ABSTRACT
Varicella-zoster virus (VZV) is a human herpesvirus, which during primary infection typically causes varicella (chicken pox) and establishes lifelong latency in sensory and autonomic ganglia. Later in life, the virus may reactivate to cause herpes zoster (HZ; also known as shingles). To prevent these diseases, a live-attenuated heterogeneous vaccine preparation, vOka, is used routinely in many countries worldwide. Recent studies of another alphaherpesvirus, infectious laryngotracheitis virus, demonstrate that live-attenuated vaccine strains can recombine in vivo, creating virulent progeny. These findings raised concerns about using attenuated herpesvirus vaccines under conditions that favor recombination. To investigate whether VZV may undergo recombination, which is a prerequisite for VZV vaccination to create such conditions, we here analyzed 115 complete VZV genomes. Our results demonstrate that recombination occurs frequently for VZV. It thus seems that VZV is fully capable of recombination if given the opportunity, which may have important implications for continued VZV vaccination. Although no interclade vaccine-wild-type recombinant strains were found, intraclade recombinants were frequently detected in clade 2, which harbors the vaccine strains, suggesting that the vaccine strains have already been involved in recombination events, either in vivo or in vitro during passages in cell culture. Finally, previous partial and complete genomic studies have described strains that do not cluster phylogenetically to any of the five established clades. The additional VZV strains sequenced here, in combination with those previously published, have enabled us to formally define a novel sixth VZV clade.
IMPORTANCE Although genetic recombination has been demonstrated to frequently occur for other human alphaherpesviruses, herpes simplex viruses 1 and 2, only a few ancient and isolated recent recombination events have hitherto been demonstrated for VZV. In the present study, we demonstrate that VZV also frequently undergoes genetic recombination, including strains belonging to the clade containing the vOKA strain.
INTRODUCTION
Varicella-zoster virus (VZV) is a human herpesvirus belonging to the genus Varicellovirus of the Alphaherpesvirinae subfamily of the order Herpesvirales. VZV has a stable prevalence in all countries worldwide, and the primary infection is almost always symptomatic. Upon initial infection, the virus typically causes chicken pox, a highly contagious disease with tropism to skin and ganglia. In temperate climates, chicken pox is most commonly observed in children and is one of the classic diseases of childhood. In contrast, in tropical regions chicken pox often occurs in adolescents or adults, typically causing more-severe symptoms (1). Primary infection is followed by the establishment of lifelong latency of the virus in dorsal root and trigeminal ganglia. The virus may then reactivate at a later stage from one or more ganglia to cause herpes zoster, affecting the corresponding dermatomes (2). The virus may also cause various neurological complications, such as facial palsy, encephalitis, meningitis, and cerebral vasculitis (3–8).
The disease burden of primary and reactivated VZV motivated the development and use of a vaccine to prevent its spread and disease. A live attenuated vaccine strain, vOka (Varivax or Varilrix) was originally developed in Japan to prevent the spread of varicella among children (9). This vaccine, a heterogeneous mixture of related haplotypes, is routinely used in many countries, including Japan, South Korea, the United States, Canada, Australia, Germany, Costa Rica, Uruguay, and Qatar. More recently, the vaccine trademarked as Zostavax was licensed for use in the prevention of herpes zoster and postherpetic neuralgia in elderly people in the United States (10) and was also approved for use in the European Union (EU) with adults aged ≥50 years (11).
Herpesviruses are double-stranded DNA viruses, and all members of the Alphaherpesvirinae subfamily share similar properties such as short replication cycles and the establishment of lifelong latency in ganglia. Three members of the Alphaherpesvirinae subfamily infect humans. These are, in addition to VZV, herpes simplex virus 1 (HSV-1) and HSV-2. VZV has a genome size of approximately 125,000 bp. Assays based on restriction fragment length polymorphism (RFLP) patterns or PCR products from variable-number tandem repeat (VNTR) regions have been frequently used to analyze genetic diversity, classify VZV genotypes, and genotype clinical VZV strains (recently reviewed in reference 12). VZV was first sequenced in 1986 (13) and remains the most widely sequenced of the herpesviruses. This has enabled analyses of complete VZV genomes and the identification of single nucleotide polymorphisms (SNPs) that allow the segregation of VZV strains into at least five distinct clades (14–17).
Several studies have demonstrated that homologous recombination contributes significantly to the evolution of many herpesviruses, including HSV-1 (12, 18–21). Recent complete genome analysis of HSV-1 strains suggests that all or most wild-type strains have recombinant mosaic genomes (22, 23). Similarly, homologous recombination has also been demonstrated for HSV-2, and it was suggested that the frequency of recombination is even higher for HSV-2 than for HSV-1, although no comparative complete genome analysis has yet been performed (22, 24). In contrast, the role, if any, of recombination in VZV remains uncertain. It has, however, been demonstrated that coinfection with more than one genotype can occur (25) and that reinfection with VZV may result in cases of herpes zoster (26). It has also been demonstrated that VZV may undergo recombination in tissue culture in vitro (27). Although Muir et al. assumed that recombination “had little effect on the evolution of the genotypes” (28), later studies demonstrated that at least some of the VZV clades emerged through recombination (14–16). In recent studies by Norberg et al. (22) and Zell et al. (29), it was demonstrated that intraclade recombination has also occurred and it was speculated that the rate of detected recombinants may increase as more genomes are sequenced. In any case, compared with other human alphaherpesviruses, only a few ancient and some isolated recent recombination events have hitherto been described for VZV. A possible explanation for the low frequency of detected recombinants for VZV may be that strains from the different clades of VZV have been geographically segregated (26, 28, 30, 31). Interestingly, however, recent studies suggest that this segregation has diminished in the face of increased human migration (32, 33), which should increase the chances of recombination between VZV strains from different clades (12, 15, 22). In addition, the introduction of VZV vaccine preparations (containing live clade 2 strains) in geographic regions where other clades dominate could be expected to further increase the extent of interclade recombination.
Recombination between wild-type and attenuated vaccine strains and the putative consequences thereof have been demonstrated and/or discussed for several viruses, such as members of the Flaviviridae family (34–36), pestiviruses (37), polioviruses (38–40), avian paramyxovirus-1 (41), myxoma viruses (42), porcine reproductive and respiratory syndrome virus (43), and herpesviruses (15, 22, 44, 45). Recombination between two live herpesvirus vaccine strains was reported for pseudorabies virus, which is an alphaherpesvirus that infects pigs (44). The authors coinfected swine with two different attenuated vaccine strains, and virus isolates derived by serial plaque purification directly from tissue homogenates were characterized as recombinant and parental strains. Another recent study of infectious laryngotracheitis virus (ILTV), an alphaherpesvirus that infects poultry, analyzed distinct viruses (referred to as genotype classes 8 and 9) that were isolated from geographically distinct areas of Australia in 2008 (45). These strains are highly virulent and have been associated with outbreaks causing mortality rates of up to 17.6% (46). Three ILTV vaccines (live attenuated strains) are available in Australia: two closely related vaccines of Australian origin, SA2 and A20 (Pfizer, Australia), and one vaccine of European origin, Serva (Intervet). The vaccines of Australian origin are closely related and classified as class 1 (47). The vaccine of European origin, however, is classified as class 7 (48), and its genome is divergent from the vaccine strains of Australian origin, having only 99.2% nucleotide sequence identity (47). Whole-genome analysis of the virulent viruses of classes 8 and 9 demonstrated that these are recombinant viruses derived from the vaccine strains of Australian and European origin (45). It is thus possible for some live attenuated herpesvirus vaccine strains to recombine in vivo to form virulent progeny that spreads through the population. The authors also studied the pathogenicity of these recombinant viruses in specific-pathogen-free chickens and concluded that each recombinant strain had distinct in vivo genotypes with significantly increased virulence or replication compared with their parental strains. These findings gave rise to concerns about the use of attenuated herpesvirus vaccines under conditions that favor recombination (45).
Although recombination has been evidenced in VZV, little is known about how frequently recombination events occur within the host population. To evaluate the likeliness of the emergence of novel interclade recombinants as a result of increased migration and/or vaccination with attenuated viruses, we need to increase our understanding of VZV recombination in general and to determine how frequently this occurs in nature. The aim of the present study was thus to perform an evolutionary analysis of VZV and thoroughly map the extent of homologous recombination between circulating VZV strains. The purpose was to evaluate whether the use of attenuated VZV vaccines might present conditions that favor recombination. We analyzed 115 complete VZV genomes, derived from GenBank or newly sequenced here, from wild-type and vaccine strains using multiple advanced algorithms in parallel for comparison. Phylogenetic analysis revealed the presence of a novel clade of VZV, which was designated clade 6. We also present data that support frequent homologous recombination of VZV and provide strong statistical support for the existence of multiple inter- and intraclade recombination events. These results may have implications for our understanding of the future evolution of VZV and the use of live VZV vaccines.
MATERIALS AND METHODS
Ethics statement.
Clinical specimens (diagnostic samples collected as part of standard clinical procedures) were independently obtained from patients with confirmed VZV infection and anonymized prior to this study. Consent for all children and adult participants was written and informed. The parent or guardian of any child participant provided informed consent on their behalf. The use of these specimens for research was approved by the East London and City Health Authority Research Ethics Committee (P/96/046: Molecular typing of cases of varicella-zoster virus). Ethical permission for collecting clinical strains in Sweden was granted by the Regional Ethical Committee in Gothenburg, Sweden (Dnr 370-06).
DNA extraction, library construction, targeted enrichment, and sequencing.
DNA was extracted from each sample using the QiaAMP DNA minikit (Qiagen) according to the manufacturer's instructions. DNA quantification was performed using a NanoDrop spectrophotometer, and samples with 260/280 ratios outside the range of 1.7 to 2.1 and/or 260/230 ratios outside the range of 1.8 to 2.2 were further purified using the Genomic DNA Clean & Concentrator kit (Zymo Research) according to the manufacturer's instructions. Whole-genome amplification using GenomiPhi V2 (GE Healthcare) was performed using 10 ng of DNA and yielded between 3,000 and 5,000 ng of DNA. Sequencing libraries were constructed using 3,000 ng of DNA as input, as per the standard SureSelect XT v1.5 protocols (Agilent). Enrichment for VZV DNA was performed as described previously (49, 50). The final sequence libraries were multiplexed and sequenced using either 2 × 150 bp or 2 × 250 bp paired-end kits across multiple runs on an Illumina MiSeq.
Genome assembly and variant calling.
Sequence data sets were demultiplexed using BaseSpace, and individual data sets were subsequently parsed through Quasr (51) for duplicate removal and read-trimming (-q 30, -l 50) and subsequently aligned against the VZV reference strain Dumas (NC_001348) using BWA (52). The resulting alignments were processed using SAMTools (17) to generate pileup files for each sample. A consensus sequence for each data set was called with the Quasr module “pileupConsensus” and a 50% frequency threshold (i.e., no ambiguities were included). Variant profiling for each data set was performed using VarScan v2.2.11 (53) with the following parameters: base call quality, ≥20; read depth, ≥50; independent reads supporting minor allele, ≥2 per strand. In addition, variant calls showing a directional strand bias of ≥0.85 were excluded from further analyses. Consensus sequences were generated for each rash sample, but iterative repeat regions R1, R2, R3, R4, and R5 (19, 20) as well as the terminal repeat region were trimmed prior to tree-building analyses.
DNA sequence analysis.
DNA sequences were aligned using Kalign (54) implemented in eBioX (55). Phylogenetic networks were constructed using SplitsTree4 (56) with Uncorrected_P characters transformation, and all gaps and repeat regions were excluded prior to analysis. The informative sites analysis was performed using SimPlot (57), and the recombination analysis was performed by using the algorithms RDP, Geneconv, Chimaera, MaxChi, BootScan, SisScan, 3Seq, and LARD implemented in RDP4 (58) with default settings.
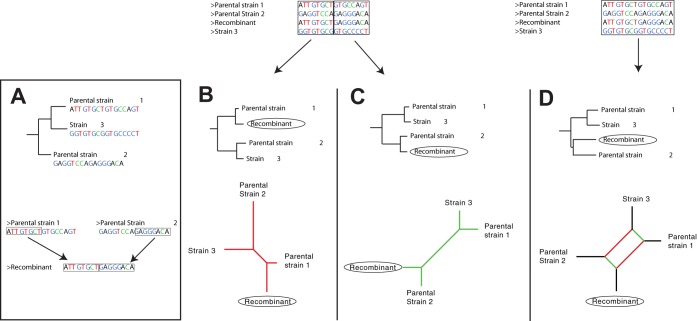
Phylogenetic analysis is the reconstruction of the evolutionary history of organisms, in this case of different VZV strains. Based on differences and similarities in their genomes, i.e., single nucleotide polymorphisms, the strains are typically arranged as external nodes in bifurcating trees, so-called phylogenetic or evolutionary trees. The strains are supposed to have descended from common ancestors that are represented in the phylogenetic tree as internal nodes to which the external nodes are attached. However, a limitation with these traditional phylogenetic trees is that they are bifurcating and assume that recombination has not contributed to the evolution. In a recombinant virus, different parts of the genome might have completely different evolutionary histories since they descend from different parental strains. At least two different phylogenetic trees are thus necessary to correctly illustrate the evolutionary history of a recombinant virus. The more recombination events that have occurred among the strains that are analyzed, the more phylogenetic trees are needed to represent the evolutionary histories of the different parts of the genomes of the strains. This quickly becomes highly impractical. To overcome this problem, the viral strains may instead be arranged in phylogenetic networks to illustrate the evolutionary relationships between them. A phylogenetic network may simply be explained as a combination of all phylogenetic trees supported by the genomic data under analysis (59), as illustrated in Fig. 1. As such, all parallel edges in a phylogenetic network thus represent conflicting phylogenetic signals. To include the possibility of recombination, we performed a phylogenetic analysis of the 115 aligned VZV genomes by constructing phylogenetic networks using SplitsTree (56).
FIG 1.
Schematic illustration demonstrating the fundamental basics of phylogenetic representations of recombination events and the limitations of traditional bifurcating phylogenetic trees. (A) Traditional phylogenetic tree illustrating the evolutionary history of three fictional small genomes, parental strain 1, parental strain 2, and strain 3 (top) and a recombination event between parental strains 1 and 2, in which the strain recombinant is created by concatenating the first genomic segment of parental strain 1 with the last genomic segment of parental strain 2 (bottom). (B and C) Rooted traditional bifurcating phylogenetic trees (top) and unrooted phylogenetic networks (bottom) based on the first (B) and last (C) parts of the genomes, respectively (i.e., before and after the recombination breakpoint). Here, all phylogenies reflect the true evolutionary history of the respective genomic segment, but none reflect the correct evolutionary history for the complete genomes. The traditional bifurcating phylogenetic tree based on the complete genome (D, top) cannot illustrate recombination events but instead places the recombinant strain separately from the other strains since it contain parts from both parental strains, which the algorithms interpret as separate evolution. Hence, this tree does not reflect the true evolutionary history of any genomic segment. The use of traditional phylogenetic trees should therefore be avoided when recombinant genomes are included in the analysis. The phylogenetic network (D, bottom), however, correctly illustrates the evolutionary relationships between the different strains. The parallel edges in the network illustrate the conflicting phylogenetic signals resulting from the multiple evolutionary histories found in the recombinant strain. In phylogenetic networks, parallel branches or internal “boxes” thus indicate that the data sets have conflicting phylogenetic signals. These conflicting signals may be a result of either convergent evolution or recombination.
Recombination analysis.
The detection of recombinant strains was carried out by analyzing conflicting phylogenetic signals (SNPs). Phylogenetically incompatible sites in a sequence alignment can be a result either of recombination or of convergent evolution on the nucleotide level, i.e., true homoplasy, where a specific nucleotide substitution independently arises at the same position in two viruses. Such recurrent mutations may arise randomly or by a selection pressure on specific biologically important sites. The challenge was therefore to detect phylogenetically conflicting sites and to deduce whether they were caused by recombination or by convergent evolution. First, we used the program SplitsTree (56) to create phylogenetic networks that illustrate the evolutionary relationships between the strains. When phylogenetic incongruences were detected, we used the phi-test, and algorithms included in the RDP4 program (RDP, Geneconv, Chimaera, MaxChi, BootScan, SisScan, 3Seq, and LARD) to perform a statistical analysis to determine whether the conflicting phylogenetic sites were the result of recombination. In addition, the informative site analysis tool implemented in the SimPlot program was used to detect and illustrate conflicting phylogenetic signals in selected strains, and putative interclade recombinants were also analyzed using the bootscan algorithm implemented in SimPlot.
Nucleotide sequence accession numbers.
Consensus sequences for all samples sequenced in this study are available in GenBank under the accession numbers shown in Table 1.
TABLE 1.
ID, accession numbers, source (body compartment), sample collection date, and pathology of the 37 strains sequenced
| Strain ID | Accession no. | Source (body compartment) | Sample collection dateb | Pathology |
|---|---|---|---|---|
| Ves/Cli/POR/Ves/5001 | KP771921 | Vesicle fluid | Not known | Varicella |
| Var/Cli/BAL/UK/2402/2009 | KP771904 | BALa specimen | 24/02/09 | Varicella |
| Var/Cli/Ves/UK/1001/2012 | KP771920 | Vesicle fluid | 10/01/12 | Varicella |
| Zos/Cli/Ves/UK/1801/2012 | KP771925 | Vesicle fluid | Not known | Zoster |
| Var/Cli/UK/BLD/1401/2012 | KP771906 | EDTA blood | 14/01/12 | Varicella (fatal) |
| Var/Cli/UK/BAL/1001/2012 | KP771905 | BAL specimen | 10/01/12 | Varicella (fatal) |
| Zos/Cli/GRE/Ves/03/2012 | KP771923 | Vesicle fluid | 01/03/12 | Zoster |
| Var/Cli/UK/Ves/1301/2013 | KP771911 | Vesicle fluid | 13/01/13 | Zoster with encephalitis |
| Var/Cli/UK/Ves/0706/2013 | KP771910 | Vesicle fluid | 07/06/13 | Zoster with encephalitis |
| Var/Cli/UK/Ves/2203/2013 | KP771912 | Vesicle fluid | 22/03/13 | Varicella |
| Var/Cli/UK/Ves/2403/2013 | KP771913 | Vesicle fluid | 24/03/13 | Varicella (fatal) |
| Var/Cli/UK/CSF/2912/2012 | KP771908 | CSF | 29/12/12 | Varicella with encephalitis |
| Var/Cli/UK/Ves/0201/2013 | KP771909 | Vesicle fluid | 02/01/13 | Varicella with encephalitis |
| Var/Cli/UK/CSF/0102/2013 | KP771907 | CSF | 01/02/13 | Encephalitis |
| DE10-1515 | KP771891 | Vesicle fluid | Not known | Zoster |
| DE10-2480 | KP771892 | Vesicle fluid | Not known | Zoster |
| DE10-2660 | KP771893 | Vesicle fluid | Not known | Zoster |
| DE10-2704 | KP771894 | Vesicle fluid | Not known | Zoster |
| DE10-3378 | KP771895 | Vesicle fluid | Not known | Zoster |
| DE10-4367 | KP771896 | Vesicle fluid | Not known | Zoster |
| DE10-4582 | KP771897 | Vesicle fluid | Not known | Zoster |
| DE10-5454 | KP771898 | Vesicle fluid | Not known | Zoster |
| DE10-567 | KP771899 | Vesicle fluid | Not known | Zoster |
| DE10-581 | KP771900 | Vesicle fluid | Not known | Zoster |
| KV6-2313 | KP771901 | CSF | Not known | Encephalitis |
| KV6-3127 | KP771902 | CSF | Not known | Encephalitis |
| KV8-1390 | KP771903 | CSF | Not known | Encephalitis |
| Zos/Cli/Ves/NIG/9 | KP771924 | Vesicle fluid | Not known | Zoster |
| Cli/UK/CSF/2909/2011 | KP771889 | CSF | 29/09/11 | Encephalitis |
| Cli/UK/CSF/3009/2011 | KP771890 | CSF | 30/09/11 | Encephalitis |
| Zos/Cli/CSF/SING/1008/2008 | KP771922 | CSF | 10/08/08 | Zoster and cerebellitis |
| Var/Cli/Ves/SING/1308/2008 | KP771919 | Vesicle fluid | 13/08/08 | Varicella |
| Var/Cli/Ves/GER/31/2005 | KP771915 | Vesicle fluid | Not known | Varicella |
| Var/Cli/Ves/GER/43/2006 | KP771916 | Vesicle fluid | Not known | Varicella |
| Var/Cli/Ves/ITA/51/2006 | KP771918 | Vesicle fluid | Not known | Varicella |
| Var/Cli/Ves/GER/63/2006 | KP771917 | Vesicle fluid | Not known | Varicella |
| Var/Cli/Ves/FRA/98/2013 | KP771914 | Vesicle | Not known | Varicella |
BAL, bronchoalveolar lavage.
Dates are in the format day/month/year.
RESULTS
Phylogenetic analysis.
We sequenced the complete genomes of 37 clinical strains (Table 1) isolated from patients in Europe and Singapore. We also accessed publicly available complete VZV genomes from GenBank. In total, 115 complete VZV genomes were aligned and analyzed. All regions containing gaps or ambiguous sites were excluded prior to any analysis.
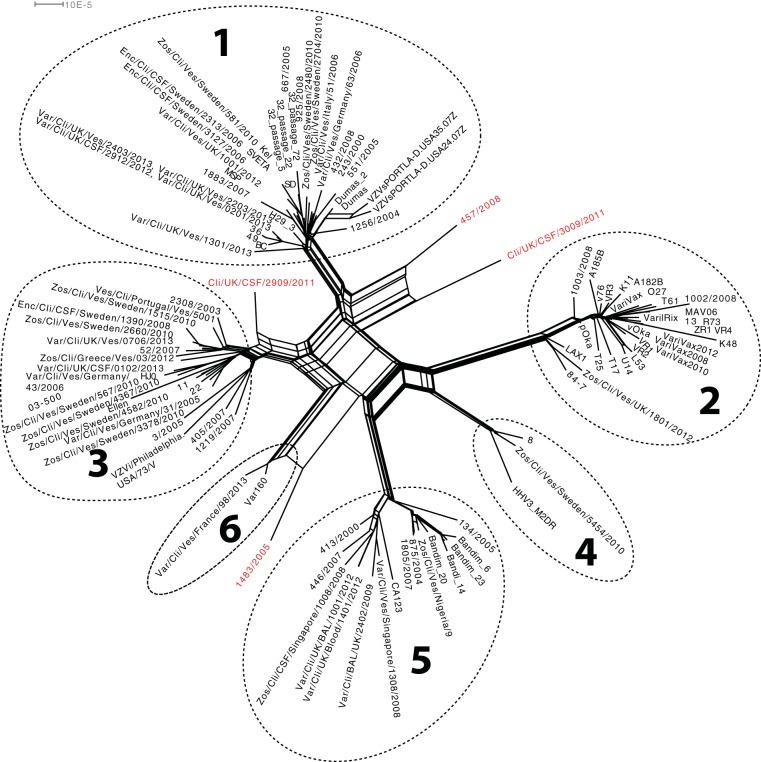
A phylogenetic network based on all strains was initially constructed. The network demonstrated a divergence into the previously described five clades designated 1 to 5 (15–17) but also showed six strains that did not cluster to any of these clades (Fig. 2.). One of the strains sequenced here, Var/Cli/Ves/France/98/2013, and strain Var160 available from GenBank clustered together and therefore fulfilled the requirements for clade designation agreed on during the VZV nomenclature meeting in 2008 (17). We designated this clade 6. Furthermore, strains Cli/UK/CSF/2909/2011 and Cli/UK/CSF/3009/2011 clustered separately as two novel clade candidates. Strains 1483/2005 and 457 also clustered separately as clade candidates as suggested previously (29).
FIG 2.
Phylogenetic network based on complete genomes of all 115 VZV strains. The strains cluster in the previously described clades 1 to 5 and a novel sixth clade, and four outliers are classified as putative interclade recombinants or novel clades (in red).
Interclade recombination.
Interclade recombinants are defined as recombinant strains containing two or more genomic segments that phylogenetically cluster closely to two or more clades, respectively, i.e., a recombinant strain that has descended from parental strains from different phylogenetic clades. If there are recombination crossovers that are present in all strains in a specific clade, we assume that these crossovers are descended from an ancient recombination event that affected a common ancestor to all strains in that clade. These recombination events are therefore classified as ancient (since the recombination event occurred prior to the divergence of that clade).
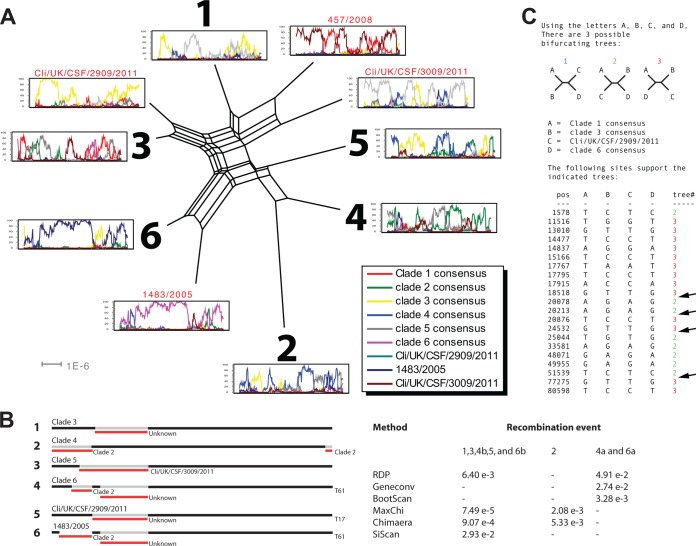
In the phylogenetic network based on all strains, there are four outliers (highlighted in red in Fig. 2) that we consider to be either interclade recombinant candidates or single representatives of novel VZV clades. To further resolve the evolutionary history of VZV and to reveal ancient interclade recombination events, we created a consensus strain for each clade. Owing to the geographical separation of strains, there might be a sampling bias favoring strains from certain clades. Here, all clades or clade candidates are treated equally to resolve the extent of interclade recombination of VZV. A new phylogenetic network was created based on the consensus sequences from all strains within each of the six clades and the four outliers. In total, 10 sequences were included in the analysis. We also performed a bootscan analysis to detect and illustrate recombination crossovers. The phi-test for statistical analysis of recombination was applied on the data set, and the informative site analysis was applied on the recent interclade recombinant candidate strain Cli/UK/CSF/2909/2011.
The topology of the network (Fig. 3A) resembles that of the network including all strains and suggests conflicting phylogenetic signals. The topology is star shaped, which is typical when there has been an evolutionary history with an exponential population growth and/or massive recombination. Furthermore, the parallel internal branches illustrate phylogenetic incongruences, which is typical when there are several recombination events and/or convergent evolution. The phi-test demonstrated high statistical significance for recombination in favor over convergent evolution (P = 0.00), and the bootscan analysis demonstrated highly shifting phylogenetic support indicating numerous recombination crossovers (Fig. 3A). The RDP4 program detected three distinct recombination events in six recombinant clades (clades 3, 4, 5, and 6 and strains Cli/UK/CSF/2909/2011 and 1483/2005) (Fig. 3B). Although the bootscan analysis of interclade recombinant candidate strain Cli/UK/CSF/2909/2011 did not demonstrate convincing evidence for recombination, the informative site analysis of the same strain demonstrated segmented differences in phylogenetic support (Fig. 3C).
FIG 3.
Phylogenetic and interclade recombination analysis. (A) Phylogenetic network based on the consensus sequence of all clades and the four outliers. A bootscan analysis was based on all genomes with respective sequence as query to illustrate shifts in phylogenetic topologies. (B) Recombinant clades suggested by the RDP program and statistical support yielded by each method for respective recombination event. (C) Simplot analysis of arbitrarily selected putative recombinant strains to illustrate phylogenetic incongruences. Arrows highlight recombination crossovers supported by two or more mutations on each side of the crossover.
Intraclade recombination.
An intraclade recombinant is defined as a recombinant strain in a specific clade that has descended from parental strains from that same clade. All intraclade recombinants are, by definition, regarded as recent recombinant strains (since the recombination event occurred after the divergence of that clade).
To estimate the frequency of intraclade recombination events of VZV, we independently analyzed strains from each clade harboring four or more strains, e.g., clades 1, 2, 3, and 5 (most methods require at least four strains in order to analyze recombination events). First, five phylogenetic networks were constructed based on the strains from each clade, respectively, to detect and illustrate the presence of conflicting phylogenetic signals. Then, a phi-test for statistical analysis of recombination was applied to each data set to determine whether there was statistical support for recombination within each clade. In a further attempt to detect and classify intraclade recombination events with statistical significance, we applied the methods RDP, Geneconv, Chimaera, MaxChi, BootScan, SisScan, 3Seq, and LARD implemented in the RDP4 program on each sequence. Finally, we analyzed the distribution of conflicting sites among selected strains using the informative site analysis tool implemented in the SimPlot program. The informative sites analysis also made it possible to estimate the frequency of recombinant candidates, (i.e., strains with conflicting phylogenetic signals but too few SNPs that differ between the putative parental strains in order to obtain a statistically significant measure of recombination).
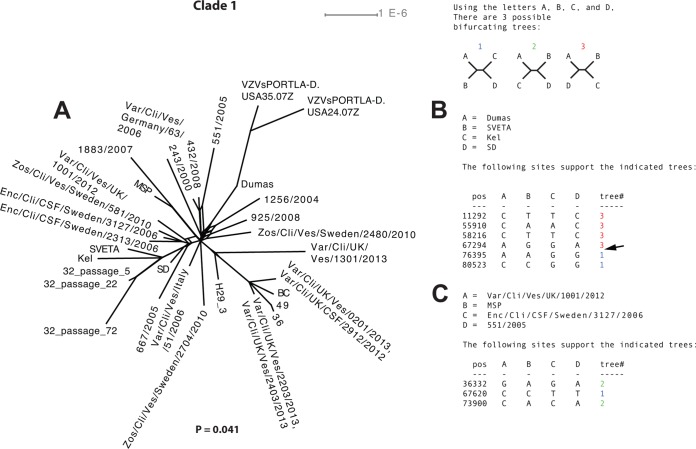
Recombination analysis of strains within clade 1.
The network based on clade 1 strains demonstrated a few internal parallel edges suggesting phylogenetic incongruences (Fig. 4A). The existence of recombinant strains was statistically supported (P = 0.041) by the phi-test, and putative recombination crossovers were detected by using the informative sites analysis tool (Fig. 4B and C). In contrast, the RDP4 program was unable to detect any recombination events among the clade 1 strains with statistical significance.
FIG 4.
Intraclade recombination analysis of clade 1. (A) Phylogenetic network based on the complete genomes of strains from clade 1. (B) Simplot analysis of arbitrarily selected putative recombinant strains from clade 1 to illustrate phylogenetic incongruences. The arrow highlights a recombination crossover supported by two or more mutations on each side of the crossover.
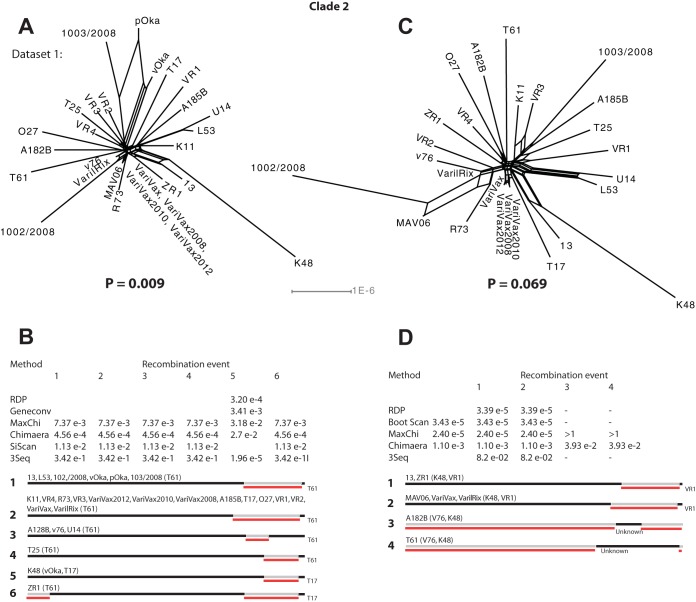
Recombination analysis of strains within clade 2.
The sequences of strains pOka and vOka from GenBank contain several ambiguous sites (denoted “N,” “Y,” etc.). As a precaution, all these sites were excluded prior to the analysis, resulting in a data set (denoted data set 1) with less information. In order to analyze additional information in the other strains, we also analyzed the original data set excluding strains pOka and vOka (denoted data set 2), i.e., two data sets were therefore analyzed in parallel for strains within clade 2.
The phylogenetic networks based on both data sets contained parallel branches demonstrating conflicting phylogenetic signals (Fig. 5A and C). The phi-test also provided high statistical significance (P = 0.018) for recombination among strains in data set 1 but not for data set 2 (P = 0.069). The RDP4 program detected several recombinant strains and recombination crossovers in both data sets (Fig. 5B and D), and recombination was statistically supported by the methods RDP, BootScan, MaxChi, Chimaera, and 3Seq. In addition, at least five recombinants and eight recombination crossovers were also detected and clearly visualized by the informative sites analysis tool (Fig. 6A to E).
FIG 5.
Intraclade recombination analysis of clade 2. (A) Phylogenetic network based on the complete genomes of strains from clade 2, excluding sites that were ambiguous in the GenBank sequences for vOka and pOka. (B) Recombinant strains among the data set analyzed in panel A suggested by the RDP program and statistical support for yielded by each method for respective recombination event. (C) Phylogenetic network based on all strains in clade 2 except for pOka and vOka. (D) Recombinant strains among the data set analyzed in panel C suggested by the RDP program and statistical support yielded by each method for respective recombination event.
FIG 6.

(A to D) Simplot analysis of arbitrarily selected putative recombinant strains from clade 2 to illustrate phylogenetic incongruences. Arrows highlight recombination crossovers supported by two or more mutations on each side of the crossover. Sites that were ambiguous in the GenBank sequences for vOka and pOka were excluded prior to the analysis.
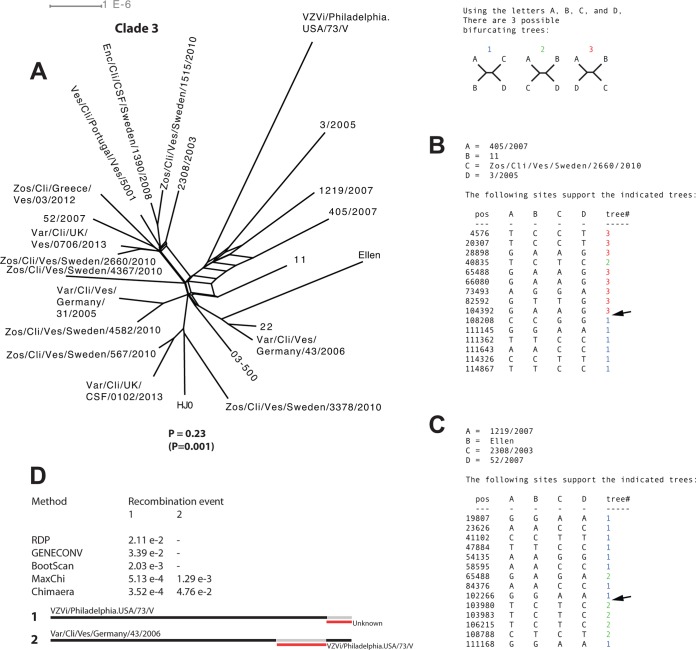
Recombination analysis of strains within clade 3.
Phylogenetic incongruences were clearly illustrated in the clade 3 network, but the phi-test showed that these were not statistically significant (P = 0.23) for recombination when all strains were analyzed (Fig. 7A). However, when putative nonrecombinants were manually identified and excluded from the analysis, the phi-test did show statistically significant (P = 0.001) signals for recombination. At least two recombinants and two recombination crossovers were independently detected by using the informative sites analysis tool (Fig. 7B and C) and the RDP4 program (Fig. 7D), respectively.
FIG 7.
Intraclade recombination analysis of clade 3. (A) Phylogenetic network based on the complete genomes of strains from clade 3. (B and C) Simplot analysis of arbitrarily selected putative recombinant strains from clade 3 to illustrate phylogenetic incongruences. Arrows highlight recombination crossovers supported by two or more mutations on each side of the crossover. (D) Recombinant strains from clade 3 suggested by the RDP program and statistical support yielded by each method for the respective recombination event.
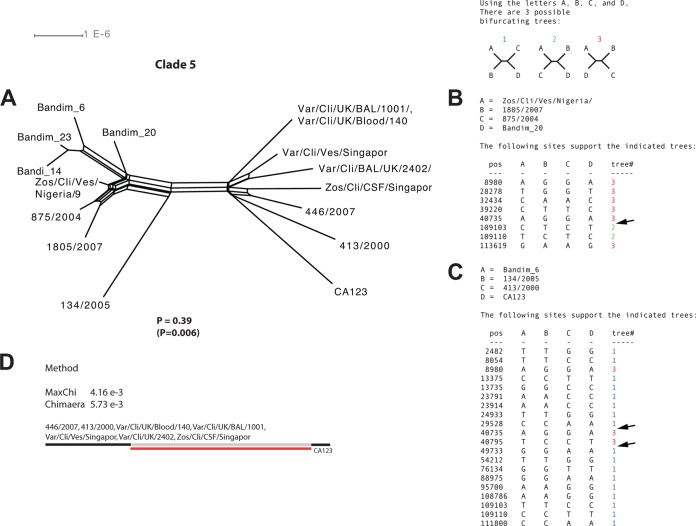
Recombination analysis of strains within clade 5.
The analysis of clade 5 strains was similar to that of clade 3 strains. The network demonstrated internal parallel branches illustrating phylogenetic incongruences (Fig. 8A). The phi-test again resulted in statistically significant recombination when putative nonrecombinants were manually identified and excluded from the analysis (P = 0.006) but not when all strains were analyzed (P = 0.39). The informative sites analysis tool identified at least two different recombination events (Fig. 8B and C), and the methods MaxChi and Chimaera implemented in the RDP4 program identified one recombination event with statistical significance (Fig. 8D).
FIG 8.
Intraclade recombination analysis of clade 5. (A) Phylogenetic network based on the complete genomes of strains from clade 5. (B and C) Simplot analysis of arbitrarily selected putative recombinant strains from clade 5 to illustrate phylogenetic incongruences. Arrows highlight recombination crossovers supported by two or more mutations on each side of the crossover. (D) Recombinant strains from clade 5 suggested by the RDP program and statistical support yielded by each method for respective recombination event.
DISCUSSION
Here, we have demonstrated that homologous recombination occurs frequently between wild-type VZV strains. We have identified putative novel interclade recombinants and well-supported frequent intraclade recombination within clades 1, 2, 3, and 5.
The VZV genome is significantly more conserved than those of HSV-1 and HSV-2. Excluding the variable repeat regions, there are approximately 30 to 200 SNP differences between any two VZV strains (60). DNA data from recombinant and parental strains may thus contain too few SNPs differing between the parental strains, resulting in no or only few parental-specific nucleotides transferred to the recombinant strains. It will hence be difficult to detect recombination events between similar parental strains or to classify them with high statistical significance. It is therefore likely that many of the strains analyzed here that contain conflicting phylogenetic signals but have putative parental strains that are too closely related to yield statistical significance for recombination are indeed true recombinants. Thus, the frequency of recombination is potentially even higher for VZV than reported here.
Little is known about when, how, and where recombination occurs, other than that recombination requires the simultaneous replication of two or more distinct viruses in the same cell. Recombination thus requires multiple infections with two or more distinct strains. It is, however, uncertain if such multiple infections occur through primary infections with heterogeneous virus populations, through simultaneous multiple primary infections from different sources, or by reinfections later in life, scenarios that all are probable. Furthermore, while a recombinant virus may be produced within a person, it is uncertain if the recombination event happens directly after infection or during latency or reactivation or what kind of mechanism may trigger recombination. Interestingly, two of the few putative interclade recombinants detected here (Cli/UK/CSF/3009/2011 and Cli/UK/CSF/2909/2011) were collected from cerebrospinal fluid (CSF) from patients with encephalitis. It may therefore be speculated that central nervous system (CNS) infections facilitate the replication of different viruses in the same cell or vice versa. Although this cannot explain all the recombination events of VZV, further studies on VZV strains collected from CSF may be warranted to better understand the underlying mechanisms of VZV recombination and of VZV CNS infections.
Clade 2, which harbors the vaccine strains, showed evidence of multiple intraclade recombination events. Although ambiguities remain about which strains are parental and which are recombinants, our data suggest that either the original vaccine strains and/or the vaccine strains isolated from lesions are intraclade recombinants. It is therefore likely that the vaccine strains have already been involved in recombination events, either in vivo or in vitro. The vaccine preparation has previously been shown to be heterogeneous (49, 61) and thus may have recombined during passaging in vitro or within the host following inoculation. In any case, it is only the detection of interclade vaccine/wild-type recombinants that can reliably reveal in vivo recombination between vaccine and wild-type strains after vaccination, since no strains from other clades are present in the vaccine batches (49).
However, vaccination with Zostavax will undoubtedly result in individuals infected with multiple strains, since most adults are already carrying latent VZV strains. Since Zostavax is based on clade 2 strains, most vaccinated persons living outside Asia (where clade 2 is predominant) will most likely also be carrying multiple strains from different clades. Despite thorough analysis, we did not, however, detect any interclade vaccine/wild-type recombinants. These results may suggest that the probability of interclade VZV vaccine/wild-type recombinants emerging is low, but this may not necessarily be the case. First, we have analyzed only a limited number of strains (n = 115). Second, as mentioned above, it is unknown when and where the actual recombination event occurs. It may thus take some time after vaccination before a recombinant emerges and before we can detect it. The best chances of detection of vaccine/wild-type recombinants may therefore be to analyze reactivating zoster in elderly patients vaccinated with Zostavax, which makes an interesting avenue for further studies.
One intriguing question is when the recombination events detected here occurred. Unfortunately, exact dating of recombination events is troublesome and requires knowledge about the parental strains involved in the process. Assuming we actually had that knowledge, the different segments in the recombinant strains retrieved from respective parental strain could be identified and compared with the corresponding regions in the recombinant strains. This would then allow us to estimate the number of substitutions that have taken place since the recombination event. Estimations of the molecular clock rate (substitutions/site/year) could then be used in combination with the number of SNPs to calculate the number of years since the recombination event. However, in this case we have no knowledge of the parental strains of the recombinants. At best, we might have DNA data from strains closely related to the parental strains, i.e., that are similar to the parental strains but differ from them by several SNPs. If we assumed that these are the true parental strains, these SNPs would mistakenly be interpreted to have originated after the recombination event. This would result in the predicted date of recombination being older than the real date. Due to these circumstances and given the uncertainties of the molecular clock rate, we have therefore not made any attempts to date single recombination events in this study, other than classifying them as recent or ancient based on the relation to clade divergence.
Several attempts have been made to distinguish recombinant clades from parental clades, i.e., which clades descend from recombinants and which are not recombinants (14–16). However, these kinds of analyses require assumptions on the total global genetic divergence and the exclusion of the possibility of other existing clades. Since we have only 115 genomes and the number of defined clades has increased as more strains are sequenced, such assumptions are likely to be erroneous. Furthermore, the recombination and parental history become increasingly complicated as more clades are included, and we cannot exclude that all clades have been involved in recombination events at some occasion. We therefore avoided distinguishing between parental and recombinant clades in the present study, but we conclude that multiple ancient interclade recombination events have occurred in the evolutionary history of VZV. We also suggest that the complexity of the recombination crossover patterns will increase as more strains are sequenced and more clades are discovered. Future studies that produce whole-genome sequences for strains from various geographic regions may shed further light on these issues.
It was suggested during the VZV nomenclature meeting in 2008 (17) that the five major VZV clades should be designated 1 to 5 and that novel strains should be designated members of an existing clade, based on phylogenetic clustering. The intention was also to introduce criteria so that each existing clade and future novel clades should have at least two whole genomes sequenced to be considered true clades. Although additional clades have been suggested (29, 62, 63), only clades 1 to 5 have hitherto fulfilled this requirement. One strain sequenced here (Var/Cli/Ves/France/98/2013), however, clusters phylogenetically with another strain (Var160; GenBank accession number KC112914.1), separately from strains from clades 1 to 5. The requirements for a sixth clade are therefore now fulfilled, and we designate this clade 6. Interestingly, these two strains that form clade 6 were collected in widely separated geographic regions. Strain Var160 was isolated from vesicle fluid from a 7-year-old boy in Mexico in 2007. Strain Var/Cli/Ves/France/98/2013, on the other hand, was isolated from vesicle fluid from a 2-year-old girl with chicken pox in France in 2013. This finding indicates that clade 6 may have a geographic distribution similar to that of clades 1 and 3, which are predominant in Europe and North America, although additional isolates are required to define its predominant geographic locations.
The high frequency of recombination detected here further supports previous suggestions that interclade recombination of VZV could dramatically increase in the future (22). Increased migration and the introduction of live vaccine strains from clade 2 in geographical regions, where strains from other clades are predominant, will undoubtedly erase previous geographical barriers between clades. The rates of interclade recombination should also increase as the geographic barriers diminish, and it is possible that most VZV strains will eventually have mosaic recombinant genomes similar to those in HSV-1 (22, 23). If so, a challenging question will be how to define a VZV clade in the future. It will be highly impractical to continue with the clade designation on a complete-genome level, since every unique interclade recombinant will cluster separately in a phylogenetic tree or network, as described for HSV-1 (22). Although it may take several decades before the majority of VZV strains are mosaic recombinants, the results from this study suggest that several wild-type recombinant strains have already emerged. Collaborative discussions about future VZV nomenclature, classification, and genotyping are thus important in the near future.
ACKNOWLEDGMENTS
We acknowledge the infrastructure support provided by MRC Centre for Molecular Medical Virology grant G0900950 and by the NIHR UCL/UCLH Biomedical Research Centre and the use of the UCL Legion High Performance Computing Facility and associated support services in the completion of this work. The work was supported by MRC grant G0700814, an MRF New Investigator Award for D.P.D., and the NIHR UCL/UCLH Biomedical Research Centre for S.K. and J.B. This work was also supported by the Swedish Medical Research Council (grant number 521-2011-3297 to T.B.) and the LUA-ALF Foundations of Sahlgren's University Hospital (grant number 145-841 to T.B.).
REFERENCES
- 1.Wharton M. 1996. The epidemiology of varicella-zoster virus infections. Infect Dis Clin North Am 10:571–581. doi: 10.1016/S0891-5520(05)70313-5. [DOI] [PubMed] [Google Scholar]
- 2.Hope-Simpson RE. 1965. The nature of herpes zoster: a long-term study and a new hypothesis. Proc R Soc Med 58:9–20. [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom T. 1996. Polymerase chain reaction for diagnosis of varicella zoster virus central nervous system infections without skin manifestations. Scand J Infect Dis Suppl 100:41–45. [PubMed] [Google Scholar]
- 4.Echevarria JM, Casas I, Tenorio A, de Ory F, Martinez-Martin P. 1994. Detection of varicella-zoster virus-specific DNA sequences in cerebrospinal fluid from patients with acute aseptic meningitis and no cutaneous lesions. J Med Virol 43:331–335. doi: 10.1002/jmv.1890430403. [DOI] [PubMed] [Google Scholar]
- 5.Frenos S, Galli L, Chiappini E, de Martino M. 2007. An increasing incidence of chickenpox central nervous system complications in children: what's happening in Tuscany? J Clin Virol 38:358–361. doi: 10.1016/j.jcv.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 6.Gilden DH, Wright RR, Schneck SA, Gwaltney JM Jr, Mahalingam R. 1994. Zoster sine herpete, a clinical variant. Ann Neurol 35:530–533. doi: 10.1002/ana.410350505. [DOI] [PubMed] [Google Scholar]
- 7.Koskiniemi M, Piiparinen H, Rantalaiho T, Eranko P, Farkkila M, Raiha K, Salonen EM, Ukkonen P, Vaheri A. 2002. Acute central nervous system complications in varicella zoster virus infections. J Clin Virol 25:293–301. doi: 10.1016/S1386-6532(02)00020-3. [DOI] [PubMed] [Google Scholar]
- 8.Persson A, Bergstrom T, Lindh M, Namvar L, Studahl M. 2009. Varicella-zoster virus CNS disease—viral load, clinical manifestations and sequels. J Clin Virol 46:249–253. doi: 10.1016/j.jcv.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M, Otsuka T, Okuno Y, Asano Y, Yazaki T. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288–1290. [DOI] [PubMed] [Google Scholar]
- 10.Oxman MN, Levin MJ, Johnson GR, Schmader KE, Straus SE, Gelb LD, Arbeit RD, Simberkoff MS, Gershon AA, Davis LE, Weinberg A, Boardman KD, Williams HM, Zhang JH, Peduzzi PN, Beisel CE, Morrison VA, Guatelli JC, Brooks PA, Kauffman CA, Pachucki CT, Neuzil KM, Betts RF, Wright PF, Griffin MR, Brunell P, Soto NE, Marques AR, Keay SK, Goodman RP, Cotton DJ, Gnann JW Jr, Loutit J, Holodniy M, Keitel WA, Crawford GE, Yeh SS, Lobo Z, Toney JF, Greenberg RN, Keller PM, Harbecke R, Hayward AR, Irwin MR, Kyriakides TC, Chan CY, Chan IS, Wang WW, Annunziato PW, Silber JL. 2005. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med 352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- 11.European Medicines Agency. 2013. Zostavax (shingles [herpes zoster] vaccine [live]): EU summary of product characteristics. European Medicines Agency, London, United Kingdom. [Google Scholar]
- 12.Norberg P. 2010. Divergence and genotyping of human alpha-herpesviruses: an overview. Infect Genet Evol 10:14–25. doi: 10.1016/j.meegid.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Davison AJ, Scott JE. 1986. The complete DNA sequence of varicella-zoster virus. J Gen Virol 67(Part 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 14.McGeoch DJ. 2009. Lineages of varicella-zoster virus. J Gen Virol 90:963–969. doi: 10.1099/vir.0.007658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norberg P, Liljeqvist JA, Bergstrom T, Sammons S, Schmid DS, Loparev VN. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J Virol 80:9569–9576. doi: 10.1128/JVI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters GA, Tyler SD, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J Virol 80:9850–9860. doi: 10.1128/JVI.00715-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breuer J, Grose C, Norberg P, Tipples G, Schmid DS. 2010. A proposal for a common nomenclature for viral clades that form the species varicella-zoster virus: summary of VZV Nomenclature Meeting 2008, Barts and the London School of Medicine and Dentistry, 24-25 July 2008. J Gen Virol 91:821–828. doi: 10.1099/vir.0.017814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowden R, Sakaoka H, Donnelly P, Ward R. 2004. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect Genet Evol 4:115–123. doi: 10.1016/j.meegid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Bowden R, Sakaoka H, Ward R, Donnelly P. 2006. Patterns of Eurasian HSV-1 molecular diversity and inferences of human migrations. Infect Genet Evol 6:63–74. doi: 10.1016/j.meegid.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Norberg P, Bergstrom T, Liljeqvist JA. 2006. Genotyping of clinical herpes simplex virus type 1 isolates by use of restriction enzymes. J Clin Microbiol 44:4511–4514. doi: 10.1128/JCM.00421-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norberg P, Bergstrom T, Rekabdar E, Lindh M, Liljeqvist JA. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol 78:10755–10764. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norberg P, Tyler S, Severini A, Whitley R, Liljeqvist JA, Bergstrom T. 2011. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PLoS One 6:e22527. doi: 10.1371/journal.pone.0022527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J Virol 88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norberg P, Kasubi MJ, Haarr L, Bergstrom T, Liljeqvist JA. 2007. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J Virol 81:13158–13167. doi: 10.1128/JVI.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlivan M, Sengupta N, Breuer J. 2009. A case of varicella caused by co-infection with two different genotypes of varicella-zoster virus. J Clin Virol 44:66–69. doi: 10.1016/j.jcv.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Quinlivan M, Hawrami K, Barrett-Muir W, Aaby P, Arvin A, Chow VT, John TJ, Matondo P, Peiris M, Poulsen A, Siqueira M, Takahashi M, Talukder Y, Yamanishi K, Leedham-Green M, Scott FT, Thomas SL, Breuer J. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J Infect Dis 186:888–894. doi: 10.1086/344228. [DOI] [PubMed] [Google Scholar]
- 27.Dohner DE, Adams SG, Gelb LD. 1988. Recombination in tissue culture between varicella-zoster virus strains. J Med Virol 24:329–341. doi: 10.1002/jmv.1890240310. [DOI] [PubMed] [Google Scholar]
- 28.Muir WB, Nichols R, Breuer J. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J Virol 76:1971–1979. doi: 10.1128/JVI.76.4.1971-1979.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zell R, Taudien S, Pfaff F, Wutzler P, Platzer M, Sauerbrei A. 2012. Sequencing of 21 varicella-zoster virus genomes reveals two novel genotypes and evidence of recombination. J Virol 86:1608–1622. doi: 10.1128/JVI.06233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaRussa PS, Gershon AA. 2001. Biologic and geographic differences between vaccine and clinical varicella-zoster virus isolates. Arch Virol Suppl 2001:41–48. doi: 10.1007/978-3-7091-6259-0_5. [DOI] [PubMed] [Google Scholar]
- 31.Loparev VN, Gonzalez A, Deleon-Carnes M, Tipples G, Fickenscher H, Torfason EG, Schmid DS. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J Virol 78:8349–8358. doi: 10.1128/JVI.78.15.8349-8358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauerbrei A, Wutzler P. 2007. Different genotype pattern of varicella-zoster virus obtained from patients with varicella and zoster in Germany. J Med Virol 79:1025–1031. doi: 10.1002/jmv.20879. [DOI] [PubMed] [Google Scholar]
- 33.Sauerbrei A, Zell R, Philipps A, Wutzler P. 2008. Genotypes of varicella-zoster virus wild-type strains in Germany. J Med Virol 80:1123–1130. doi: 10.1002/jmv.21178. [DOI] [PubMed] [Google Scholar]
- 34.Holmes EC, Worobey M, Rambaut A. 1999. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol 16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- 35.Seligman SJ, Gould EA. 2004. Live flavivirus vaccines: reasons for caution. Lancet 363:2073–2075. doi: 10.1016/S0140-6736(04)16459-3. [DOI] [PubMed] [Google Scholar]
- 36.Norberg P, Roth A, Bergstrom T. 2013. Genetic recombination of tick-borne flaviviruses among wild-type strains. Virology 440:105–116. doi: 10.1016/j.virol.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Becher P, Orlich M, Thiel HJ. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J Virol 75:6256–6264. doi: 10.1128/JVI.75.14.6256-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuervo NS, Guillot S, Romanenkova N, Combiescu M, Aubert-Combiescu A, Seghier M, Caro V, Crainic R, Delpeyroux F. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J Virol 75:5740–5751. doi: 10.1128/JVI.75.13.5740-5751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahourou G, Guillot S, Le Gall O, Crainic R. 2002. Genetic recombination in wild-type poliovirus. J Gen Virol 83:3103–3110. http://vir.sgmjournals.org/content/83/12/3103.long. [DOI] [PubMed] [Google Scholar]
- 40.Liu HM, Zheng DP, Zhang LB, Oberste MS, Kew OM, Pallansch MA. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant polioviruses in China. J Virol 77:10994–11005. doi: 10.1128/JVI.77.20.10994-11005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong YL, Padhi A, Hudson PJ, Poss M. 2010. The effect of vaccination on the evolution and population dynamics of avian paramyxovirus-1. PLoS Pathog 6:e1000872. doi: 10.1371/journal.ppat.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camus-Bouclainville C, Gretillat M, Py R, Gelfi J, Guerin JL, Bertagnoli S. 2011. Genome sequence of SG33 strain and recombination between wild-type and vaccine myxoma viruses. Emerg Infect Dis 17:633–638. doi: 10.3201/eid1704.101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, Baoli S, Yingzuo B. 2012. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol 86:9543. doi: 10.1128/JVI.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dangler CA, Henderson LM, Bowman LA, Deaver RE. 1993. Direct isolation and identification of recombinant pseudorabies virus strains from tissues of experimentally co-infected swine. Am J Vet Res 54:540–545. [PubMed] [Google Scholar]
- 45.Lee SW, Markham PF, Coppo MJ, Legione AR, Markham JF, Noormohammadi AH, Browning GF, Ficorilli N, Hartley CA, Devlin JM. 2012. Attenuated vaccines can recombine to form virulent field viruses. Science 337:188. doi: 10.1126/science.1217134. [DOI] [PubMed] [Google Scholar]
- 46.Devlin JM, Hartley CA, Gilkerson JR, Coppo MJ, Vaz P, Noormohammadi AH, Wells B, Rubite A, Dhand NK, Browning GF. 2011. Horizontal transmission dynamics of a glycoprotein G deficient candidate vaccine strain of infectious laryngotracheitis virus and the effect of vaccination on transmission of virulent virus. Vaccine 29:5699–5704. doi: 10.1016/j.vaccine.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Lee SW, Devlin JM, Markham JF, Noormohammadi AH, Browning GF, Ficorilli NP, Hartley CA, Markham PF. 2011. Comparative analysis of the complete genome sequences of two Australian origin live attenuated vaccines of infectious laryngotracheitis virus. Vaccine 29:9583–9587. doi: 10.1016/j.vaccine.2011.10.055. [DOI] [PubMed] [Google Scholar]
- 48.Blacker HP, Kirkpatrick NC, Rubite A, O'Rourke D, Noormohammadi AH. 2011. Epidemiology of recent outbreaks of infectious laryngotracheitis in poultry in Australia. Aust Vet J 89:89–94. doi: 10.1111/j.1751-0813.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 49.Depledge DP, Kundu S, Jensen NJ, Gray ER, Jones M, Steinberg S, Gershon A, Kinchington PR, Schmid DS, Balloux F, Nichols RA, Breuer J. 2014. Deep sequencing of viral genomes provides insight into the evolution and pathogenesis of varicella zoster virus and its vaccine in humans. Mol Biol Evol 31:397–409. doi: 10.1093/molbev/mst210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Depledge DP, Palser AL, Watson SJ, Lai IY, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson SJ, Welkers MR, Depledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P. 2013. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 368:20120205. doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lassmann T, Sonnhammer EL. 2005. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrio AM, Lagercrantz E, Sperber GO, Blomberg J, Bongcam-Rudloff E. 2009. Annotation and visualization of endogenous retroviral sequences using the Distributed Annotation System (DAS) and eBioX. BMC Bioinformatics 10(Suppl 6):S18. doi: 10.1186/1471-2105-10-S6-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 57.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 60.Tyler SD, Peters GA, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. 2007. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359:447–458. doi: 10.1016/j.virol.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 61.Gomi Y, Imagawa T, Takahashi M, Yamanishi K. 2000. Oka varicella vaccine is distinguishable from its parental virus in DNA sequence of open reading frame 62 and its transactivation activity. J Med Virol 61:497–503. doi:. [DOI] [PubMed] [Google Scholar]
- 62.Loparev V, Martro E, Rubtcova E, Rodrigo C, Piette JC, Caumes E, Vernant JP, Schmid DS, Fillet AM. 2007. Toward universal varicella-zoster virus (VZV) genotyping: diversity of VZV strains from France and Spain. J Clin Microbiol 45:559–563. doi: 10.1128/JCM.01738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loparev VN, Rubtcova EN, Bostik V, Tzaneva V, Sauerbrei A, Robo A, Sattler-Dornbacher E, Hanovcova I, Stepanova V, Splino M, Eremin V, Koskiniemi M, Vankova OE, Schmid DS. 2009. Distribution of varicella-zoster virus (VZV) wild-type genotypes in northern and southern Europe: evidence for high conservation of circulating genotypes. Virology 383:216–225. doi: 10.1016/j.virol.2008.10.026. [DOI] [PubMed] [Google Scholar]