ABSTRACT
Neuraminidase (NA), an influenza virus envelope glycoprotein, removes sialic acid from receptors for virus release from infected cells. For this study, we used a baculovirus-insect cell expression system to construct and purify recombinant NA (rNA) proteins of H5N1 (A/Vietnam/1203/2004) and pandemic H1N1 (pH1N1) (A/Texas/05/2009) influenza viruses. BALB/c mice immunized with these proteins had high titers of NA-specific IgG and NA-inhibiting (NI) antibodies against H5N1, pH1N1, H3N2, and H7N9 viruses. H5N1 rNA immunization resulted in higher quantities of NA-specific antibody-secreting B cells against H5N1 and heterologous pH1N1 viruses in the spleen. H5N1 rNA and pH1N1 rNA immunizations both provided complete protection against homologous virus challenges, with H5N1 rNA immunization providing better protection against pH1N1 virus challenges. Cross-reactive NI antibodies were further dissected via pH1N1 rNA protein immunizations with I149V (NA with a change of Ile to Val at position 149), N344Y, and I365T/S366N NA mutations. The I365T/S366N mutation of pH1N1 rNA enhanced cross-reactive NI antibodies against H5N1, H3N2, and H7N9 viruses. It is our hope that these findings provide useful information for the development of an NA-based universal influenza vaccine.
IMPORTANCE Neuraminidase (NA) is an influenza virus enzymatic protein that cleaves sialic acid linkages on infected cell surfaces, thus facilitating viral release and contributing to viral transmission and mucus infection. In currently available inactivated or live, attenuated influenza vaccines based on the antigenic content of hemagglutinin proteins, vaccine efficacy can be contributed partly through NA-elicited immune responses. We investigated the NA immunity of different recombinant NA (rNA) proteins associated with pH1N1 and H5N1 viruses. Our results indicate that H5N1 rNA immunization induced more potent cross-protective immunity than pH1N1 rNA immunization, and three mutated residues, I149V, I365T, and S366N, near the NA enzyme active site(s) are linked to enhanced cross-reactive NA-inhibiting antibodies against heterologous and heterosubtypic influenza A viruses. These findings provide useful information for the development of an NA-based universal influenza vaccine.
INTRODUCTION
Members of the Orthomyxoviridae family, influenza A viruses are enveloped viruses containing a single-strand, 8-segment, negative-sense RNA genome typically encoding 11 or 12 viral proteins (1, 2). Influenza A virus subtypes have been classified based on the antigenic properties of hemagglutinin (HA) and neuraminidase (NA) glycoproteins, designated H1 to H16 and N1 to N9, respectively. Besides these subtypes, H17N10 and H18N11 viruses from fruit bats were also identified (3, 4). According to phylogenetic analyses, subtypes N1 to N9 can be classified as belonging to group 1 (including N1, N4, N5, and N8) or group 2 (including N2, N3, N6, N7, and N9) (5). To date, N1, N2, N7, and N9 subtypes are known to trigger human epidemics (6, 7). N3 and N8 subtypes only cause limited sporadic human infections, as reported for H7N3 in Mexico (8) and Canada (9) and for H10N8 in China (10).
NA, an enzymatic protein with a complex tetrameric structure (11), is capable of cleaving sialic acid linkages on cell surfaces, thereby facilitating viral release from infected cells (12). NA also contributes to viral transmission and infection by destroying decoy receptors on cilia, mucins, and cellular glycocalyx (12–15). NA immunogenicity was first observed in human subjects immunized with an NA-specific inactivated vaccine (16). The use of recombinant NA (rNA) proteins expressed in yeast (17) or insect cells (18) elicits protection against lethal virus challenges in immunized mice. Ferrets immunized with rNA proteins exhibit a distinctive type of protection in addition to that provided by HA immunization alone (19). NA-inhibiting (NI) antibodies are known to limit virus spreading and to mitigate clinical symptoms of influenza A virus infection (19). Mice immunized with a reverse-genetic reassortant H1N1 virus containing seasonal influenza virus NA exhibit cross-reactive NI antibodies and reduced mortality from pandemic H1N1 (pH1N1) virus challenges (20). Live attenuated influenza vaccines for seasonal H1N1, H3N2, and pH1N1 strains have been reported as inducing cross-reactive NI antibodies to H5N1 viruses in ferrets (21), and NI antibodies elicited by a seasonal trivalent influenza vaccine have been reported as providing cross-protective immunity against lethal H5N1 challenges, also in ferrets (22). Furthermore, NA-based virus-like particles (VLPs) containing NA and matrix proteins M1 and M2 have been shown to elicit more potent NI antibodies and to confer cross-protective immunity against H5N1 and pH1N1 viral challenges in mice (23). NI antibodies have also been detected in humans vaccinated with an H5N1 inactivated vaccine (24), as well as in humans exposed to natural infections (25). NA immunogenicity and cross-protection mechanisms remain unclear.
For this study, we used a baculovirus-insect cell expression system to obtain soluble rNA proteins from H5N1 and pH1N1. Two-dose active immunizations in mice were used to evaluate the immunogenicities of H5N1 rNA and pH1N1 rNA. In addition to detecting NA-specific total IgG and IgG subtypes, NI antibodies in sera, and antibody-secreting B cells (ASCs) in splenocytes, we also observed protective immunity following live-virus challenges. Our data indicate that H5N1 rNA and pH1N1 rNA immunizations provided complete protection against homologous virus challenges, with H5N1 rNA immunization eliciting more potent cross-reactive NI antibodies and inducing better protection against heterologous pH1N1 virus challenges. The residues at positions 365 and 366 near the NA enzyme active site were observed associating with enhanced cross-reactive NI antibodies against heterologous and heterosubtypic influenza viruses. It is our hope that these findings provide useful information for the development of an NA-based universal influenza vaccine.
MATERIALS AND METHODS
Recombinant NA protein expression and purification.
cDNA from the NA genes of A/Vietnam/1203/2004 (H5N1) (GI 145284408) and A/Texas/05/2009 (pH1N1) (GI 255602223) was separately synthesized with insect cell-optimized codon sequences from Genomics, Inc. The coding sequences of the NA H5N1 and pH1N1 ectodomains, together with additional N-terminal sequences containing gp67 signal peptides (MLLVNQSHQGFNKEHTSKMVSAIVLYVLLAAAAHSAFA), six-His residues (HHHHHH), tetrameric human vasodilator-stimulated phosphoprotein (hvsp) domains (SSSDYSDLQRVKQELLEEVKKELQKVKEEIIEAFVQELRKRGS), and a thrombin cleavage site (LVPRGS), were cloned into pFastBac expression vectors (26). Next, rNA proteins were produced using a Bac-to-Bac insect cell expression system (Invitrogen) according to the manufacturer's instructions. Briefly, Sf9 cells were infected with recombinant baculoviruses expressing the NA ectodomains of H5N1 and pH1N1 for 48 h prior to collecting supernatants for additional rNA protein purification using nickel-chelated resin affinity chromatography (Tosoh). H5N1 rNA and pH1N1 rNA purity was confirmed by Coomassie blue staining. Anti-His horseradish peroxidase (HRP)-conjugated antibodies (Affymetrix) were used for Western blotting characterization.
Production and purification of H5N1 and H7N9 VLPs.
H5N1 and H7N9 VLPs were produced using the procedures described in reference 27. Briefly, the H5 HA gene of A/Thailand/1(KAN-1)/2004 (H5N1), the H7 HA gene of A/Shanghai/2/2013 (H7N9), and the M1 gene of A/WSN/1933 (H1N1) were cloned into separate pFastBacDual vectors (Invitrogen). The N1 NA gene of A/Vietnam/1203/2004 (H5N1), the N9 NA gene of A/Shanghai/2/2013 (H7N9), and the M2 gene of A/WSN/1933 (H1N1) were cloned into separate vectors. For H5N1 VLP production, Sf9 cells were coinfected with Bac-H5HA-M1 and Bac-N1NA-M2 recombinant baculovirus at a multiplicity of infection (MOI) of 3 or 1, respectively. Culture supernatants were harvested and concentrated 72 h postinfection. VLPs were further purified using a 20% sucrose solution and centrifugation at 33,000 rpm for 3 h. H5N1 VLPs were obtained and stored at 4°C until used for NI assays. For H7N9 VLPs, Sf9 cells were coinfected with Bac-H7HA-M1 and Bac-N9NA-M2 recombinant baculovirus at an MOI of 3 or 1, respectively, for 72 h. The subsequent steps were the same as for H5N1 VLP production.
Mouse immunizations.
BALB/c mice (6 to 8 weeks old) purchased from the Taiwan National Laboratory Animal Center were immunized twice intramuscularly with a 3-week interval with 2 or 20 μg of rNA proteins plus 10 μg CpG and 10% poly(ethylene glycol)-block-poly(lactide-co-epsilon-caprolactone) (PEGb-PLACL), squalene, and Span85 (PELC) emulsion as described in a previous report (28). Serum samples were collected 2 weeks after the second inoculation; splenocytes were harvested and isolated 1 week later. Procedures involving live animals were performed according to guidelines established by the Laboratory Animal Center of National Tsing Hua University (NTHU). All animal use protocols were reviewed and approved by the NTHU Institutional Animal Care and Use Committee (IACUC) (approval no. 10002).
Viral challenges.
For the two-dose immunization strategy, BALB/c mice (6 to 8 weeks old) were placed in one of 5 groups, with each group consisting of 5 mice immunized twice with a 3-week interval with either 2 or 20 μg of H5N1 rNA or pH1N1 rNA proteins plus CpG and PELC or with phosphate-buffered saline (PBS). Three weeks after the second inoculations, all mice were intranasally challenged with 10 50% murine lethal doses (MLD50) of the H5N1 (NIBRG-14 [RG-14]), pH1N1 (A/California/07/2009 [CA/09]), or H7N9 (A/Taiwan/01/2013 [TW/13]) virus. PBS-immunized mice were used as a mock control. Survival rates and body weights were recorded daily for 14 days. For mouse virus challenges, the experimental procedures were reviewed and approved by the IACUC of Academia Sinica, Taiwan. According to IACUC guidelines, a weight loss of 25% or more was established as an endpoint.
Enzyme-linked immunosorbent assays (ELISAs).
Individual wells in 96-well plates were coated with purified rNA proteins (100 μl at 2 μg/ml) and held overnight at 4°C, washed 3 times with PBST (0.05% Tween 20 in PBS), and blocked with blocking buffer (1% bovine serum albumin [BSA] in PBS) for at least 1 h. Next, 100 μl of 2-fold serially diluted serum samples were added and held at room temperature (RT) for 1 h, followed by 3 additional washes with PBST. HRP-conjugated goat anti-mouse IgG antibodies (Bethyl Laboratories, Inc.) were added to each well, the plates were incubated for 1 h, and the wells washed 3 times with PBST. Anti-NA IgG titers were determined by adding TMB substrate (3,3′,5,5′-tetramethyl benzidine; Biolegend), holding for 15 min at RT, and stopping the reaction with 2 N H2SO4. Endpoint titers were determined as the reciprocals of the most-diluted serum concentrations giving a mean optical density (OD) at 450 nm of more than 0.2.
NI assay.
To determine the optimal concentrations of viruses or VLPs for the subsequent NI assays, the NA activities induced by each virus or VLPs as determined from ELISA data were plotted in GraphPad Prism 5.0 and fit to a nonlinear curve as previously described (29). The amounts of each virus or VLP reacted with fetuin and agglutinin resulted in OD values of around 2.0, half the maximal OD value in the ELISAs. Quantities of 1 μg of VLPs (H5N1 or H7N9) or 105 PFU of viruses (pH1N1 or H3N2) were used for NI serological assays. NI antibodies were measured using a previously described fetuin-based assay procedure (30). Briefly, 96-well plates were coated with 50 μg/ml fetuin (Sigma) and held overnight at 4°C before being washed 3 times with PBST and blocked with blocking buffer for 2 h. Twofold serially diluted serum samples in blocking buffer were incubated with equal volumes containing 1 μg VLPs (H5N1 or H7N9) or 105 PFU viruses (pH1N1 or H3N2) for 1 h at 37°C, added to the fetuin-coated plates, held for another 1 h at 37°C, and then washed 3 times with PBST. Peroxidase-labeled peanut agglutinin (100 μl at 2.5 μg/ml) (Sigma) was added to each well, incubated for 1 h at RT, and washed 3 times with PBST. The NA activity levels of viruses (pH1N1 or H3N2) and VLPs (H5N1 or H7N9) were determined by adding TMB substrate (Biolegend), holding for 15 min at RT, and stopping the reaction with 2 N H2SO4. Plates were read with an ELISA reader (Tecan) at an OD of 450 nm. The corresponding 50% inhibitory concentrations (IC50s) were defined as the reciprocal serum dilutions inhibiting 50% of viral NA enzyme activity.
NA-specific antibody-secreting B cells.
Splenocytes were collected from each group of rNA-immunized or PBS-immunized mice 3 weeks after the second inoculations. Multiscreen 96-well filtration plates (Millipore) were coated with rNA proteins (1 μg per well) and incubated overnight at 4°C. Plates were blocked with 200 μl/well of complete RPMI 1640 (10% fetal bovine serum [FBS], 1× penicillin/streptomycin, 1× sodium pyruvate, 1× nonessential amino acids [NEAA], and 100 μM β-mercaptoethanol) and held for 1 h at RT. Splenocytes (2 × 105) diluted in complete RPMI 1640 were added to individual plates and incubated for 48 h at 37°C. After 3 washes with PBST, HRP-conjugated anti-mouse IgG was added to each well and held for 2 h at RT. After 3 PBST and 2 PBS washes, 3-amino-9-ethylcarbazole (AEC) substrate was added to each plate and the plates held at RT for approximately 10 to 60 min before the reactions were stopped with double-distilled water. Immunospots for each immunized group were determined using an enzyme-linked immunosorbent spot assay (ELISPOT) plate reader (CTL, Inc.).
Statistical analysis.
All results were analyzed using two-tailed Student's t tests. Asterisks in the figures indicate statistical significance as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RESULTS
H5N1 rNA and pH1N1 rNA proteins were expressed and purified using a baculovirus-insect cell expression system.
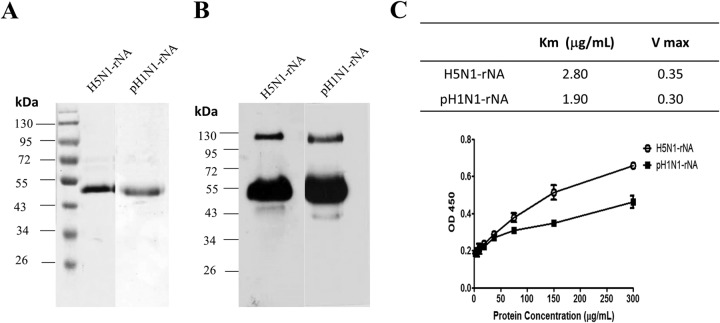
Soluble H5N1 rNA and pH1N1 rNA were produced from the culture supernatants of Sf9 insect cells infected by recombinant baculoviruses and purified using Ni-chelated affinity chromatography, and their production yields were approximately 0.5 mg/liter and 0.25 mg/liter, respectively, with 80 to 90% purity (Fig. 1A). According to the results from SDS-PAGE gels with Coomassie blue staining (Fig. 1A) and Western blotting (Fig. 1B), purified H5N1 rNA and pH1N1 rNA proteins had molecular masses of 53 kDa each. Based on Eadie-Hofstee measurements, the Km values of H5N1 rNA and pH1N1 rNA were 2.80 and 1.90, respectively (Fig. 1C), indicating enzyme activity of insect cell-expressed H5N1 rNA and pH1N1 rNA.
FIG 1.
Expression and characterization of soluble recombinant NA (rNA) proteins. The cDNA coding sequences of NA ectodomains (A/Vietnam/1203/2004 [H5N1] or A/Texas/05/2009 [pH1N1]) with additional N-terminal sequences of 6×His residues and hvsp sequences were constructed and expressed using a baculovirus-insect cell expression system. (A and B) Purified rNA proteins (2 μg) were confirmed by Coomassie blue staining (A) and Western blotting (B). (C) Fetuin-based assays were used to determine rNA protein enzymatic activity.
NA-specific IgG antibodies were induced by H5N1 rNA and pH1N1 rNA immunizations.
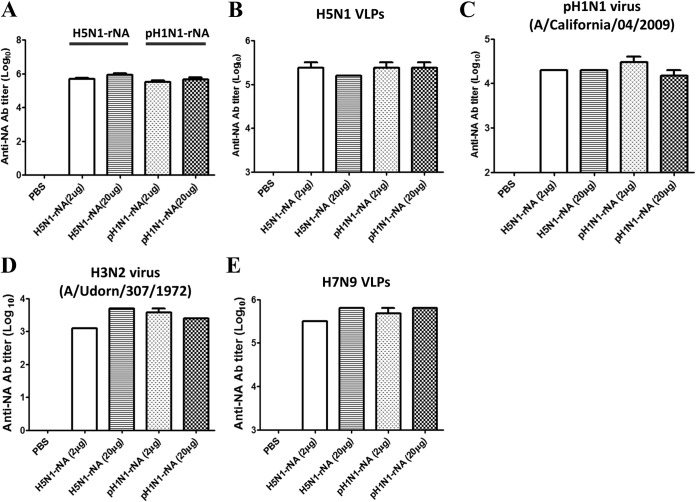
Groups of five female BALB/c mice were immunized intramuscularly with two doses of H5N1 rNA or pH1N1 rNA proteins (2 or 20 μg per dose) with a 3-week interval between doses. Antisera were collected 2 weeks after the second immunization. Data for NA-specific IgG titers against the same immunogens (H5N1 rNAs or pH1N1 rNA proteins) in each group are presented in Fig. 2A. We used an anti-mouse N1 NA-specific antibody as a positive control and PBS-immunized sera as a negative control (NA-specific IgG titers were 103.5 to 104 and undetectable, respectively). As shown by the results in Fig. 2A, mice immunized with 20 μg of H5N1 rNA or pH1N1 rNA exhibited slightly higher NA-specific total IgG titers than mice immunized with 2 μg of H5N1 rNA or pH1N1 rNA. We also performed ELISAs with H5N1 VLPs (Fig. 2B) and the pH1N1 (A/California/04/2009) virus (Fig. 2C). Our data indicate that at either 2 or 20 μg, both H5N1 rNA and pH1N1 rNA cross-reacted with H5N1 VLPs and the pH1N1 virus to induce similar total IgG titers (Fig. 2B and C). Similar cross-reactive results were also found for the H3N2 virus (A/Udorn/307/1972) and H7N9 VLPs (Fig. 2D and E). No significant differences were noted for either IgG1 or IgG2a subclass titers in the same immunization groups. The single exception was significantly greater IgG2a for mice immunized with 20 μg H5N1 rNA (data not shown).
FIG 2.
NA-specific IgG antibodies (Ab) against different viruses induced by rNA immunization. Female BALB/c mice were intramuscularly immunized with 2 or 20 μg of H5N1 rNA or pH1N1 rNA plus 10% PELC and 10 μg CpG adjuvant twice with a 3-week interval; serum samples were collected 14 days after the second inoculation. Briefly, 96-well plates were coated with 2 μg/ml of recombinant N1 NA proteins (A), 10 μg/ml of H5N1 VLPs (B), 20 μg/ml of the pH1N1 virus (C), 20 μg/ml of the H3N2 virus (D), or 10 μg/ml of H7N9 VLPs (E) and held overnight at 4°C. ELISAs with H5N1 rNA or pH1N1 rNA mouse antisera or anti-mouse N1 NA-specific antibodies (Abcam) were performed to measure NA-specific total IgG titers (0.2 endpoint). Data are expressed as means ± standard deviations (SD) for three experiments.
NI antibodies are elicited by H5N1 rNA and pH1N1 rNA immunizations.
To measure NI antibody titers, 2-fold serially diluted serum samples from each immunization group were mixed with 1 μg of H5N1 VLPs, 105 PFU of the pH1N1 (A/California/04/2009) virus, 105 PFU of the H3N2 (A/Udorn/307/1972) virus, or 1 μg of H7N9 VLPs and then examined using fetuin-based assays. The percentages of NA inhibition and NI antibody titers against H5N1 and pH1N1 in each immunization group are shown in Fig. 3A, B, E, and F. Those results indicate dose dependency, with the NI curves from all rNA immunization groups being significantly higher than the curves for the PBS immunization group. NA inhibition percentages and NI antibody titers were also demonstrated for the H3N2 virus and H7N9 VLPs, with similar results but lower NI antibody titers (Fig. 3C, D, G, and H). Regarding the IC50s, similar NI antibody titer ranges were observed against the homologous viruses, 3.7 to 3.8 for H5N1 rNA immunization against H5N1 and 4.3 to 4.4 for pH1N1 rNA immunization against pH1N1 (Fig. 3I). However, significant differences were noted among the six immunization groups in terms of NI antibody titers against the H5N1, pH1N1, H3N2, and H7N9 heterologous viruses. Both H5N1 rNA immunization groups (2 and 20 μg) expressed higher heterologous NI antibody titers against pH1N1 than were elicited by the two pH1N1 rNA immunization groups against H5N1 viruses. In contrast, H5N1 rNA and pH1N1 rNA elicited lower NI antibody titers, with IC50s of less than 2.32 for all groups immunized against the H3N2 and H7N9 viruses (Fig. 3I). Our data indicate that the effect of H5N1 rNA immunization was limited to more potent heterologous NI antibodies against pH1N1 viruses.
FIG 3.
NA-inhibiting-antibody curves and the corresponding IC50s against different virus strains produced by immunizing mice with H5N1 rNA and pH1N1 rNA. Fetuin-based assays were used to measure NA-inhibiting-antibody titers as reductions in the NA enzymatic activities of the pH1N1 or H3N2 viruses or H5N1 or H7N9 VLPs. (A to H) H5N1 rNA inhibition curves against homologous H5N1 VLPs (A), heterosubtypic pH1N1 viruses (B), the H3N2 viral strain (C), or H7N9 VLPs (D) are plotted. pH1N1 rNA inhibition curves are plotted against the homologous pH1N1 viral strain (E), heterosubtypic H5N1 VLPs (F), the H3N2 viral strain (G), or H7N9 VLPs (H). (I) Corresponding IC50s were determined as the 50% reduction in the NA enzymatic activities of the homologous strains (white bars) or heterosubtypic viruses (filled bars). Integrated data are expressed as means ± SD from three experiments. *, P < 0.05; **, P < 0.01.
Antibody-secreting B cells detected in spleen.
To measure anti-NA IgG-secreting B cells elicited by H5N1 rNA or pH1N1 rNA, splenocytes were collected from immunized mice 3 weeks after their second immunization, reacted with 1 μg H5N1 rNA or pH1N1 rNA protein per well, and examined using ELISPOT assays. The results shown in Fig. 4 indicate that the numbers of spots against homologous viruses due to H5N1 rNA immunization were slightly higher than those resulting from pH1N1 rNA immunization. Significantly higher numbers of spots against H5N1 and pH1N1 heterologous viruses were only noted in the 20-μg H5N1 rNA immunization group, which also exhibited higher quantities of ASCs in splenocytes against homologous and heterologous H5N1 and pH1N1 viruses.
FIG 4.
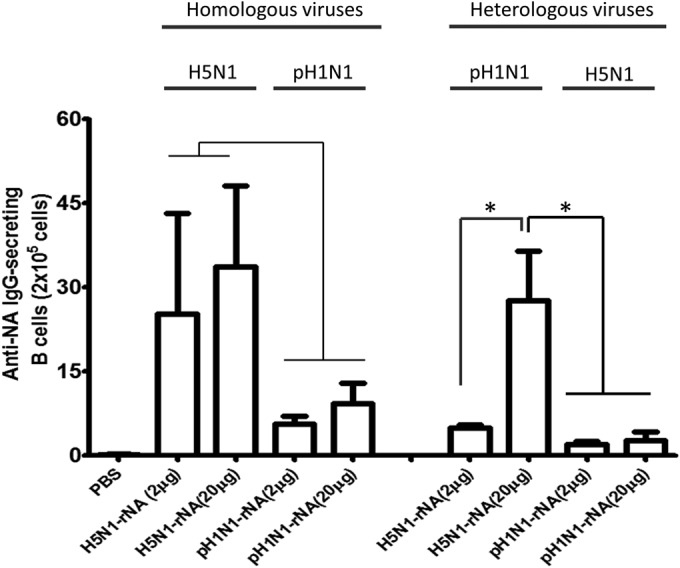
Detection of antibody-secreting B cells in mouse spleens induced by H5N1 rNA and pH1N1 rNA proteins. ELISPOT assays were used to measure IgG-secreting B cell numbers. Splenocytes collected from mice immunized with H5N1 rNA (2 × 105) were cocultured with 1 μg of H5N1 rNA or pH1N1 NA proteins and held for 48 h at 37°C. Splenocytes from mice immunized with pH1N1 rNA were incubated for 48 h at 37°C with 1 μg of pH1N1 rNA or H5N1 NA proteins. Immunospots were determined by ELISPOT assays. Data are expressed as the means ± SD from at least two experiments. *, P < 0.05.
Protective immunity against H5N1, pH1N1, and H7N9 virus challenges.
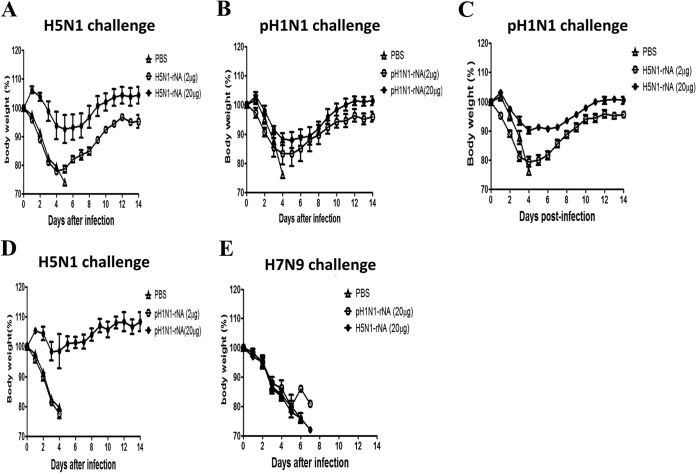
To assess protective immunity triggered by rNA immunizations, mice immunized with 2 or 20 μg of H5N1 rNA or pH1N1 rNA proteins were challenged with 10 MLD50 of H5N1 (RG-14), pH1N1 (CA/09), or H7N9 (TW/13) viruses 3 weeks after their second immunizations. According to the results shown in Fig. 5A and B, with the exceptions of an 80% survival rate for mice immunized with 2 μg of pH1N1 rNA and a 0% rate for the PBS control mice, all immunization groups had 100% survival rates after homologous H5N1 or pH1N1 viral challenges. Significantly smaller body weight losses were observed for mice in the 20-μg H5N1 rNA and 20-μg pH1N1 rNA immunization groups challenged with homologous H5N1 or pH1N1 than for those in the 2-μg H5N1 rNA and 2-μg pH1N1 rNA immunization groups (Fig. 6A and B). In terms of cross-protection levels, mice receiving either 2- or 20-μg H5N1 rNA inoculations exhibited complete protection against pH1N1 viral challenges (Fig. 5C), with significantly smaller body weight losses noted in the 20-μg group (Fig. 6C). A 60% survival rate (Fig. 5D) and faster weight recovery following challenge with the heterologous H5N1 virus were observed in the 20-μg pH1N1 rNA immunization group (Fig. 6D). No protection against the heterosubtypic H7N9 virus challenge was observed in the 20-μg H5N1 rNA and 20-μg pH1N1 rNA immunization groups (Fig. 5E and 6E).
FIG 5.
H5N1 rNA and pH1N1 rNA protective immune responses against different virus challenges. (A and C) Mice were intramuscularly immunized with 2 or 20 μg of H5N1 rNA twice with a 3-week interval and intranasally challenged with 10 MLD50 of the homologous H5N1 (RG-14) virus (A) or heterologous pH1N1 (CA/09) (C) at the end of week 6. Shown are survival curves for mice immunized with H5N1 rNA over a 14-day period after challenge. (B and D) Mice immunized with 2 or 20 μg of pH1N1 rNA were challenged with 10 MLD50 of the homologous pH1N1 (CA/09) virus strain (B) or heterologous H5N1 (RG-14) virus (D) at the end of week 6. Shown are survival curves for mice immunized with pH1N1 rNA over a 14-day period after challenge. (E) Mice were intramuscularly immunized with 20 μg of H5N1 rNA or pH1N1 rNA twice with a 3-week interval and intranasally challenged with 10 MLD50 of heterosubtypic H7N9 (TW/13) viruses 3 weeks after the second inoculations. Survival rates over the 14 days after the viral challenges are shown.
FIG 6.
Body weight recovery tied to H5N1 rNA and pH1N1 rNA immunization responses against different virus challenges. (A and C) Mice were intramuscularly immunized with 2 or 20 μg of H5N1 rNA twice with a 3-week interval and intranasally challenged with 10 MLD50 of the homologous H5N1 (RG-14) virus (A) or heterologous pH1N1 (CA/09) (C) at the end of week 6. Shown are body weight losses over a 14-day period for mice immunized with H5N1 rNA. (B and D) Mice immunized with 2 or 20 μg of pH1N1 rNA were challenged with 10 MLD50 of the homologous pH1N1 (CA/09) virus strain (B) or heterologous H5N1 (RG-14) virus (D) at the end of week 6. Shown are body weight losses over a 14-day period for mice immunized with pH1N1 rNA. (E) Mice were intramuscularly immunized with 20 μg of H5N1 rNA or pH1N1 rNA twice with a 3-week interval and intranasally challenged with 10 MLD50 of heterosubtypic H7N9 (TW/13) viruses 3 weeks after the second inoculation. Body weights were measured for the 14 days after each viral challenge. Body weight loss of >25% was used as an endpoint.
Cross-reactive NI antibodies elicited by pH1N1 rNA mutant proteins.
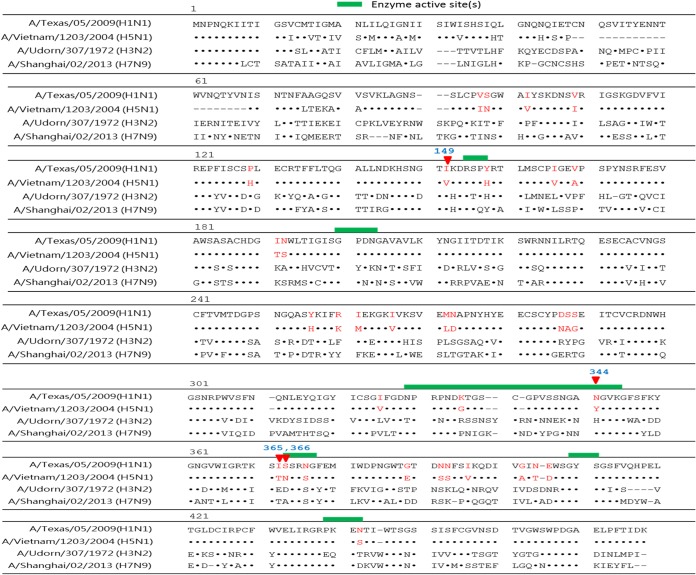
To add detail to our investigation of cross-reactive NI epitopes, we aligned the amino acid sequences of A/Vietnam/1203/2004 (H5N1), A/Texas/05/2009 (pH1N1), A/Udorn/307/1972 (H3N2), and A/Shanghai/02/2013 (H7N9), as shown in Fig. 7. Based on the alignment analysis, we identified 34 different amino acids (Fig. 7, red highlighting) in the NA ectodomains of A/Vietnam/1203/2004 (H5N1) and A/Texas/05/2009 (pH1N1) sequences (94.6% identical). Previous reports have identified influenza NA enzyme catalytic sites at residues 118 and 119, 151 and 152, 198, 224, 227, 243, 274, 276 and 277, 292, 330, 350, and 425 (11, 30); therefore, we targeted residues 149, 344, 365, and 366, all located close to NA enzyme active sites and contributing to NI antibody elicitation. These four target residues (with changes of Ile to Val at position 149 [I149V], N344Y, and I365T/S366N) are shaded in blue and the enzyme active sites are shown as red sticks in a three-dimensional (3-D) NA structure diagram in Fig. 8. Site-directed mutagenesis at these four residues produced the three mutant pH1N1 rNA proteins with mutations I149V, N344Y, or I365T/S366N (Fig. 9). The results from an analysis of antisera from the wild type and the three mutant pH1N1 rNAs show that the I149V and I365T/S366N proteins elicited more NI antibodies against the homologous pH1N1 strain (Fig. 10A), with the I365T/S366N protein eliciting more potent cross-reactive NI antibodies against the H5N1 (Fig. 10B), H3N2 (Fig. 10C), and H7N9 viruses (Fig. 10D). The corresponding IC50s calculated from NI response curves indicate that the I149V and I365T/S366N mutant proteins resulted in increased NI antibody titers against the homologous pH1N1 viruses and that all three mutant proteins resulted in increased NI antibody titers against heterosubtypic H3N2 and H7N9 viruses (Fig. 10E). The I365T/S366N mutation of pH1N1 rNA induced the highest NI antibody titers against the homologous, heterologous, and heterosubtypic strains.
FIG 7.
Sequence alignments on influenza A virus NA proteins. The full lengths of four influenza NA sequences, from A/Texas/05/2009 (pH1N1), A/Vietnam/1203/2004 (H5N1), A/Udorn/307/1972 (H3N2), and A/Shanghai/02/2013 (H7N9), were aligned using the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and Vector NTI software. Identical amino acids are indicated by dots, and noncorresponding amino acid sites are shown by dashes. Identical amino acids (94.6%) comprising 34 different residues between the NA ectodomain of A/Texas/05/2009 (pH1N1) and A/Vietnam/1203/2004 (H5N1) are shown by red highlighting. The four target residues selected from those 34 (I149V, N344Y, I365T, and S366N) are highlighted in blue.
FIG 8.
Three-dimensional neuraminidase structure diagram. The 3-D protein structures of the tetrameric H5N1 rNA proteins were modified using the crystal structure of NA from A/Vietnam/1203/04 strain (PDB 2HTY). The top and side views of the monomeric structures were diagramed by using PyMOL software, and the enzyme active sites are shown as red sticks. The four target residues (I149V, N344Y, and I365T/S366N) are shaded in blue.
FIG 9.
Construction, expression, and characterization of WT and mutant pH1N1 rNAs. (A) The wild and mutant pH1N1 rNAs were constructed by a site-directed mutagenesis strategy and expressed using a baculovirus-insect cell expression system. (B and C) Purified rNA proteins (2 μg) were confirmed by Coomassie blue staining (B) and Western blotting (C).
FIG 10.
NA-inhibiting-antibody curves and the corresponding IC50s against different virus strains produced by immunizing mice with the WT and mutant pH1N1 rNAs. Fetuin-based assays were used to measure NA-inhibiting-antibody titers as reductions in the NA enzymatic activities of the pH1N1 or H3N2 viruses or H5N1 or H7N9 VLPs. (A to D) Inhibition curves for the WT pH1N1 rNA or mutant pH1N1 rNAs (I149V, N344Y, or I365T/S366N) against homologous pH1N1 viruses (A), the heterosubtypic H5N1 VLPs (B), the H3N2 viral strain (C), or H7N9 VLPs (D) are plotted. (E) The corresponding IC50s were determined as the 50% reduction in the NA enzymatic activities of the homologous strains (white bars) or heterosubtypic viruses (filled bars). Integrated data are expressed as means ± SD from three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
NA-based influenza vaccines are attractive because of the relatively smaller number of changes in NA antigens compared to the changes in HA antigens in host immune systems (31, 32). For this study, we constructed and purified H5N1 rNA and pH1N1 rNA proteins from Sf9 insect cells and found that mice immunized with H5N1 rNA and pH1N1 rNA proteins exhibited higher quantities of NA-specific total IgG, IgG1, IgG2a subclass, and NI antibodies, increased numbers of ASCs in the spleen, and better protective immunity against live-virus challenges. We also found that H5N1 rNA immunization induced more potent cross-reactive NI antibodies and protective immunity against pH1N1 viruses than did pH1N1 rNA immunization against H5N1 viruses. Cross-reactive NI epitopes were further dissected by immunization using pH1N1 rNA proteins with I149V, N344Y, and I365T/S366N NA mutations. The I365T/S366N mutation of pH1N1 rNA was found to increase cross-reactive NI antibodies against heterologous and heterosubtypic influenza A viruses.
H5N1 rNA and pH1N1 rNA proteins expressed in baculovirus-infected Sf9 cells and purified by affinity chromatography displayed similar enzymatic activities. The Km value of pH1N1 rNA was slightly lower (higher affinity) than that of H5N1 rNA (Fig. 1C). Note that only NA ectodomain proteins (amino acids [aa] 62 to 447 for H5N1 and aa 82 to 467 for pH1N1) were encoded for soluble rNA expression. The coding sequences did not include N-terminal stalk regions; that is, H5N1 lacked aa 38 to 57 and pH1N1 lacked aa 38 to 77 (Fig. 7). Each NA monomer consists of six identical β sheets (β) plus several strands (S) and loops (L). Influenza NA enzyme catalytic sites are located at the upper corners of each monomer (tetrameric structure); amino acids encircling active catalytic sites are highly conserved among various influenza A and B virus strains (11, 33). A comparison of the NA sequences of H5N1 (A/Vietnam/1203/2004) and pH1N1 (A/Texas/05/2009), based on X-ray crystallographic diffraction data for NA active sites from A/Tokyo/3/67, did not identify amino acid differences at residues 118 or 119 (on β1L01), 151 or 152 (on β1L23 [L23 denotes a loop between strands 2 and 3 within a sheet]); 198 (β2L23), 224 or 227 (β3L01), 243 (β3L23), 274, 276, or 277 (β4L01), 292 (β1L23), 330 or 350 (β5L01), or 425 (β6S2) (11). However, we did find four amino acid differences between pH1N1 rNA and H5N1 rNA (I149V, N344Y, I365T, and S366N) close to enzymatic catalytic pockets consisting of the β1L23 loop (151 and 152) and the β5L01 loop (330 and 350).
NI antibodies are important for NA-based vaccine development. Other researchers have reported that NI antibodies induced by pH1N1 immunizations are cross-reactive with H5N1 viruses in mice (34) and ferrets (21, 22). Our results indicate that the NI antibodies induced by pH1N1 rNA immunizations (20 μg), with an IC50 of 2.78, resulted in a 60% survival rate after H5N1 virus challenges. In contrast, no protection against H5N1 virus challenges was noted following immunizations with 2 μg of pH1N1 rNA, with an IC50 of 2.47 (Fig. 3I, 5D, and 6D), while 100% protection against pH1N1 virus challenges was observed in mice immunized with either 2 or 20 μg of H5N1 rNA (IC50s of 3.53 and 3.75, respectively) (Fig. 3I, 5C, and 6C). The NI antibodies induced by 2- or 20-μg H5N1 rNA immunizations had IC50s of 1.92 and 2.30, respectively, against the H3N2 virus, and the NI antibodies induced by 2- or 20-μg pH1N1 rNA immunizations had IC50s of 2.03 and 2.32, respectively (Fig. 3I). The NI antibodies induced by 2- or 20-μg H5N1 rNA immunizations had IC50s of 1.45 and 2.26 against the H7N9 virus, respectively, and the NI antibodies induced by 2- or 20-μg pH1N1 rNA immunizations had IC50s of 1.31 and 2.17, respectively. In other words, H5N1 rNA immunizations induced more potent cross-reactive NI antibodies and protective immunity against heterologous pH1N1 viruses compared to the pH1N1 rNA protective immunity against heterologous H5N1 viruses. These results are in agreement with those from studies of pH1N1 rNA (A/California/04/2009) immunization against an H5N1 (A/Turkey/1/2005) virus strain (NI antibody titer of 2.67) (19) and of NA antisera induced by live, attenuated pH1N1 (A/California/7/2009) that cross-reacted with various H5N1 strains in ferrets (NI antibody titers of ∼2.2 to 2.3) (21). However, in neither case was any protection observed against the heterosubtypic H7N9 virus challenges (Fig. 3I, 5E, and 6E). Again, our findings are similar to those of a recent report using rNA immunizations that showed no protection against heterosubyptic influenza virus challenges (29).
Conserved influenza NA epitopes are of great interest to researchers working on a universal influenza vaccine. According to one report, 20 amino acid residues in 7 upper surface loops (195 to 202, 216 to 231, 243 to 251, 316 to 353, 364 to 374, 398 to 407, and 428 to 439) close to enzymatically active sites may be key to eliciting cross-reactive NI antibodies between H1N1 (A/Beijing/262/95) and H5N1 (A/Hong Kong/483/97) (35). Another research team reported that N1 conserved epitopes at NA residues 273, 338, and 339 induced cross-reactive NA-specific antibodies and provided protection against seasonal H1N1, 1918 H1N1, 2009 pandemic H1N1 (pH1N1), and H5N1 avian influenza viruses (34). To date, the only NA epitopes that are universally conserved among all influenza A viruses have been reported at NA residues 222 to 230 for the formation of active sites at residues 224 and 227 (β3L01) (36). In the present study, we found four amino acid residues close to active enzyme sites in pH1N1 (A/Texas/05/2009) and H5N1 (A/Vietnam/1203/2004): I149V, N344Y, I365T, and S366N (Fig. 8). We also investigated three mutant pH1N1 rNA proteins (I149V, N344Y, and I365T/S366N mutants) by using mutagenesis to map these amino acids that may contribute to cross-reactive NI antibody elicitation. Our results indicate that the pH1N1 rNA (I365T/S366N) mutant protein was the most active in eliciting increased NI antibody titers against homologous, heterologous, and heterosubtypic viruses (Fig. 10). These findings provide useful information for the development of an NA-based universal influenza vaccine.
ACKNOWLEDGMENTS
This work was supported by Ministry of Science and Technology, Taiwan (grants MOST103-2312-B-007-009 and MOST104-2321-B-007-005), and National Tsing Hua University (grant 103N2703E1).
We thank Ting-Hsuan Chen for providing H7N9 VLPs for NI serological assays.
REFERENCES
- 1.Medina RA, Garcia-Sastre A. 2011. Influenza A viruses: new research developments. Nat Rev Microbiol 9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Hale BG, Xu K, Sun B. 2013. Viral and host factors required for avian H5N1 influenza A virus replication in mammalian cells. Viruses 5:1431–1446. doi: 10.3390/v5061431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. 2014. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 6.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 7.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2012. Household preparedness for public health emergencies—14 states, 2006-2010. MMWR Morb Mortal Wkly Rep 61:713–719. [PubMed] [Google Scholar]
- 9.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T, Bi Y, Tian H, Li X, Liu D, Wu Y, Jin T, Wang Y, Chen Q, Chen Z, Chang J, Gao GF, Xu B. 2014. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis 20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colman PM, Varghese JN, Laver WG. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 12.Marcelin G, Sandbulte MR, Webby RJ. 2012. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol 22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilyushina NA, Bovin NV, Webster RG. 2012. Decreased neuraminidase activity is important for the adaptation of H5N1 influenza virus to human airway epithelium. J Virol 86:4724–4733. doi: 10.1128/JVI.06774-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Liu Q, Bawa B, Qiao C, Qi W, Shen H, Chen Y, Ma J, Li X, Webby RJ, Garcia-Sastre A, Richt JA. 2012. The neuraminidase and matrix genes of the 2009 pandemic influenza H1N1 virus cooperate functionally to facilitate efficient replication and transmissibility in pigs. J Gen Virol 93:1261–1268. doi: 10.1099/vir.0.040535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 17.Martinet W, Saelens X, Deroo T, Neirynck S, Contreras R, Min Jou W, Fiers W. 1997. Protection of mice against a lethal influenza challenge by immunization with yeast-derived recombinant influenza neuraminidase. Eur J Biochem 247:332–338. doi: 10.1111/j.1432-1033.1997.00332.x. [DOI] [PubMed] [Google Scholar]
- 18.Johansson BE, Brett IC. 2008. Recombinant influenza B virus HA and NA antigens administered in equivalent amounts are immunogenically equivalent and induce equivalent homotypic and broader heterovariant protection in mice than conventional and live influenza vaccines. Hum Vaccin 4:420–424. doi: 10.4161/hv.4.6.6201. [DOI] [PubMed] [Google Scholar]
- 19.Bosch BJ, Bodewes R, de Vries RP, Kreijtz JH, Bartelink W, van Amerongen G, Rimmelzwaan GF, de Haan CA, Osterhaus AD, Rottier PJ. 2010. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J Virol 84:10366–10374. doi: 10.1128/JVI.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcelin G, DuBois R, Rubrum A, Russell CJ, McElhaney JE, Webby RJ. 2011. A contributing role for anti-neuraminidase antibodies on immunity to pandemic H1N1 2009 influenza A virus. PLoS One 6:e26335. doi: 10.1371/journal.pone.0026335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Kim L, Subbarao K, Jin H. 2012. The 2009 pandemic H1N1 virus induces anti-neuraminidase (NA) antibodies that cross-react with the NA of H5N1 viruses in ferrets. Vaccine 30:2516–2522. doi: 10.1016/j.vaccine.2012.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockman S, Brown LE, Barr IG, Gilbertson B, Lowther S, Kachurin A, Kachurina O, Klippel J, Bodle J, Pearse M, Middleton D. 2013. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J Virol 87:3053–3061. doi: 10.1128/JVI.02434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu CY, Yeh YC, Chan JT, Yang YC, Yang JR, Liu MT, Wu HS, Hsiao PW. 2012. A VLP vaccine induces broad-spectrum cross-protective antibody immunity against H5N1 and H1N1 subtypes of influenza A virus. PLoS One 7:e42363. doi: 10.1371/journal.pone.0042363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritz R, Sabarth N, Kiermayr S, Hohenadl C, Howard MK, Ilk R, Kistner O, Ehrlich HJ, Barrett PN, Kreil TR. 2012. A Vero cell-derived whole-virus H5N1 vaccine effectively induces neuraminidase-inhibiting antibodies. J Infect Dis 205:28–34. doi: 10.1093/infdis/jir711. [DOI] [PubMed] [Google Scholar]
- 25.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, Belmont JW. 2013. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J Infect Dis 207:974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. 2008. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei HJ, Chang W, Lin SC, Liu WC, Chang DK, Chong P, Wu SC. 2011. Fabrication of influenza virus-like particles using M2 fusion proteins for imaging single viruses and designing vaccines. Vaccine 29:7163–7172. doi: 10.1016/j.vaccine.2011.05.077. [DOI] [PubMed] [Google Scholar]
- 28.Lin SC, Huang MH, Tsou PC, Huang LM, Chong P, Wu SC. 2011. Recombinant trimeric HA protein immunogenicity of H5N1 avian influenza viruses and their combined use with inactivated or adenovirus vaccines. PLoS One 6:e20052. doi: 10.1371/journal.pone.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6(2):e02556–14. doi: 10.1128/mbio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambre CR, Terzidis H, Greffard A, Webster RG. 1990. Measurement of anti-influenza neuraminidase antibody using a peroxidase-linked lectin and microtitre plates coated with natural substrates. J Immunol Methods 135:49–57. doi: 10.1016/0022-1759(90)90255-T. [DOI] [PubMed] [Google Scholar]
- 31.Eichelberger MC, Wan H. 2015. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386:275–299. [DOI] [PubMed] [Google Scholar]
- 32.Johansson BE, Cox MM. 2011. Influenza viral neuraminidase: the forgotten antigen. Expert Rev Vaccines 10:1683–1695. doi: 10.1586/erv.11.130. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Zhang F, Wang M, Xu C, Song J, Zhou J, Lin X, Zhang Y, Wu X, Tan W, Lu J, Zhao H, Gao J, Zhao P, Wang Y. 2010. Characterization of neuraminidases from the highly pathogenic avian H5N1 and 2009 pandemic H1N1 influenza A viruses. PLoS One 5:e15825. doi: 10.1371/journal.pone.0015825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, Liu F, Zeng H, Sheu T, Achenbach JE, Veguilla V, Gubareva LV, Garten R, Smith C, Yang H, Stevens J, Xu X, Katz JM, Tumpey TM. 2014. Evaluation of the antigenic relatedness and cross-protective immunity of the neuraminidase between human influenza A (H1N1) virus and highly pathogenic avian influenza A (H5N1) virus. Virology 454–455:169–175. doi: 10.1016/j.virol.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li C, Wang J, He R, Brown EG, Li X. 2013. The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288:18283–18289. doi: 10.1074/jbc.M113.468884. [DOI] [PMC free article] [PubMed] [Google Scholar]