig. 3E). Inoculation of the two mutants with the G857A or A3621G silent mutation resulted in 20% or 10% survival of mice, respectively, yet 80% or 100% of infected mice showed clinical signs of diseases, respectively (data not shown). All surviving mice except for one mouse challenged with the A3564G mutant had detectable neutralizing antibodies and anti-N IgG at 21 dpi (data not shown). Inoculation of all other mutants resulted in 100% death of infected mice. At 3 dpi, the mean body weights of the mice challenged with the U46C (I9T), U795C (Y259H), G857A, A2259U (I747L), or A3564G (R1182G) mutant were not significantly decreased compared to those of the mice in the group challenged with RST-MP12-M (Fig. 3F). Taken together, our results clearly indicate that the U795C [Y259H (Gn)] and A3564G [R1182G (Gc)] mutations independently contribute to MP-12 M-segment attenuation. While it appears that the other individual M-segment mutations do not contribute to the overall attenuation of MP-12, we could not exclude the possibility of a synergistic attenuation effect.
FIG 3.
Mice challenged with rZH501 with a single mutation in the MP-12 M segment. Survival curves for outbred CD1 mice (n = 10 per group) challenged with a 1 × 103-PFU dose (i.p.) of rZH501 with one mutation in the MP-12 M segment are shown. (A) Mutations U46C (I9T) and G69A (V17I) in the 78-kDa protein; (B) mutations U795C (Y259H) and G857A in Gn; (C) mutations A2259U (I747L) and A3564G (R1182G) in Gc; (D) mutations A3621G, A3644G, and A3660U in the 5′ UTR. P values obtained by the log-rank test (versus the results for parental strain rZH501) are shown (*, P < 0.05; **, P < 0.01). (E) Clinical signs of diseases in mice challenged with the U795C [Y259H (Gn)] or A3564G [R1182G (Gc)] mutant. Gray shading, period of disease in parental strain rZH501-infected mice. Anti-N IgG was not detectable in a surviving mouse challenged with the A3564G mutant (#). (F) Change in body weight (in percent) at 3 dpi compared to the body weight at 0 dpi. The bars represent the mean and standard error. P values obtained by the Mann-Whitney test (versus the results for RST-MP12-M) are shown (*, P < 0.05; **, P < 0.01). The dotted line indicates a 100% change in body weight. ns, not significant.
Single point mutations in the MP-12 L segment do not alter the mortality of mice infected with ZH501.
Next, we analyzed the effect of individual MP-12 L-segment mutations on the attenuation of the virulence of strain ZH501. Mice were challenged with rZH501 or 1 of 10 rZH501 mutants with L-segment point mutations, i.e., C351U, U507C, U533C (V172A), G2130A, G2925A, G3104A (R1029K), A3198G, G3750A (M1244I), G4368U, or A5208G (Fig. 1A). While the survival curves for mice challenged with the C351U, U533C (V172A), G2130A, G3104A (R1029K), and A5208G mutants were significantly different from those for mice infected with parental strain rZH501 (log-rank test), none of the mice survived after challenge with those L-segment mutants (Fig. 4A and B). Among the three nucleotide mutations that resulted in an amino acid substitution, U533C (V172A) appeared to delay the onset of disease in some infected mice; i.e., death occurred at 5 dpi (10% of mice), 6 dpi (10%), 8 dpi (40%), 9 dpi (10%), and 10 dpi (30%) (Fig. 4C). However, the G3104A (R1029K) or G3750A (M1244I) mutation did not delay the death of infected mice; i.e., 20%, 50%, and 20% of mice challenged with the R1029K mutant were dead at 3 dpi, 6 dpi, and 7 dpi, respectively, and 20%, 20%, 20%, 30%, and 10% of mice challenged with the M1244I mutant were dead at 3 dpi, 4 dpi, 5 dpi, 7 dpi, and 8 dpi, respectively (data not shown). Compared to the changes in the body weights of mice challenged with RST-MP12-L, mice challenged with all L-segment single mutants tested showed a significant decrease in body weight at 3 dpi (Fig. 4D). Those results indicate that none of the L-segment single mutations independently confers MP-12 L-segment attenuation but that a combination of mutations may play a role in attenuation.
FIG 4.
Mice challenged with rZH501 with a single MP-12 L-segment mutation. Survival curves for outbred CD1 mice (n = 10 per group) challenged with a 1 × 103-PFU dose (i.p.) of rZH501 with one of the MP-12 L-segment mutations are shown. (A) C351U, U507C, U533C (V172A), G2130A, G2925A, and G3104A (R1029K) mutants; (B) A3198G, G3750A (M1244I), C4368U, and A5208G mutants. P values obtained by the log-rank test (versus the results for parental strain rZH501) are shown (*, P < 0.05; **, P < 0.01). (C) Clinical signs of disease in mice challenged with the U533C (V172A) mutant. Gray shading, period of disease in parental strain rZH501-infected mice. (D) Change in body weight (in percent) at 3 dpi compared to the body weight at 0 dpi. The bars represent the mean and standard error. P values obtained by the Mann-Whitney test (versus the results for RST-MP12-L) are shown (*, P < 0.05; **, P < 0.01). The dotted line indicates a 100% change in body weight.
The Y259H (Gn) or R1182G (Gc) mutation decreases viral replication in mice.
Because the U795C [Y259H (Gn)] or A3564G [R1182G (Gc)] mutation in rZH501 decreased the rate of mortality in mice, we hypothesized that rZH501 with either mutation would fail to replicate efficiently in vivo. To confirm our hypothesis, mice (n = 3 per group) were either mock infected or infected with 1 × 103 PFU (i.p.) of rZH501 or rZH501 with either Y259H or R1182G and euthanized at 3 dpi. For mice infected with rZH501, we could determine levels of viremia of 1 × 103 to 1.5 × 104 PFU/ml at 3 dpi (Fig. 5A). As for the mutant viruses, two out of three mice infected with the R1182G (Gc) mutant had levels of viremia of 1 × 102 to 4.5 × 103 PFU/ml (detection limit of the assay, 1 × 102 PFU/ml), while none of the mice infected with the Y259H (Gn) mutant developed any detectable viremia (Fig. 5A). Viral S-segment RNA and N mRNA levels in the liver and spleen were measured using droplet digital PCR (Fig. 5B and C). The mean RNA copy number of rZH501 was 2.1 × 106 or 2.9 × 104 per 1 μg total RNA in liver or spleen, respectively. On the other hand, the mean RNA copy number of the Y259H mutant was 9.7 × 103 or 8.5 × 101 per 1 μg total RNA in the liver or spleen, respectively, while that of the R1182G mutant was 4.9 × 104 or 4.6 × 102 per 1 μg total RNA in the liver or spleen, respectively. These data indicate that rZH501 with either the Y259H or R1182G mutation accumulates less viral RNA in the liver and spleen than parental strain rZH501. To further determine the amount of tissue injury caused by virus infection, the alanine aminotransferase (ALT), albumin, alkaline phosphatase (ALP), amylase, total calcium, creatinine, globulin, glucose, phosphorus, K+, Na+, total bilirubin, total protein, and blood urea nitrogen levels in heparinized blood samples were analyzed by use of the VetScan comprehensive diagnostic profile (Abaxis). The ALT levels in all samples from mice infected with rZH501 were outside the detection range (Fig. 5D). In blood derived from parental strain rZH501-infected mice, the mean ALP (Fig. 5E) and total bilirubin (Fig. 5F) levels were increased by 297% and 411%, respectively, over the levels in the blood of the mock-infected controls (71). Meanwhile, the mean levels of ALP and total bilirubin were similar between mice infected with the Y259H and R1182G mutants and mock-infected control mice, while the mean ALT level in samples from mice infected with the R1182G mutant were increased by 363% over that in samples from the mock-infected control mice, which exceeds the physiologically normal ALT level (71) (Fig. 5D to F). At 3 dpi, liver tissues from mice infected with the Y259H or R1182G mutant were histologically less affected (Fig. 5G) than those from parental strain rZH501-infected mice (Fig. 2) and contained fewer hepatocytes with detectable viral antigens. Taken together, at 3 dpi both the Y259H and R1182G rZH501 mutants replicated less efficiently in the liver and spleen than parental strain rZH501. Further, these data suggest that the attenuation induced by the R1182G mutation is weaker than that induced by the Y259H mutation and hepatocellular necrosis is ongoing in mice infected with the R1182G mutant.
FIG 5.
Viral replication and blood clinical chemistry at 3 dpi. Outbred CD1 mice (n = 3 per group) were challenged with a 1 × 103-PFU dose (i.p.) of parental strain rZH501, phosphate-buffered saline (mock-infected controls), or rZH501 with the U795C (Y259H) or A3564G (R1182G) mutation. At 3 dpi, the mice were euthanized and whole blood, sera, or total RNA from livers or spleens was analyzed. (A) Virus titer determined by plaque assay. The detection limit (102 PFU/ml) is shown by a dotted line. (B and C) Viral RNA copy numbers per 1 μg of total RNA in the liver (B) and spleen (C) at 3 dpi were determined by droplet digital PCR using a TaqMan probe detecting the RVFV S segment at the N ORF. (D to F) VetScan comprehensive diagnostic profile (Abaxis) test results for ALT (D), ALP (E), and total bilirubin (F) using heparinized whole-blood samples obtained at 3 dpi. The bars in panels A to F represent means and standard errors. (G) Immunohistochemistry of livers from mice infected with the U795C [Y259H (Gn)] (left) or A3564G [R1182G (Gc)] (right) mutant. Arrows, localization of RVFV N antigens.
Increased attenuation occurs with a combination of Y259H (Gn), R1182G (Gc), and R1029K (L).
Since virus with individual mutations in the MP-12 S, M, or L segment by itself was not fully attenuated, a combination of MP-12 mutations is probably required for full attenuation. Therefore, we next evaluated whether an increase in the attenuation of rZH501 could be achieved by a combination of the Y259H (Gn) and R1182G (Gc) mutations and selected L-segment mutations. We selected the following three L-segment mutations: V172A (which caused a delayed onset of disease and a statistically significant difference in survival curve compared to that for mice infected with rZH501), R1029K (which caused disease progression similar to that caused by rZH501 but a statistically significant difference in survival curve compared to that for mice infected with rZH501), and A3198G (which caused disease progression similar to that in mice infected with rZH501 and resulted in a rate of survival similar to that for mice infected with rZH501). Namely, we tested the attenuation of rZH501 with the following combinations of MP-12 mutations: Y259H (Gn) and R1182G (Gc); Y259H (Gn), R1182G (Gc), and V172A (L); Y259H (Gn), R1182G (Gc), and R1029K (L); or Y259H (Gn), R1182G (Gc), and A3198G (L) (Fig. 6). The death rate among mice infected with rZH501 with the Y259H (Gn) and R1182G (Gc) mutations was 60%, which was similar to the rate of death for mice infected with RST-MP12-M (Fig. 1B). The attenuation of rZH501 with the Y259H (Gn), R1182G (Gc), and V172A (L) mutations or the Y259H (Gn), R1182G (Gc), and A3198G (L) mutations was not further increased compared with that of rZH501 with the Y259H (Gn) and R1182G (Gc) mutations (i.e., death rates were 70% or 90%, respectively); rather, the number of mice with detectable clinical signs of disease was increased: 60% in mice infected with rZH501 with the Y259H and R1182G mutations, 100% in mice infected with rZH501 with the Y259H, R1182G, and V172A mutations, and 90% in mice infected with rZH501 with the Y259H, R1182G, and A3198G mutations (Fig. 6). However, all mice infected with rZH501 with the Y259H (Gn), R1182G (Gc), and R1029K (L) mutations survived without detectable clinical signs of disease for 21 days of observation (Fig. 6A). All the survivors showed detectable neutralizing antibody titers (PRNT80) and anti-N IgG at 21 dpi (Fig. 6D and E). These results suggest that the two M-segment mutations (Y259H and R1182G) and R1029K (L) in combination are major contributors to the attenuation of rZH501. Next, we analyzed the replication kinetics of rZH501, rZH501 with the Y259H and R1182G mutations, rZH501 with the Y259H, R1182G, and R1029K mutations, or rMP-12 in type I interferon (IFN)-incompetent Vero cells or type I IFN-competent mouse Hepa1-6 cells. Though all viruses replicated efficiently in Vero cells (Fig. 7A), the replication kinetics of rMP-12 were less efficient than those of rZH501 or the two rZH501 mutants in Hepa1-6 cells (Fig. 7B). On the other hand, there was no detectable difference in the replication kinetics between the Y259H and R1182G mutant and the Y259H, R1182G, and R1029K mutant in Vero or Hepa1-6 cells. Plaques of rZH501 and rMP-12 were similar and were heterogeneous in size. Meanwhile, the plaque sizes of the Y259H and R1182G mutant and the Y259H, R1182G, and R1029K mutant were also heterogeneous, yet the average size of the plaques was approximately 50% smaller than those of rZH501 and rMP-12 (Fig. 7B). The results indicated that MP-12 mutations other than Y259H (Gn), R1182G (Gc), and R1029K (L) also play a role in the MP-12 phenotype.
FIG 6.
Mice challenged with rZH501 with a combination of MP-12 M- and L-segment mutations. (A) Survival curves for outbred CD1 mice (n = 10 per group) challenged with a 1 × 103-PFU dose (i.p.) of rZH501 with a combination of mutations: Y259H (Gn) and R1182G (Gc); Y259H (Gn), R1182G (Gc), and V172A (L); Y259H (Gn), R1182G (Gc), and R1029K (L); and Y259H (Gn), R1182G (Gc), and A3198G (L). P values obtained by the log-rank test (versus the results for mice challenged with Y259H and R1182G) are shown (*, P < 0.05; **, P < 0.01). (B) Clinical signs of disease in mice challenged with rZH501 with combinations of MP-12 mutations, i.e., Y259H (Gn) and R1182G (Gc); Y259H (Gn), R1182G (Gc), and V172A (L); and Y259H (Gn), R1182G (Gc), and A3198G (L). (C) Change in body weight (in percent) at 3 dpi compared to the body weight at 0 dpi. The Mann-Whitney test (versus the results for the Y259H-R1182G mutant) was performed, yet no statistically significant differences were found. ns, not significant. The dotted line indicates a 100% change in body weight. (D and E) PRNT80 (D) or anti-N IgG titers (E) at 21 dpi are shown. The bars in panels C to E represent means and standard errors.
FIG 7.
Viral replication kinetics and plaque phenotypes of rZH501 mutants. (A) Viral replication kinetics (multiplicity of infection, 0.01) in Vero cells or Hepa1-6 cells for rMP-12, rZH501, or the mutants with the Y259H (Gn) and R1182G (Gc) mutations or the Y259H (Gn), R1182G (Gc), and R1029K (L) mutations. (B) Plaque images (top) and plaque sizes (bottom) of rMP-12, rZH501, and the mutants. The graph represents the average + standard error of plaque diameters for 23 randomly selected plaques. **, statistically significant difference by Student's unpaired t test (P < 0.0001) versus the results for rZH501.
A combination of Y259H (Gn), R1182G (Gc), and R1029K (L) is not the sole requirement for MP-12 attenuation.
Though a combination of Y259H (Gn), R1182G (Gc), and R1029K (L) was sufficient to completely attenuate rZH501, it is still possible that the combination of mutations is not essential for the entire attenuation of rZH501. Thus, we tested whether reversion of Y259H (Gn), R1182G (Gc), and R1029K (L) affected the full attenuation of rMP-12. We tested rMP-12 with the following reversion mutations: H259Y (Gn) only; G1182R (Gc) only; K1029R (L) only; H259Y (Gn) and G1182R (Gc); and H259Y (Gn), G1182R (Gc), and K1029R (L). Infection of mice with rMP-12 with either the H259Y (Gn) mutation only, the G1182R (Gc) mutation only, or a combination of the H259Y (Gn) and G1182R (Gc) mutations resulted in the death 10% of the infected mice, while all mice infected with rMP-12 with the K1029R reversion survived (Fig. 8A). On the other hand, 20% of the mice challenged with rMP-12 with a combination of the H259Y (Gn), G1182R (Gc), and K1029R (L) mutations died (Fig. 8A and B). Though we could observe an increased rate of death for mice infected with a mutant with a combination of H259Y (Gn), G1182R (Gc), and K1029R (L), 80% of challenged mice still survived the infection, and no significant weight loss was observed at 3 dpi (Fig. 8C). The surviving mice showed detectable neutralizing antibodies and anti-N IgG at 21 dpi, confirming that the mice became infected with the challenge viruses (data not shown). Our results indicate that mutations other than Y259H (Gn), R1182G (Gc), and R1029K (L) also contribute to the attenuation of MP-12 and that a combination of Y259H (Gn), R1182G (Gc), and R1029K (L) is not the sole requirement for MP-12 attenuation.
FIG 8.
Mice challenged with rMP-12 with M- and L-segment reversion mutations. (A) Survival curves for outbred CD1 mice (n = 10 per group) challenged with a 1 × 103-PFU dose (i.p.) of rMP-12 with a reversion mutation(s): K1029R(L) only; H259Y (Gn) only; G1182R (Gc) only; H259Y (Gn) and G1182R(Gc); or H259Y (Gn), G1182R (Gc), and K1029R (L). Survival curves were not statistically significantly different by the log-rank test (versus the results for the K1029R mutant). (B) Clinical signs of disease in mice challenged with rMP-12 with a reversion mutation(s), i.e., H259Y (Gn) only; G1182R (Gc) only; H259Y (Gn) and G1182R (Gc); or H259Y (Gn), G1182R (Gc), and K1029R (L). Gray shading, period of disease in parental strain rZH501-infected mice. There were no detectable clinical signs of disease in mice infected with the K1029R (L) mutant (not shown). (C) Change in body weight (in percent) at 3 dpi compared to the body weight at 0 dpi. The Mann-Whitney test (versus the results for the K1029R mutant) was performed, yet no statistically significant differences were found. ns, not significant. The dotted line indicates a 100% change in body weight. The bars represent the mean and standard error.
DISCUSSION
As of 2015, MP-12 and the rZH501 NSs/NSm double deletion mutant (72) were excluded from the select agent rule of HHS and USDA in the United States and considered candidate live-attenuated vaccines for RVF in the United States. The safety of strain MP-12 has been demonstrated by various challenge studies in nonhuman primates and ruminants (26–30). On the other hand, MP-12 NSs still retains functions similar to those of pathogenic RVFV strains, such as the ability to shut off host transcription, inhibit IFN-β gene induction, and degrade PKR and TFIIH p62 (48, 49, 53, 64, 67). However, a recombinant ZH548 strain with the MP-12 NSs was shown to prolong the survival time of infected mice, indicating that MP-12 NSs is partially attenuated (61). Meanwhile, the mechanism of MP-12 full attenuation has not yet been characterized in detail. Since the MP-12 vaccine is conditionally licensed for veterinary use in the United States, it is important to understand the impact of individual mutations on MP-12 attenuation so that it may be used to vaccinate animals in the field (under BSL-1 conditions). In this study, we generated various rZH501 reassortants or mutants to test the attenuation effects of mutations in MP-12. As a result, we found that the attenuation of MP-12 occurs through the S, M, or L segment, yet none of those segments is fully attenuated. Replacement of any single segment of ZH501 with that of MP-12 (i.e., RST-MP12-S, RST-MP12-M, or RST-MP12-L) showed virulent phenotypes in mice. The attenuation strength of the individual MP-12 genome segments in the ZH501 backbone was M > L > S. We found that two mutations in the surface glycoproteins, Y259H (Gn) and R1182G (Gc), independently contribute to attenuation of the M segment. On the other hand, other M-segment mutations significantly altered the survival curves, yet the onset of disease was still observed from 3 to 7 dpi, which is similar to the time of onset of disease caused by parental strain rZH501. Our previous study found that the I9T (78-kDa protein), V17I (78-kDa protein), and I747L (Gc) mutations are present in genetic subpopulations of parental strain ZH548 (25). Thus, Y259H (Gn) and R1182G (Gc) are the only amino acid substitutions unique to the strain MP-12 M segment. However, the attenuation of the MP-12 M segment is partial, and the full attenuation of MP-12 requires additional mutations. We found that none of the individual L-segment mutations of MP-12 alters the rate of mortality caused by rZH501 in mice, though the V172A (L) mutations delayed the onset of clinical signs of disease. We then selected three L-segment mutations, V172A, R1029K, and A3198G, to further characterize the synergistic effect with the M-segment mutations Y259H (Gn) and R1182G (Gc). Only the combination of Y259H (Gn), R1182G (Gc), and R1029K (L) resulted in the increased attenuation of rZH501 compared to that achieved with a combination of two M-segment mutations, Y259H (Gn) and R1182G (Gc). These results indicate that R1029K in combination with the Y259H (Gn) and R1182G (Gc) mutations contributes to the attenuation. Though Y259H (Gn), R1182G (Gc), and R1029K (L) were identified to be involved in the attenuation of MP-12, it is possible that those three mutations are sufficient, yet not essential, for the full attenuation of MP-12. Thus, we evaluated the synergistic attenuation effects of mutations other than Y259H (Gn), R1182G (Gc), and R1029K (L) by introducing the reversion mutations H259Y (Gn), G1182R (Gc), and K1029R (L) into rMP-12. Importantly, infection with MP-12 with those three reversion mutations resulted in only 20% mortality among infected mice, indicating that the combination of Y259H (Gn), R1182G (Gc), and R1029K (L) is not the sole requirement for the attenuation of MP-12.
Strains ZH501 and ZH548 were isolated from different patients in the same RVFV outbreak in Egypt in 1977 and 1978, and the strain ZH501 genome encodes 15 silent mutations and 5 amino acid substitutions compared to the sequence of ZH548, i.e., 4 mutations in the S segment (U369C, G461A, A599G, and C716U), 6 mutations in the M segment (U715A [Leu to Gln], A1824G [Ile to Val], U2711C, C2981U, U3655C, and A3678G), and 10 mutations in the L segment (A822C, C1057U, C1173U, G1427A [Ser to Asn], A3385G [Ser to Gly], U3768C, C4413U, G5406A, A5783G [Lys to Arg], and G6303A). Because they have the same genetic lineage (11, 73), 7 of those unique ZH501 mutations (U369C, G461A, and C716U in the S segment; U715A in the M segment; and G1427A, U3768C, and G6303A) were also found in subpopulations with the ZH548 genetic background (25). The locations of the mutations in MP-12 are distinct from those in ZH501, and thus, we used ZH501 as a backbone to screen for the attenuation caused by MP-12 mutations. We cannot exclude the possibility that a single MP-12 mutation other than U795C [Y259H (Gn)] or A3564G [R1182G (Gc)] specifically attenuates strain ZH548. Meanwhile, it does not affect the safety of the MP-12 vaccine, because multiple mutations attenuating MP-12 clearly prevent the reversion to virulence.
Mice are extremely susceptible to RVFV infection, and the LD50 for strain ZH501 is 0.5 to 3 PFU (i.p.) (24, 69). Mice were therefore used as a model to evaluate MP-12 mutations. Even though mice are highly susceptible to RVFV infection, the pathology is not identical to that in human or ruminant RVFV infections, e.g., rapid neuroinvasion and a lack of febrile illness occur in mice but not commonly in humans and ruminants (9, 74). In our model, major lesions could be detected in the liver and spleen at 3 and 4 dpi and in the brain at 5 dpi or later in mice infected with RVFV (Fig. 2). We confirmed that rZH501 with either the Y259H (Gn) or R1182G (Gc) mutation failed to replicate efficiently in the liver or spleen at 3 dpi. Thus, those attenuating mutations affect viral replication at least in those target organs at early stages of infection. We also observed a relatively small reduction in body weight at 3 dpi for mice infected with mutants with one of the following mutations: I9T (78-kDa protein), Y259H (Gn), G857A (Gn), I747L (Gc), R1182G (Gc), V172A (L), and G2130A (L). Thus, in addition to Y259H (Gn) or R1182G (Gc), those mutations might play a role in the attenuation of rZH501 at an early stage of infection. Assuming that all mice succumb to infection if viral neuroinvasion occurs, a specific combination of mutations, e.g., Y259H (Gn), R1182G (Gc), and R1029K (L), plays a role in decreasing the neuroinvasiveness. We also observed that different MP-12 L mutations (V172A or A3198G) in combination with Y259H and R1182G slightly increase the neuroinvasiveness of rZH501. The mechanism of RVFV neuroinvasion is, however, poorly understood. We previously reported that rMP-12 carrying the Toscana virus (TOSV) NSs increases the neuroinvasion of rMP-12 in mice (death rate, ∼36%) (68). TOSV NSs inhibits the IFN-β promoter and promotes the degradation of PKR, yet it does not suppress host general transcription (75). This indicates that the quality of the host antiviral response in target cells influences the neuroinvasion of RVFV. It should be noted that the majority of humans or ruminants infected with RVFV develop febrile illness rather than lethal encephalitis. If attenuation of RVFV is tested in humans or adult ruminants, temperature sensitivity is another important factor for MP-12 attenuation. A previous study using a reassortant RVFV between strains MP-12 and ArD38661 (an RVFV isolate recovered in Senegal) indicated the presence of temperature sensitivity in the MP-12 M and L segments (60). It will be important to identify temperature-sensitive mutations for a MP-12 vaccine and their contribution to vaccine attenuation in ruminants or humans.
Our results indicate that the risk that MP-12 will revert to virulence is low, which is supported by the finding that multiple attenuation mutations are effective in combination. On the other hand, the partial attenuation of the MP-12 S, M, or L segment leads to the creation of reassortant RVFV strains which have moderately virulent phenotypes. Though the natural reassortment between strain Smithburn and wild-type RVFV has been indicated (12), it is difficult to predict the outcome of MP-12 vaccination. In addition to the risk of reassortment, it was also indicated that MP-12 may retain teratogenicity in the fetuses of ewes vaccinated in early gestation (34) or may be involved with hepatocellular necrosis in cattle (35). These observations are not fully consistent with the findings of safety evaluations of MP-12 (26–29), and it might be possible that the different immune statuses of the vaccinated animals affected the outcome of MP-12 vaccination. Thus, surveillance of MP-12-vaccinated ruminants and the mosquito populations surrounding the area where animals are vaccinated will be important in the future. As part of a long-term plan that extends beyond the period of conditional licensing of MP-12, a further improved live-attenuated RVF vaccine, e.g., recombinant MP-12 lacking NSs or NSm or with additional stable attenuation mutations fully attenuating each S, M, and L segment without decreasing the efficacy of the vaccine, should be developed (76–78).
ACKNOWLEDGMENTS
We thank C. J. Peters and J. C. Morrill at The University of Texas Medical Branch at Galveston (UTMB) for their helpful information on the study of the MP-12 vaccine.
This study was supported by NIH grant R01 AI087643 (to T.I. and A.N.F.) and funding from The Sealy Center for Vaccine Development at UTMB (to T.I.). T.E.H. was partially funded through Training in Emerging Infectious Diseases and Biodefense grant T32 AI007536 and a postdoctoral fellowship through the James W. McLaughlin Fellowship Fund at UTMB.
REFERENCES
- 1.Swanepoel R, Coetzer JAW. 2004. Rift Valley fever, p 1037–1070. In Coetzer JAW, Tustin RC (ed), Infectious diseases of livestock with special reference to southern Africa, 2nd ed Oxford University Press, Cape Town, South Africa. [Google Scholar]
- 2.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmaljohn C, Nichol ST. 2007. Bunyaviridae, p 1741–1789. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Linthicum KJ, Davies FG, Kairo A, Bailey CL. 1985. Rift Valley fever virus (family Bunyaviridae, genus Phlebovirus). Isolations from Diptera collected during an inter-epizootic period in Kenya. J Hyg (Lond) 95:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies FG, Linthicum KJ, James AD. 1985. Rainfall and epizootic Rift Valley fever. Bull World Health Organ 63:941–943. [PMC free article] [PubMed] [Google Scholar]
- 6.Anyamba A, Small JL, Britch SC, Tucker CJ, Pak EW, Reynolds CA, Crutchfield J, Linthicum KJ. 2014. Recent weather extremes and impacts on agricultural production and vector-borne disease outbreak patterns. PLoS One 9:e92538. doi: 10.1371/journal.pone.0092538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digoutte JP, Peters CJ. 1989. General aspects of the 1987 Rift Valley fever epidemic in Mauritania. Res Virol 140:27–30. doi: 10.1016/S0923-2516(89)80081-0. [DOI] [PubMed] [Google Scholar]
- 8.LaBeaud AD, Kazura JW, King CH. 2010. Advances in Rift Valley fever research: insights for disease prevention. Curr Opin Infect Dis 23:403–408. doi: 10.1097/QCO.0b013e32833c3da6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikegami T, Makino S. 2011. The pathogenesis of Rift Valley fever. Viruses 3:493–519. doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubney R, Hudson JR. 1931. Enzootic hepatitis or Rift Valley fever: an undescribed virus disease of sheep cattle and man from east Africa. J Pathol Bacteriol 34:545–579. doi: 10.1002/path.1700340418. [DOI] [Google Scholar]
- 11.Bird BH, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. 2007. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J Virol 81:2805–2816. doi: 10.1128/JVI.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grobbelaar AA, Weyer J, Leman PA, Kemp A, Paweska JT, Swanepoel R. 2011. Molecular epidemiology of Rift Valley fever virus. Emerg Infect Dis 17:2270–2276. doi: 10.3201/eid1712.111035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll SA, Reynes JM, Khristova ML, Andriamandimby SF, Rollin PE, Nichol ST. 2011. Genetic evidence for Rift Valley fever outbreaks in Madagascar resulting from virus introductions from the East African mainland rather than enzootic maintenance. J Virol 85:6162–6167. doi: 10.1128/JVI.00335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley DM, Rinderknecht JL, Nipp TL, Clarke NP, Snowder GD, National Center for Foreign Animal and Zoonotic Disease Defense Advisory Group on Rift Valley Fever . 2011. Potential effects of Rift Valley fever in the United States. Emerg Infect Dis 17:e1. doi: 10.3201/eid1708.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dar O, Hogarth S, McIntyre S. 2013. Tempering the risk: Rift Valley fever and bioterrorism. Trop Med Int Health 18:1036–1041. doi: 10.1111/tmi.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson CB, Cole KS, Roy CJ, Perlin DS, Byrne G. 2013. Challenges and practices in building and implementing biosafety and biosecurity programs to enable basic and translational research with select agents. J Bioterror Biodef Suppl 3:12634. doi: 10.4172/2157-2526.S3-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shurtleff AC, Garza N, Lackemeyer M, Carrion R Jr, Griffiths A, Patterson J, Edwin SS, Bavari S. 2012. The impact of regulations, safety considerations and physical limitations on research progress at maximum biocontainment. Viruses 4:3932–3951. doi: 10.3390/v4123932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smithburn KC. 1949. Rift Valley fever; the neurotropic adaptation of the virus and the experimental use of this modified virus as a vaccine. Br J Exp Pathol 30:1–16. [PMC free article] [PubMed] [Google Scholar]
- 19.Ikegami T, Makino S. 2009. Rift valley fever vaccines. Vaccine 27(Suppl 4):D69–D72. doi: 10.1016/j.vaccine.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall R, Gibbs CJ Jr, Aulisio CG, Binn LN, Harrison VR. 1962. The development of a formalin-killed Rift Valley fever virus vaccine for use in man. J Immunol 89:660–671. [PubMed] [Google Scholar]
- 21.Eddy GA, Peters CJ, Meadors G, Cole FE Jr. 1981. Rift Valley fever vaccine for humans. Contrib Epidemiol Biostat 3:124–141. [Google Scholar]
- 22.Pittman PR, Liu CT, Cannon TL, Makuch RS, Mangiafico JA, Gibbs PH, Peters CJ. 1999. Immunogenicity of an inactivated Rift Valley fever vaccine in humans: a 12-year experience. Vaccine 18:181–189. doi: 10.1016/S0264-410X(99)00218-2. [DOI] [PubMed] [Google Scholar]
- 23.Meegan JM. 1979. The Rift Valley fever epizootic in Egypt 1977-78. 1. Description of the epizootic and virological studies. Trans R Soc Trop Med Hyg 73:618–623. doi: 10.1016/0035-9203(79)90004-X. [DOI] [PubMed] [Google Scholar]
- 24.Caplen H, Peters CJ, Bishop DH. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol 66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 25.Lokugamage N, Freiberg AN, Morrill JC, Ikegami T. 2012. Genetic subpopulations of Rift Valley fever ZH548, MP-12 and recombinant MP-12 strains. J Virol 86:13566–13575. doi: 10.1128/JVI.02081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrill JC, Carpenter L, Taylor D, Ramsburg HH, Quance J, Peters CJ. 1991. Further evaluation of a mutagen-attenuated Rift Valley fever vaccine in sheep. Vaccine 9:35–41. doi: 10.1016/0264-410X(91)90314-V. [DOI] [PubMed] [Google Scholar]
- 27.Morrill JC, Mebus CA, Peters CJ. 1997. Safety of a mutagen-attenuated Rift Valley fever virus vaccine in fetal and neonatal bovids. Am J Vet Res 58:1110–1114. [PubMed] [Google Scholar]
- 28.Morrill JC, Mebus CA, Peters CJ. 1997. Safety and efficacy of a mutagen-attenuated Rift Valley fever virus vaccine in cattle. Am J Vet Res 58:1104–1109. [PubMed] [Google Scholar]
- 29.Morrill JC, Jennings GB, Caplen H, Turell MJ, Johnson AJ, Peters CJ. 1987. Pathogenicity and immunogenicity of a mutagen-attenuated Rift Valley fever virus immunogen in pregnant ewes. Am J Vet Res 48:1042–1047. [PubMed] [Google Scholar]
- 30.Morrill JC, Peters CJ. 2003. Pathogenicity and neurovirulence of a mutagen-attenuated Rift Valley fever vaccine in rhesus monkeys. Vaccine 21:2994–3002. doi: 10.1016/S0264-410X(03)00131-2. [DOI] [PubMed] [Google Scholar]
- 31.Morrill JC, Peters CJ. 2011. Mucosal immunization of rhesus macaques with Rift Valley fever MP-12 vaccine. J Infect Dis 204:617–625. doi: 10.1093/infdis/jir354. [DOI] [PubMed] [Google Scholar]
- 32.Morrill JC, Peters CJ. 2011. Protection of MP-12-vaccinated rhesus macaques against parenteral and aerosol challenge with virulent Rift Valley fever virus. J Infect Dis 204:229–236. doi: 10.1093/infdis/jir249. [DOI] [PubMed] [Google Scholar]
- 33.Bouloy M, Flick R. 2009. Reverse genetics technology for Rift Valley fever virus: current and future applications for the development of therapeutics and vaccines. Antiviral Res 84:101–118. doi: 10.1016/j.antiviral.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter P, Erasmus BJ, Vorster JH. 2002. Teratogenicity of a mutagenised Rift Valley fever virus (MVP 12) in sheep. Onderstepoort J Vet Res 69:95–98. [PubMed] [Google Scholar]
- 35.Wilson WC, Bawa B, Drolet BS, Lehiy C, Faburay B, Jasperson DC, Reister L, Gaudreault NN, Carlson J, Ma W, Morozov I, McVey DS, Richt JA. 2014. Evaluation of lamb and calf responses to Rift Valley fever MP-12 vaccination. Vet Microbiol 172:44–50. doi: 10.1016/j.vetmic.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Reguera J, Weber F, Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog 6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez N, Muller R, Prehaud C, Bouloy M. 1995. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol 69:3972–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikegami T, Peters CJ, Makino S. 2005. Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol 79:5606–5615. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. 2008. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J Virol 82:10341–10348. doi: 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huiskonen JT, Overby AK, Weber F, Grunewald K. 2009. Electron cryo-microscopy and single-particle averaging of Rift Valley fever virus: evidence for GN-GC glycoprotein heterodimers. J Virol 83:3762–3769. doi: 10.1128/JVI.02483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman MB, Freiberg AN, Holbrook MR, Watowich SJ. 2009. Single-particle cryo-electron microscopy of Rift Valley fever virus. Virology 387:11–15. doi: 10.1016/j.virol.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerrard SR, Nichol ST. 2002. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J Virol 76:12200–12210. doi: 10.1128/JVI.76.23.12200-12210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerrard SR, Nichol ST. 2007. Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology 357:124–133. doi: 10.1016/j.virol.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozach PY, Kuhbacher A, Meier R, Mancini R, Bitto D, Bouloy M, Helenius A. 2011. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10:75–88. doi: 10.1016/j.chom.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Harmon B, Schudel BR, Maar D, Kozina C, Ikegami T, Tseng CT, Negrete OA. 2012. Rift Valley fever virus strain MP-12 enters mammalian host cells via caveolae-mediated endocytosis. J Virol 86:12954–12970. doi: 10.1128/JVI.02242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J Virol 75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vialat P, Billecocq A, Kohl A, Bouloy M. 2000. The S segment of Rift Valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol 74:1538–1543. doi: 10.1128/JVI.74.3.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. doi: 10.1016/S0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- 49.Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of Rift Valley fever virus promotes post-translational downregulation of the TFIIH subunit p62. J Virol 85:6234–6243. doi: 10.1128/JVI.02255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kainulainen M, Habjan M, Hubel P, Busch L, Lau S, Colinge J, Superti-Furga G, Pichlmair A, Weber F. 2014. Virulence factor NSs of Rift Valley fever virus recruits the F-box protein FBXO3 to degrade subunit p62 of general transcription factor TFIIH. J Virol 88:3464–3473. doi: 10.1128/JVI.02914-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog 4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog 5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol 83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Won S, Ikegami T, Peters CJ, Makino S. 2007. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol 81:13335–13345. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terasaki K, Won S, Makino S. 2013. The C-terminal region of Rift Valley fever virus NSm protein targets the protein to the mitochondrial outer membrane and exerts anti-apoptotic function. J Virol 87:676–682. doi: 10.1128/JVI.02192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kading RC, Crabtree MB, Bird BH, Nichol ST, Erickson BR, Horiuchi K, Biggerstaff BJ, Miller BR. 2014. Deletion of the NSm virulence gene of Rift Valley fever virus inhibits virus replication in and dissemination from the midgut of Aedes aegypti mosquitoes. PLoS Negl Trop Dis 8:e2670. doi: 10.1371/journal.pntd.0002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crabtree MB, Kent Crockett RJ, Bird BH, Nichol ST, Erickson BR, Biggerstaff BJ, Horiuchi K, Miller BR. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: potential role for NSm in mosquito infection. PLoS Negl Trop Dis 6:e1639. doi: 10.1371/journal.pntd.0001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weingartl HM, Zhang S, Marszal P, McGreevy A, Burton L, Wilson WC. 2014. Rift Valley fever virus incorporates the 78 kDa glycoprotein into virions matured in mosquito C6/36 cells. PLoS One 9:e87385. doi: 10.1371/journal.pone.0087385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saluzzo JF, Smith JF. 1990. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine 8:369–375. doi: 10.1016/0264-410X(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 61.Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol 47:613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 63.Morrill JC, Ikegami T, Yoshikawa-Iwata N, Lokugamage N, Won S, Terasaki K, Zamoto-Niikura A, Peters CJ, Makino S. 2010. Rapid accumulation of virulent Rift Valley fever virus in mice from an attenuated virus carrying a single nucleotide substitution in the mRNA. PLoS One 5:e9986. doi: 10.1371/journal.pone.0009986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalveram B, Lihoradova O, Indran SV, Ikegami T. 2011. Using reverse genetics to manipulate the NSs gene of the Rift Valley fever virus MP-12 strain to improve vaccine safety and efficacy. J Vis Exp 2011:e3400. doi: 10.3791/3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikegami T, Won S, Peters CJ, Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol 80:2933–2940. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Won S, Ikegami T, Peters CJ, Makino S. 2006. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J Virol 80:8274–8278. doi: 10.1128/JVI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lihoradova OA, Indran SV, Kalveram B, Lokugamage N, Head JA, Gong B, Tigabu B, Juelich TL, Freiberg AN, Ikegami T. 2013. Characterization of Rift Valley fever virus MP-12 strain encoding NSs of Punta Toro virus or sandfly fever Sicilian virus. PLoS Negl Trop Dis 7:e2181. doi: 10.1371/journal.pntd.0002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Indran SV, Lihoradova OA, Phoenix I, Lokugamage N, Kalveram B, Head JA, Tigabu B, Smith JK, Zhang L, Juelich TL, Gong B, Freiberg AN, Ikegami T. 2013. Rift Valley fever virus MP-12 vaccine encoding Toscana virus NSs retains the neuroinvasiveness in mice. J Gen Virol 94:1441–1450. doi: 10.1099/vir.0.051250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Battles JK, Dalrymple JM. 1988. Genetic variation among geographic isolates of Rift Valley fever virus. Am J Trop Med Hyg 39:617–631. [DOI] [PubMed] [Google Scholar]
- 70.Smith DR, Steele KE, Shamblin J, Honko A, Johnson J, Reed C, Kennedy M, Chapman JL, Hensley LE. 2010. The pathogenesis of Rift Valley fever virus in the mouse model. Virology 407:256–267. doi: 10.1016/j.virol.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Quimby FW, Luong RH. 2007. Clinical chemistry of the laboratory mouse, p 171–216. In Fox JG, Barthold SW, Davisson MT, Newcomer EC, Quimby FW, Smith AL (ed), The mouse in biomedical research, 2nd ed, vol III Normative biology, husbandry, and models; Academic Press, Burlington, MA. [Google Scholar]
- 72.Bird BH, Maartens LH, Campbell S, Erasmus BJ, Erickson BR, Dodd KA, Spiropoulou CF, Cannon D, Drew CP, Knust B, McElroy AK, Khristova ML, Albarino CG, Nichol ST. 2011. Rift Valley fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J Virol 85:12901–12909. doi: 10.1128/JVI.06046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikegami T. 2012. Molecular biology and genetic diversity of Rift Valley fever virus. Antiviral Res 95:293–310. doi: 10.1016/j.antiviral.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray KK, Worthy MN, Juelich TL, Agar SL, Poussard A, Ragland D, Freiberg AN, Holbrook MR. 2012. Chemotactic and inflammatory responses in the liver and brain are associated with pathogenesis of Rift Valley fever virus infection in the mouse. PLoS Negl Trop Dis 6:e1529. doi: 10.1371/journal.pntd.0001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalveram B, Ikegami T. 2013. Toscana virus NSs protein promotes degradation of double-stranded RNA-dependent protein kinase. J Virol 87:3710–3718. doi: 10.1128/JVI.02506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrill JC, Laughlin RC, Lokugamage N, Pugh R, Sbrana E, Weise WJ, Adams LG, Makino S, Peters CJ. 2013. Safety and immunogenicity of recombinant Rift Valley fever MP-12 vaccine candidates in sheep. Vaccine 31:559–565. doi: 10.1016/j.vaccine.2012.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morrill JC, Laughlin RC, Lokugamage N, Wu J, Pugh R, Kanani P, Adams LG, Makino S, Peters CJ. 2013. Immunogenicity of a recombinant Rift Valley fever MP-12-NSm deletion vaccine candidate in calves. Vaccine 31:4988–4994. doi: 10.1016/j.vaccine.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lihoradova O, Ikegami T. 2014. Countermeasure development for Rift Valley fever: deletion, modification or targeting of major virulence factor. Future Virol 9:27–39. doi: 10.2217/fvl.13.117. [DOI] [PMC free article] [PubMed] [Google Scholar]