ABSTRACT
The human herpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are associated with Hodgkin's lymphoma (HL) and Primary effusion lymphomas (PEL), respectively, which are B cell malignancies that originate from germinal center B cells. PEL cells but also a quarter of EBV-positive HL tumor cells do not express the genuine B cell receptor (BCR), a situation incompatible with survival of normal B cells. EBV encodes LMP2A, one of EBV's viral latent membrane proteins, which likely replaces the BCR's survival signaling in HL. Whether KSHV encodes a viral BCR mimic that contributes to oncogenesis is not known because an experimental model of KSHV-mediated B cell transformation is lacking. We addressed this uncertainty with mutant EBVs encoding the KSHV genes K1 or K15 in lieu of LMP2A and infected primary BCR-negative (BCR−) human B cells with them. We confirmed that the survival of BCR– B cells and their proliferation depended on an active LMP2A signal. Like LMP2A, the expression of K1 and K15 led to the survival of BCR− B cells prone to apoptosis, supported their proliferation, and regulated a similar set of cellular target genes. K1 and K15 encoded proteins appear to have noncomplementing, redundant functions in this model, but our findings suggest that both KSHV proteins can replace LMP2A's key activities contributing to the survival, activation and proliferation of BCR– PEL cells in vivo.
IMPORTANCE Several herpesviruses encode oncogenes that are receptor-like proteins. Often, they are constitutively active providing important functions to the latently infected cells. LMP2A of Epstein-Barr virus (EBV) is such a receptor that mimics an activated B cell receptor, BCR. K1 and K15, related receptors of Kaposi's sarcoma-associated herpesvirus (KSHV) expressed in virus-associated tumors, have less obvious functions. We found in infection experiments that both viral receptors of KSHV can replace LMP2A and deliver functions similar to the endogenous BCR. K1, K15, and LMP2A also control the expression of a related set of cellular genes in primary human B cells, the target cells of EBV and KSHV. The observed phenotypes, as well as the known characteristics of these genes, argue for their contributions to cellular survival, B cell activation, and proliferation. Our findings provide one possible explanation for the tumorigenicity of KSHV, which poses a severe problem in immunocompromised patients.
INTRODUCTION
The Epstein-Barr virus (EBV) and the Kaposi's sarcoma-associated herpesvirus (KSHV) are lymphotropic human herpesviruses linked to the development of Hodgkin's lymphoma (HL) and primary effusion lymphoma (PEL), respectively. Hodgkin-Reed-Sternberg (HRS) cells, the genuine tumor cells of HL, originate from germinal center (GC) B cells (1) and PEL cells originate from GC or post-GC B cells (2). In 25% of cases of classical HL, crippling mutations in the immunoglobulin genes (Ig) of HRS cells destroy the coding capacity of a functional B cell receptor (BCR) (3). HRS cells with such destructive Ig mutations are EBV-positive (EBV+), supporting an essential role of EBV in HL lymphomagenesis (1). EBV+ HRS cells express the viral protein latent membrane protein 2A (LMP2A), which can functionally replace the BCR because Ig-negative B cells can only survive if infected with EBV but not with an LMP2A-negative EBV mutant (4). BCR and LMP2A both contain an immunoreceptor tyrosine-based activation motif (ITAM) (5, 6), which is central for the induction of BCR signaling cascades. In contrast to the BCR, LMP2A is thought to be constitutively active and independent of any ligand or antigen encounter (references 7 and 8 and references therein). EBV also encodes LMP1, a constitutive CD40 receptor mimic that provides a second survival signal for B cells. LMP1 together with LMP2A activates HRS cells, protects them from apoptosis, and induces their proliferation (1, 9).
PEL tumor cells are infected with KSHV, and ca. 80% are EBV+, although LMP1 and LMP2A are barely expressed (10, 11). PEL cells have Ig rearrangements but usually lack B cell-typical surface markers, including the BCR (12, 13). Reports of BCR−, KSHV+/EBV− PEL cells (14, 15) raised the question of whether KSHV itself encodes a BCR mimic. The K1 and K15 KSHV proteins are likely candidates because they are transmembrane proteins with cytoplasmic domains, which could activate certain signaling pathways similar to EBV's latent membrane proteins. For example, K1 encodes an ITAM similar to LMP2A but has a genomic location homologous with EBV's LMP1 (see reference 16 for a recent review). K15, in turn, has a genomic location like that of LMP2A but lacks an ITAM and recruits signaling mediators such as LMP1 (17). In a recombinant herpesvirus saimiri chimera and in transgenic mice, K1 is oncogenic in vivo (18, 19). In addition, K1 protein downregulates BCR surface expression (20), whereas K15 blocks BCR-induced Ca2+-influx antagonizing BCR signaling (21) similar to LMP2A (22).
EBV infects quiescent primary human B cells, induces their proliferation, and establishes a latent infection in them, which emerge as growth-transformed lymphoblastoid cell lines (LCLs) in vitro. A comparable model is not available for the study of KSHV, which has been an obstacle to our understanding of KSHV-induced B cell lymphomas (23; see reference 24 for a recent review). We therefore introduced the K1 or K15 genes in lieu of LMP2A into mutant EBV strains and tested their phenotypes in infected primary human B cells in order to analyze the contribution of the KSHV genes to B cell growth transformation in a tractable experimental setting.
MATERIALS AND METHODS
Ethics statement.
The human material used in the present study has been obtained in accordance with the Declaration of Helsinki, stems from anonymous healthy donors, and therefore does not require the approval of the board of the local ethics committee.
Isolation and separation of human primary B lymphocytes.
Anonymous adenoid tissue samples from routine adenoidectomies were provided by the Department of Otorhinolaryngology, Klinikum Grosshadern, Ludwig Maximilians University of Munich, and Dritter Orden Clinic, Munich-Nymphenburg, Germany. Human primary B cells from adenoids were prepared as described previously (25). To isolate BCR− and BCR+ B cells, the cells were labeled with α-CD3-PE (Immunotools), α-λ-FITC, and α-κ-APC light chain antibodies (Invitrogen) and sorted with a fluorescence-activated cell sorter (FACS) Aria III instrument (Becton Dickinson). BCR+ B cells were defined as CD3− and λ+ or κ+ lymphocytes, and BCR− B cells were defined as CD3− and both λ− and κ− lymphocytes. BCR+ and BCR− lymphocytes are termed λ+/κ+ and λ−/κ−, respectively, throughout the manuscript.
Cell lines and culture conditions.
The B-cell line Raji and the EBV-negative derivative of the Daudi B-cell line are described (26, 27). The single cell LCL clone 16 was described previously (28), is derived from an EBV-infected patient and does not express a functional BCR. Primary B cells infected with EBV stocks were cultivated in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 μg of streptomycin/ml, 100 U of penicillin/ml, 1 mM sodium pyruvate, 100 nM sodium selenite, 50 μM β-mercaptoethanol, 250 μM α-tocopherol, 10 μg of ciprofloxacin/ml, and 1 μg of cyclosporine/ml. Primary B cells infected with EBV were kept at a reduced oxygen level adjusted to 5%.
Construction of mutant EBV strains.
EBV mutants were derived from p2089, which comprises the B95.8 EBV genome cloned onto an F-factor plasmid in Escherichia coli (29). p2089 was genetically modified in E. coli by homologous recombination with the galK-based recombineering system (30). The ΔLMP2A EBV (p2525) was described previously (4). The mutant EBVs that express the KSHV genes K1 and K15 were constructed essentially as described in detail recently (31, 32). In p4082 and p3998, the cDNAs of KSHV K1 and K15 P type were inserted in between nucleotide coordinates 166100 to 166458 and coordinates 166103 to 166458 of the B95.8 reference EBV genome, respectively, replacing the first exon of LMP2A in the p2089 maxi-EBV plasmid (Fig. 1A). The EBV plasmid DNAs were prepared from E. coli by two sequential rounds of CsCl-ethidium bromide density ultracentrifugation and carefully analyzed on agarose gels after cleavage with several restriction enzymes (AgeI, BamHI, MluI, and XhoI). The modified loci and flanking regions were confirmed by extensive DNA sequencing in the E. coli derived EBV DNAs covering >6 kbp in each of the two maxi-EBV plasmids p3998 and p4082.
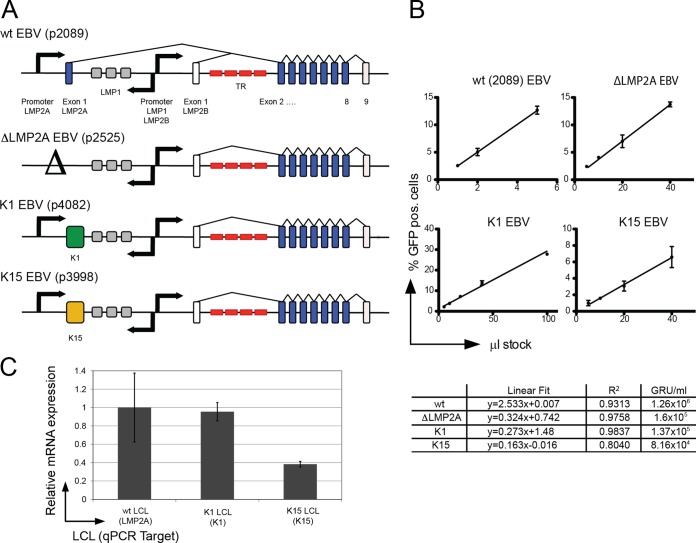
FIG 1.
Mutant EBVs encoding K1 and K15 in lieu of LMP2A. (A) Genomic structure of the LMP2A locus in mutant EBVs. Nine exons encode the LMP2A gene, which flank both sides of the terminal repeats (TR) of EBV (red). LMP2A's first exon encodes the signaling domain. LMP2A and LMP2B share exons 2 to 9. The first exon of LMP2B and exon nine of LMP2A and LMP2B are noncoding (white); protein encoding exons are shown in blue. Separate promoters (arrows) drive LMP2A and LMP2B expression; the locus in wt EBV (p2089) reflects this situation (top). In the ΔLMP2A mutant, the promoter and the first exon of LMP2A were deleted, but LMP2B is not affected (4). In the K1 and K15 EBV mutants, the first exon of LMP2A was exactly replaced by the cDNAs coding for K1 or K15 P-type. The LMP2A promoter controls the expression of K1 and K15 genes. (B) Titration and linear regression of four virus stocks determine their virion concentrations. A total of 5 × 104 Raji cells were infected with the indicated volumes of virus stocks. The percentage of GFP-positive cells was assessed by FACS analysis 3 days p.i. Shown are the linear regressions, goodness of fit (R2) and the calculated concentration of GRU per ml. (C) LMP2A, K1, and K15 transcript levels in established LCLs. LCLs were established by infection of primary adenoid B cells with wt EBV, K1 EBV, and K15 EBV. Steady-state mRNA levels were determined after cDNA synthesis and subsequent qPCR analysis. The levels of LMP2A, K1, and K15 cDNAs were normalized to the housekeeper CLK2. LMP2A cDNA levels were set to 1. The mean and the standard deviation of two independent experiments are shown. The steady-state levels of LMP2A and K1 mRNAs in bulk LCLs were similar, whereas the level of K15 mRNA was reduced by a factor of 2.6 compared to LMP2A transcripts.
Generation and quantitation of viral stocks.
Viral stocks were generated in stably transfected single cell clones of HEK293 producer cells and quantified as described in detail (25, 31–33). Briefly, we scrutinized the virus concentrations of the four different virus stocks (Fig. 1B). All recombinant EBVs used here were engineered to carry the egfp gene. Infectious units were defined with the aid of the Raji cell line, which is our model B cell for virus quantitation. Raji cells turn green fluorescent protein (GFP)-positive upon infection and allow the direct assessment of the concentration of infectious EBV virions as green Raji units (GRU) per milliliter of virus stock, as described earlier (34).
Quantitative reverse transcription-PCR (RT-PCR).
RNA was isolated using the RNeasy Mini Kit (Qiagen) and digested with DNase I (Invitrogen) prior to cDNA synthesis with the SuperScript III first-strand synthesis System (Invitrogen) and oligo(dT) as primers. cDNAs were quantified with a LightCycler 480 instrument and SYBR green I (Roche Diagnostics). The crossing point (Cp) values were corrected for primer efficiencies. Relative expression levels were assessed with CDC-like kinase 2 gene (Clk2) as housekeeping gene to calculate Cp(target)/Cp(reference) ratios. The following primer pairs were used: LMP2A (5′-CGTCACTCGGACTATCAACC-3′ and 5′-TACAGGCAGGCATACTGGAT-3′), K1 (5′-GGAGTGATTTCAACGCCTTA-3′ and 5′-GCCATGTAATCCAAATGCTC-3′), K15 (5′-GGGCCCTACTGGTATGTTTT-3′ and 5′-GGATGAAGGCCATTTAGGAT-3′), and Clk2 (5′-CTGACACATACAGACCTCAAGCCTG-3′ and 5′-ACCAGCCCAACTCAAGGATGAC-3′).
Phospho-specific flow cytometry.
5 × 105 cells were washed in phosphate-buffered saline (PBS), pelleted, and recovered in 160 μl of PBS and 25 μg of goat α-hIgG/M antibody (Jackson Research)/ml for 15 min on ice. Signaling was induced at 37°C in a water bath for 10 min. A total of 160 μl of prewarmed BD Cytofix fixation buffer was added to the cell solution for 10 min at 37°C. The cells were pelleted, diluted in 50 μl of supernatant, permeabilized in 1 ml of ice-cold methanol for 30 min on ice, washed twice in Phosflow buffer (BD Biosciences), and stained with phospho-specific α-pSyk-Alexa 647 or α-pPLCγ2-Alexa 647 antibodies (BD Biosciences) for 30 min at room temperature.
Western blot immunodetection.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 2 mM EDTA, Complete protease inhibitor [Roche]). Protein concentration was determined by Bradford analysis. All Western blots were performed according to standard protocols, using SDS-PAGE, nitrocellulose membranes (Hybond ECL; GE Healthcare, catalog no. RPN3032D), 5% nonfat milk in PBS-Tween for blocking, and antibody dilutions and ECL Western blotting detection reagents (GE Healthcare, catalog no. RPN2106). The following primary antibodies were used: rabbit anti-Syk (Cell Signaling, catalog no. 2712), rabbit anti-PLCγ2 (Cell Signaling, catalog no. 3872), and mouse anti-α-tubulin (Santa-Cruz, catalog no. 23948). The following secondary antibodies were used: anti-rabbit HRP-conjugated antibody (Cell Signaling, catalog no. 7074) and anti-mouse HRP-conjugated antibody (Cell Signaling, catalog no. 7076).
Calcium flux analysis.
Cells were loaded with 3 μM Indo-1 AM (Invitrogen) at 37°C for 30 min, washed once, and diluted in 2 ml of RPMI 1640 on ice. A total of 106 cells in 200 μl were prewarmed at 37°C for 20 min, and the baseline violet/blue ratio of Indo-1-loaded cells was measured for 1 min on a FACS LSR II flow cytometer (Becton Dickinson). Subsequently, the BCR was cross-linked with 25 μg of goat α-hIgG/A/M F(ab′)2 fragment (Jackson Research)/ml, the cells were quickly mixed, and the Ca2+-influx was measured based on the change in the violet/blue ratio over 5 min.
Cellular targets of LMP2A, K1, or K15.
A total of 4 × 106 B cells each were infected with the four virus stocks (wt EBV, K1 EBV, K15 EBV and ΔLMP2A EBV). On day 8 postinfection (p.i.), cells with characteristics of lymphoblastoid cells were identified by forward and sideward scatter criteria and physically sorted on a FACS Aria III instrument (Becton Dickinson). Four biological replicates were conducted with B cells from four different donors. Cellular RNA samples were prepared with the RNeasy Mini Kit (Qiagen), and their quality was analyzed on an Agilent 2000 Bioanalyzer. RNA was converted to cDNA by using an Ambion WT expression kit, and the DNA was fragmented and labeled with an Affymetrix GeneChip WT terminal labeling and hybridization kit. Hybridization to GeneChip Human Gene 1.0 ST arrays (Affymetrix) and scanning of the arrays were performed according to the manufacturer's protocol.
Microarray data preprocessing and bioinformatic analysis.
Affymetrix CEL files were preprocessed using the R packages affy, hugene10sttranscriptcluster.db, and genefilter provided by bioconductor.org (35, 36). Microarray data sets were normalized using the robust multiarray average (RMA). Probe sets were filtered using the nsFilter (available with the R genefilter package). We performed a principal component analysis (PCA) for quality control and outlier detection (37). A PCA plot (see Fig. 6A) shows that the first two principal components cluster the data into biological interpretable groups. No experimental or technical outlier could be detected. To decompose the gene expression data matrix into independent source profiles, we performed independent component analysis (ICA) using the JADE algorithm (38). Hierarchical clustering of the selected genes was performed using Euclidean distance and average linking.
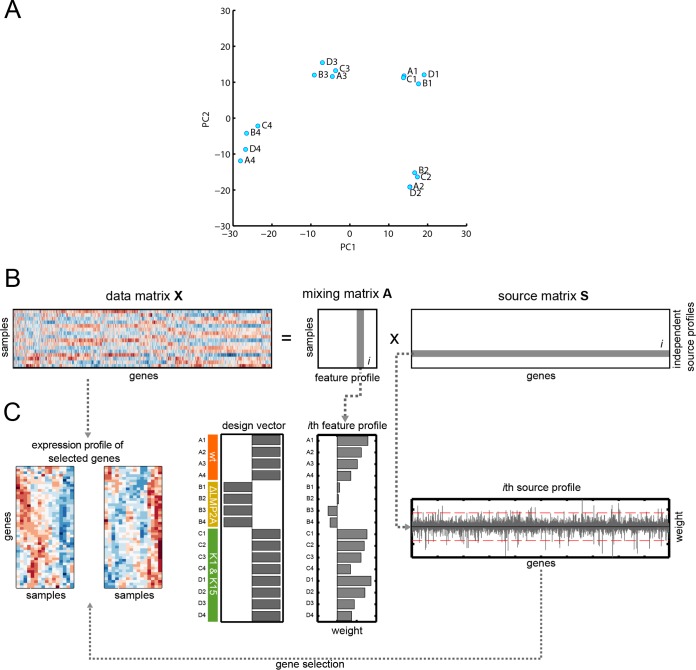
FIG 6.
Principal component analysis (PCA) and schematic sketch of individual steps of the independent component analysis (ICA). (A) The PCA plot of the first two principal components is shown. The analysis reveals that the measured gene expression profiles were highly biased by specific effects arising from the four individual donors of the B lymphocytes. (B) We used ICA to decompose a data matrix (X) into a matrix of independent source profiles (S) and a mixing matrix (A). The ith column of the mixing matrix forms a called feature profile that contains the mixing coefficients denoting the contribution weight of the corresponding ith source profile to the respective samples. A sample here corresponds to a single microarray measurement. (C) We selected the feature profile that displays weights that correspond to a specific experimental setup. Here, we wanted to identify a source profile that contains genes showing a maximal difference between presence or absence of LMP2A-mediated regulation. To do so, we identified the feature profile that displays strongest correlation to a design vector. The corresponding source profile was used subsequently to identify genes of interest by thresholding the source profile weights (red dashed lines). Gene expression profiles for the two identified clusters were derived from the original data matrix.
NCBI GEO accession number.
Microarray data sets were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE53914.
RESULTS
Construction of mutant EBVs encoding K1 and K15 in lieu of LMP2A.
Wild-type (wt) EBV rescued infected BCR− GC B cells from apoptosis (28, 39, 40), but LMP2A-knockout (ΔLMP2A) EBV did not (4), indicating that LMP2A can functionally replace the BCR in primary human B cells. LMP2A is encoded by nine exons and shares exons two to nine with LMP2B (Fig. 1A). The two KSHV proteins K1 and K15 partially resemble LMP2A in structure and function (18, 41), but it was not known whether they also act similarly. To address this question, two mutant EBV strains were constructed on the basis of the recombinant EBV plasmid p2089 (29), which encompasses the entire genome of the B95.8 prototype EBV strain. To express K1 or K15 under the control of the LMP2A promoter (Fig. 1A), the first exon of LMP2A was replaced by the cDNAs coding for K1 or K15 P-type (45-kDa isoform) to yield K1 EBV (p4082) or K15 EBV (p3998), respectively.
The two EBV plasmids p4082 and p3998 were engineered in E. coli, and plasmid DNAs were purified, carefully scrutinized by restriction fragment analysis and extensive DNA sequencing, and stably introduced into HEK293 cells. Successfully transfected cells were selected, and clonal cell lines were established as described previously (31). The genetic composition of the EBV genomes in the different producer cell lines was carefully evaluated and confirmed by Southern blotting hybridization with total cellular DNA and appropriate EBV-specific probes. DNA sequencing of PCR-amplified EBV DNA from the producer cell lines covering all critical regions of the mutant EBVs confirmed the integrity of the mutated loci in EBV. The potential producer cell lines were tested for virus release upon transient transfection with two expression plasmids encoding the viral genes BZLF1 and BALF4 essentially as described previously (31).
Analysis of the concentration of viral stocks.
We routinely analyzed the concentration of infectious virus particles in our viral stocks with the aid of Raji cells. All recombinant EBVs used here are engineered to carry the egfp gene, and infectious units are defined with the aid of these cells, which turn GFP-positive upon infection, allowing the direct assessment of the concentration of infectious EBV virions as green Raji units (GRU) per milliliter by FACS analysis as described previously (33, 34, 42). As shown in Fig. 1B, the concentration of infectious recombinant EBVs was determined by regression assays measuring the fraction of GFP-positive cells 3 days after infection. The Raji cell line is our preferred target cell to determine the concentration of recombinant EBV stocks, because Raji cells deliver consistent and very reproducible results in infection experiments with EBV. The definition of GRU was introduced about 15 years ago and has evolved as a standard approach in the field, but it underestimates the concentration of biologically active, infectious Epstein-Barr virions by at least a factor of 10 (see below).
K1 and K15 are expressed by the K1 EBV or K15 EBV mutants.
We established lymphoblastoid cell lines (LCLs) with BCR+ B cells infected with the different viruses and measured the transcripts of LMP2A, K1, and K15 in these cell lines by quantitative RT-PCR. The K1 and K15 genes are expressed from the LMP2A promoter (Fig. 1A) and showed steady-state transcript levels, which were similar (K1 LCL) or lower (K15 LCL) compared to transcripts of LMP2A in LCLs infected with wt EBV (wt LCL) (Fig. 1C). We failed to detect K1 or K15 protein in the LCLs infected with either K1 EBV or K15 EBV in several attempts at immunoprecipitation and/or Western blotting, presumably because the available K1 and K15 specific antibodies are not sensitive enough to detect the two KSHV proteins. We therefore assumed that both KSHV proteins are expressed at low levels in our LCLs compared to KSHV-infected endothelial cells, which we used as positive controls.
Assessment of optimal B cell infections.
In order to optimize the success of EBV infection in the experiments throughout the manuscript, we assessed the optimal ratio of EBV and human primary B lymphocytes. We infected primary B lymphocytes purified with different multiplicities of infection (MOIs) of a wt 2089 EBV stock and measured the absolute number of emerging B blasts in the LCL gate over time, as shown in Fig. 2A, using calibrated APC beads (Calibrite [Becton Dickinson]) as a volume standard. The concentration of infectious particles in this virus stock had been determined with the aid of Raji cells as described above. In three independent experiments, an MOI of 0.1 was found to be optimal, because this dose yielded the highest numbers of lymphoblasts within the first 2 weeks p.i., as illustrated in Fig. 2B. Consequently, we chose an MOI of 0.1 for all subsequent experiments.
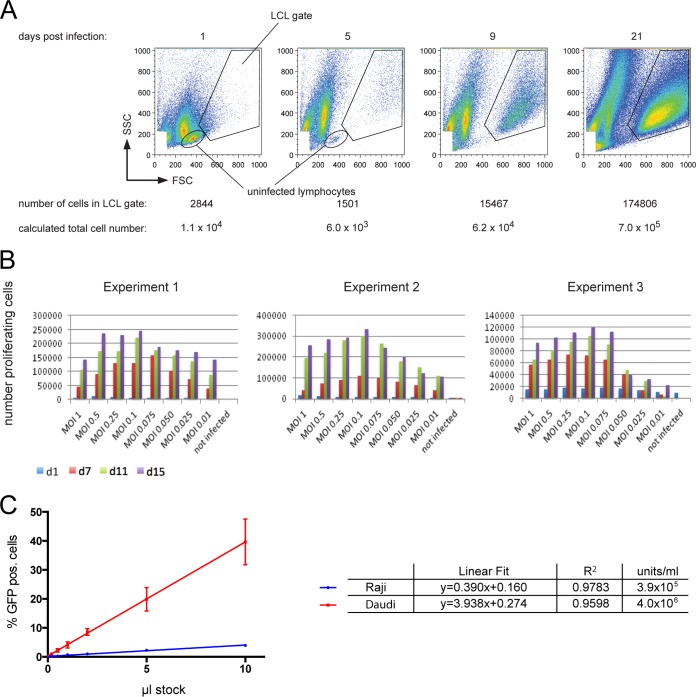
FIG 2.
Parameters and conditions optimally supporting EBV infection of primary B lymphocytes. (A) Determination of the number of proliferating, growth transformed cells after EBV infection. EBV-infected primary B lymphocytes increase in size and granularity to become B blasts, which can be detected in flow cytometry. The FSC (forward scatter) represents the size and the SSC (sideward scatter) represents the granularity of a cell. In contrast to noninfected, primary B cells, which exhibit low FSC/SSC signals, EBV-infected lymphoblasts locate to the “LCL gate” as indicated as early as 5 days p.i. In the experimental results shown, 6 × 105 adenoid B cells were infected with wt EBV (MOI = 0.1 GRU). At 1 day p.i., infected cells were supplied with fresh medium and divided into four fractions at a concentration of 1.5 × 105 cells/ml each. At 1, 5, 9, and 21 days p.i., one fraction was quantitatively analyzed by flow cytometry as indicated, and the total number of growth-transformed cells was calculated with the aid of a known number of APC beads, which provide a volume standard for sample normalization. The figure shows an example of a typical experiment with BCR+ B cells infected with wt EBV. (B) Infection of primary B lymphocytes with different MOIs and cellular outgrowth over time. Unsorted B cells were infected with the indicated wt 2089 EBV. The different MOI values are designated in the figure. At the given time points, the absolute numbers of activated and proliferating cells in the LCL gate were determined by flow cytometry as in panel A. Three independent experiments are shown. (C) Titration of a wt EBV stock sample comparing the infectivity of Epstein-Barr virions in Raji versus EBV-negative Daudi cells. A total of 5 × 104 Raji cells or 2 × 105 Daudi cells were infected with the indicated volumes of a wt 2089 virus stock. Percentage of GFP-positive cells was assessed by FACS analysis at 3 days p.i. For comparison, the GFP values were adjusted to 105 cells total. Shown are the linear regressions and the coefficient of determination (R2) derived from three independent experiments.
K1 and K15 can partially replace LMP2A.
BCR− B cells are proapoptotic because they lack the survival signals normally provided by the BCR. Upon infection, the wt 2089 EBV strain encoding LMP2A, which can substitute BCR's functions, can rescue these cells from apoptosis, as reported previously by us (28) and with B95.8 EBV by others (39, 40). We postulated that the KSHV proteins K1 and K15 might also functionally replace LMP2A. In order to challenge this hypothesis, we established mutant EBVs encoding K1 or K15 in lieu of LMP2A (Fig. 1A).
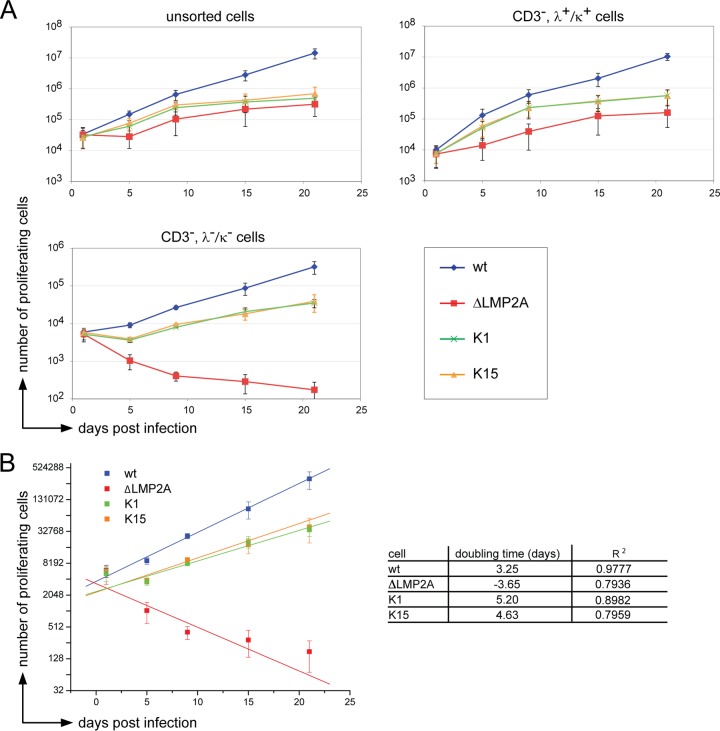
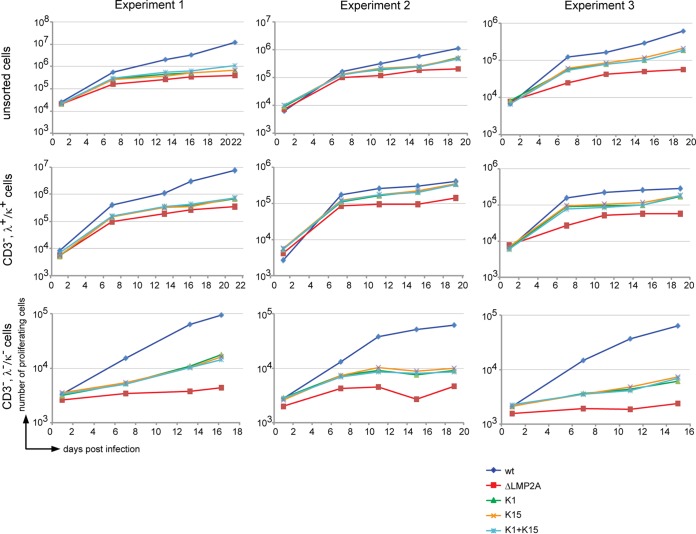
Unsorted, BCR+ (CD3−, λ+/κ+), or BCR− (CD3−, λ−/κ−) primary B cells were infected with wt 2089 EBV, ΔLMP2A EBV, K1 EBV, or K15 EBV. The proliferation of unsorted or BCR+ B cells did not depend on LMP2A, K1, or K15 because the endogenous BCR provides the necessary survival signals in contrast to BCR− B cells (Fig. 3A). As expected, ΔLMP2A EBV-infected BCR− B cells did not proliferate (Fig. 3A, bottom panel) but presumably went into apoptosis, as reflected by a negative doubling time of −3.65 days (Fig. 3B). K1 EBV or K15 EBV-infected BCR− B cells were rescued from apoptosis with doubling times of 5.2 and 4.65 days, respectively, and longer than 2089 wt EBV-infected BCR− B cells (3.25 days). We concluded from this set of experiments that K1 and K15 could partially replace LMP2A's function in this infection model.
FIG 3.
The KSHV genes K1 and K15 partially rescue BCR− B cells from apoptosis. (A) Unsorted, sorted BCR+ (CD3−, λ+/κ+) and BCR− (CD3−, λ−/κ−) B cells were infected with the indicated EBV strains. At the given time points, the absolute numbers of activated proliferating cells in the LCL gate was determined by flow cytometry (Fig. 2A). The means and standard deviations of three independent experiments are shown. (B) The numbers of BCR− cells (y axis, log2 scale) in the LCL FACS gate after infection with four different EBV strains was plotted over time (x axis). The doubling time is the slope of the linear fit function.
Dual infection with K1 EBV and K15 EBV revealed no cooperativity of the two KSHV genes.
The KSHV proteins K1 and K15 differ in their molecular design and in the cellular signaling mediators they recruit. It thus appears that the two proteins might serve different and yet additive or synergistic roles in B cells or in other cells infected with KSHV. We therefore sought to determine whether BCR− B cells dually infected with K1 EBV and K15 EBV might benefit from possible cooperative functions of K1 and K15. We chose to infect the primary B cells with virus stocks at concentrations that ensured doubly infected cells under optimal conditions. Two additional findings support the settings, which we chose for these experiments.
(i) We learned from quantitative PCR analysis that the experimental definition of GRU underestimates the concentration of physical particles by several logs (33). In addition, Raji cells are readily infected with EBV but less so than other cells. For example, EBV-negative Daudi cells (27) are far more permissive and better targets than are Raji cells. A careful titration of a wt 2089 EBV stock on both cell lines revealed that an infection of Raji cells underestimates the number of infectious Epstein-Barr virions present in the viral stocks by a factor of 10 (Fig. 2C). However, comparing Raji and EBV-negative Daudi cells also revealed that the latter are less consistent with respect to EBV infection, as indicated by a generally higher standard deviation of the slope of linear regression and slightly inferior goodness of fit. EBV-negative Daudi cells were reported to contain a variable fraction of apoptotic cells and are more difficult to maintain (27), which might explain our observation. Taken together, an MOI of 0.1 ensures that each B-lymphocyte is infected with at least one functionally intact Epstein-Barr virion on average.
(ii) Infections of B lymphocytes with an MOI of 0.25 were well tolerated and provided cell numbers very close to the optimal virus dose, i.e., an MOI of 0.1 (Fig. 2B), indicating that there is little viral interference if any. We therefore hypothesized that infection with two virus stocks with an MOI of 0.1 each is tolerated and should yield a major fraction of doubly infected primary B lymphocytes on average according to the model of Gaussian normal distribution.
Therefore, we infected unsorted primary B cells, sorted BCR+ (CD3−, λ+, or κ+) or BCR− (CD3−, λ−, and κ−) B cells with our four single virus stocks or with both K1 EBV and K15 EBV as indicated in Fig. 4. In three independent experiments with B cells from three different donors, the proliferation rate of BCR+ or BCR− B cells singly or dually infected with K1 EBV and K15 EBV did not differ. This finding did not prove but did suggest that K1 and K15 have similar, noncomplementing functions. In this model and under our experimental conditions, K1 and K15 did not act additively or synergistically in infected primary human B cells. In contrast to similar infection experiments shown in Fig. 3, we had to compromise on the purity of sorted BCR− primary B lymphocytes in order to obtain sufficient numbers of B cells to be used in the three experiments shown in Fig. 4. As a consequence, BCR− (CD3−, λ−, and κ−) B cells infected with ΔLMP2A EBV appear to be contaminated with a low fraction of BCR+ cells, which obscured the cellular phenotype reported earlier by us (4) and shown in Fig. 3A (bottom panel).
FIG 4.
High-titer double infection with K1 EBV and K15 EBV stocks revealed no cooperativity of the two KSHV genes. In the three individual experiments shown, unsorted, sorted BCR+ (CD3−, λ+/κ+), or BCR− (CD3−, λ−/κ−) B cells were infected with wt EBV, ΔLMP2A, K1, or K15 mutant EBV strains at 0.1 GRU per cell. In coinfection experiments, all B cells were dually infected with K1 and K15 mutant EBVs at this virus dose. At the indicated time points p.i., the absolute numbers of cells in the LCL gate were determined by flow cytometry as described in Fig. 2A.
K1 and K15 do not block BCR signaling.
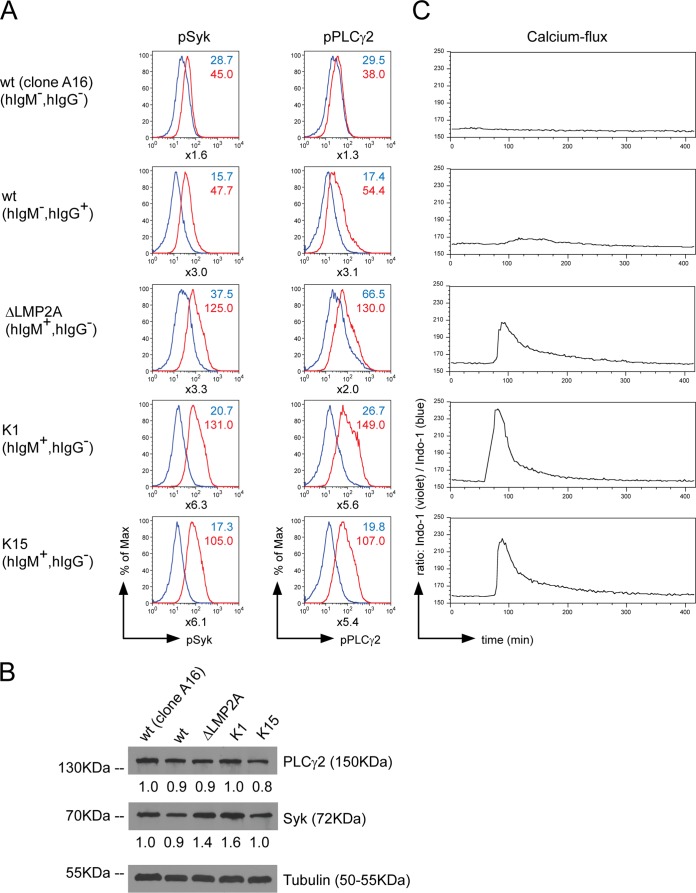
K1 and K15 were reported to interfere with BCR signaling upon ectopic expression in B cells (20, 21, 41; see reference 16 for a recent review). The influence of LMP2A, K1, and K15 on BCR-induced phosphorylation of Syk and PLCγ2, two key signal mediators of BCR, as well as calcium influx, were investigated in established LCLs, which had been generated by infecting adenoid B cells with wt 2089 EBV, ΔLMP2A EBV, K1 EBV, or K15 EBV stocks. The BCR of the LCLs was cross-linked for 10 min with an antibody directed against surface immunoglobulin or the cells were left untreated. The LCL clone A16, which is BCR negative and infected with wt EBV (28) was included as negative control. Intracellular pSyk and pPLCγ2 levels were detected with suitable antibodies as shown in Fig. 5A. In BCR+ LCLs infected with wt EBV, ΔLMP2A EBV, K1 EBV, or K15 EBV strains, BCR cross-linking induced Syk and PLCγ2 phosphorylation, indicating that neither LMP2A nor K1 and K15 inhibited phosphorylation of BCR signaling molecules. The BCR− wt EBV-infected LCLs (clone A16) did not show a significant increase in Syk and PLCγ2 phosphorylation after antibody-mediated BCR activation, as expected. In addition, the total levels of Syk and PLCγ2 in the five LCLs investigated in Fig. 5A were analyzed by Western blotting immunodetection revealing similar levels (Fig. 5B). This finding indicates that the changes in the phosphorylation levels of Syk and PLCγ2 do not reflect differences in the steady-state levels of these proteins.
FIG 5.
The KSHV proteins K1 and K15 do not inhibit BCR signaling. LCLs were established by infection of primary B cells with wt EBV, ΔLMP2A EBV, K1 EBV, and K15 EBV. An LCL (clone 16) generated from infection with wt 2089 EBV and unable to express a BCR was included as negative control. (A) The LCLs were cross-linked with 25 μg of α-hIgG/M antibody/ml for 10 min or left untreated. Intracellular phosphorylated Syk (pSyk) and PLCγ2 (pPLCγ2) were stained with phosphospecific antibodies. Histograms of flow cytometry analysis are shown. Numbers next to the graphs display the mean fluorescence intensities for uninduced (blue) and cross-linked (red) cells. Numbers below the graphs indicate the fold increases in the mean fluorescence intensities of the pSyk and pPLCγ2 signals after BCR stimulation. (B) Western blot immunodetection of the total levels of Syk and PLCγ2 in the LCLs used in panel A. Three independent experiments were carried out and Western blot bands corresponding to Syk and PLCγ2 were quantified and normalized against tubulin levels. The protein levels of clone A16 were set to one. (C) The LCLs were loaded with 3 μM Indo-1, and the violet/blue ratio of Indo-1 was measured over time. After 1 min of baseline measurement, the BCR was cross-linked with 25 μg of α-hIgG/A/M F(ab)2 fragment/ml.
Next, we analyzed calcium influx as a second function of BCR-emanating signals in this model (Fig. 5C). BCR was cross-linked with an α-hIgG/A/M F(ab)2 antibody fragment. The clone A16 LCL did not show an increase in cytoplasmic calcium levels after antibody-mediated BCR cross-linking and served as a negative control. LCLs infected with wt EBV but not ΔLMP2A EBV showed impaired calcium mobilization after BCR cross-linking as reported (22), but BCR cross-linking in LCLs generated with K1 EBV or K15 EBV induced calcium influx, suggesting that K1 and K15 proteins did not block BCR-activated calcium signaling in this model.
LMP2A, K1 and K15 control a related set of cellular genes in newly infected B cells.
Phenotypes of B cells infected with wt EBV, K1 EBV, or K15 EBV were remarkably similar but differed from ΔLMP2A EBV-infected cells in the experiments shown in Fig. 3 and 4. We hypothesized that LMP2A, together with K1 and K15 could regulate related sets of downstream cellular genes. Therefore, we infected unsorted B cells with the four virus stocks, wt EBV, K1 EBV, K15 EBV or ΔLMP2A EBV, adjusted to yield similar rates of EBV-activated and growth-transformed B cells. At 8 days p.i. activated B cells in the lymphoblast gate were collected by FACS sorting, and their RNAs were analyzed with the aid of the Affymetrix GeneChip Human Gene 1.0 ST Array covering the whole transcriptome of approximately 29,000 annotated human genes. B cell preparations from four different donors were infected and analyzed accordingly.
A standard principal component analysis (PCA) showed that the measured gene expression profiles were highly biased by specific effects arising from the four individual donors of the B lymphocytes (Fig. 6A). Thus, we applied ICA (43) to the data, a method able to uncover processes that are hidden by superimposing processes. ICA is a general computational method that has been further developed to disclose complex biological processes and mechanisms from microarray data in models, which study conditions of differentially expressed genes (43, 44). ICA is an unsupervised, hypothesis-free approach capable of extracting microarray data to yield statistically independent components, e.g., genes that define novel biological processes or regulatory networks (45).
ICA can yield modes of gene expression indicative of the underlying biological processes but it can also be applied to group samples into functional categories identifying regulatory networks (44, 46). In comparison to PCA, wherein the data are transformed to linearly uncorrelated principal components, ICA searches for statistically independent sources. In brief and as shown in Fig. 6B, in a matrix (X) of gene expression data where each row corresponds to a sample and each column is the expression profile of a gene, the goal is to find a matrix (S) of source profiles that reflects the underlying biological processes. Under the assumption that these processes are statistically independent in the first approximation and following a linear mixing model, ICA can be used to find a mixing matrix (A) such that X = AS (45, 47). The columns of the mixing matrix form so-called feature profiles that display the contribution of the corresponding source to the samples (Fig. 6C). A specific source profile of interest can now be identified by comparing a design vector reflecting a desired quality available from the experimental design with the corresponding feature profile (44).
Subsequent clustering, another type of unsupervised analysis, groups genes that appear to contribute to the same independent source profile assuming that these profiles represent biological processes. Here, we were interested to identify the source profile characteristic of ΔLMP2A EBV-infected primary B cells and the set of genes that mainly reflects a differential response to the LMP2A mediated signal. We therefore identified the feature profile with the strongest correlation to our experimental setup (Fig. 6B and C).
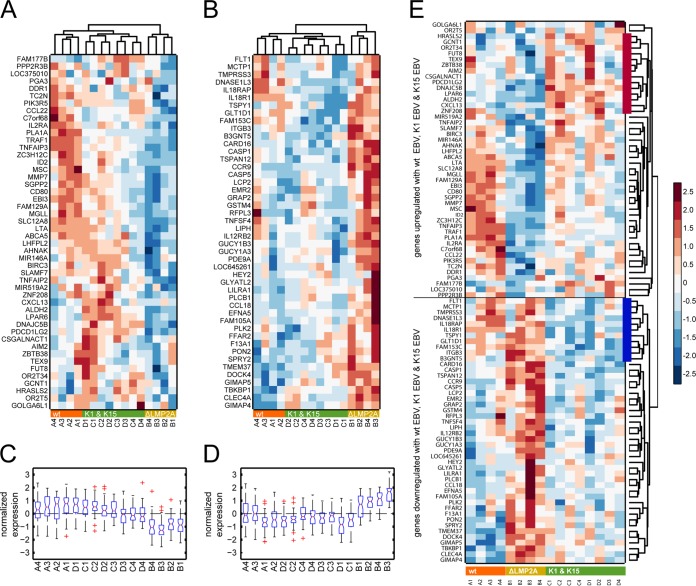
It has been shown that meaningful biological information can be deduced from source profiles by selecting its most active genes (46). From the related source profile, we identified two clusters of genes with a source expression below or above the mean plus 3.5 times the standard deviation (see Fig. 6C). A stringent threshold of 3.5 was chosen in order to obtain two clusters of about 50 genes each.
The heat map in Fig. 7A shows cellular genes upregulated in B cells infected with wt EBV, K1 EBV, or K15 EBV compared to genes in ΔLMP2A EBV-infected cells. Vice versa, genes in the heat map in Fig. 7B were upregulated in cells infected with ΔLMP2A EBV. One-way hierarchical clustering of the 16 samples yielded two dendrograms in Fig. 7A and B that clearly separated the four samples infected with ΔLMP2A from the 12 samples with wt EBV-, K1 EBV-, and K15 EBV-infected cells. Profiles of genes regulated in K1 EBV- and K15 EBV-infected cells were very similar; their relations merely reflected individual, donor-specific characteristics, as revealed by the indexed pairs in Fig. 7 (C1 and D1 from donor 1, C2 and D2 from donor 2, etc.). Figure 7A and B clearly indicated that K1- or K15-infected samples were more closely related to wt EBV-infected cells but only distantly related to ΔLMP2A-infected cells.
FIG 7.
ICA and subsequent sample clustering identifies genes coregulated by LMP2A, K1, and K15. (A) Heat map displaying genes upregulated during wt (strain 2089) EBV, K1 EBV, or K15 EBV infection compared to ΔLMP2A EBV infection. (B) Genes upregulated in ΔLMP2A EBV-infected cells. After ICA, samples were clustered using two-way hierarchical clustering. The dendrograms on top of the heat maps displays sample similarities separating the four ΔLMP2A samples from wt EBV, K1 EBV, and K15 EBV samples. The indices A1, A2 … to … D3, D4 denote the 16 samples, where characters and numbers indicate the four different virus stocks and four individual B cell donors, respectively. The term prefixes “A” and “B” indicate wt EBV- and ΔLMP2A EBV-infected B cell samples, respectively. Likewise, the term prefixes “C” and “D” indicate samples infected with K1 and K15 EBV. Infected B cells were purified from adenoids of four different donors (donors 1 to 4). Individual gene expression profiles were normalized by setting the mean as 0 and the standard deviation as 1. Relative expression values were depicted as a color scale in which blue and red denote down- and upregulated gene expression, respectively. (C and D) Box plots show the distributions of the normalized gene expression of the 16 samples. In panel C, gene expression of the clustered genes in cells infected with ΔLMP2A shows remarkable downregulation compared to wt EBV, K1 EBV, and K15 EBV, whereas the genes in panel D show the reverse situation for their expression profiles. (E) One-way hierarchical clustering of the combined genes (A and B) reveals coregulated clusters that depend on LMP2A. A heat map of the combined gene clusters shown in panels A and B was produced. The dendrogram indicates two major groups encompassing genes that are upregulated in wt EBV-, K1 EBV-, and K15 EBV-infected cells but downregulated in ΔLMP2A EBV-infected cells and vice versa. Additional groups that encompass genes commonly upregulated or downregulated in B cells infected with either K1 EBV or K15 EBV (red and blue bars, respectively) are readily visible.
Box plots with the distributions of gene expression of the 16 samples supported this interpretation and revealed a remarkable downregulation (Fig. 7C) or upregulation (Fig. 7D) of the expression of clustered genes in cells infected with ΔLMP2A EBV compared to wt EBV, K1 EBV, and K15 EBV.
We combined the two clusters and performed a one-way cluster analysis of ∼100 cellular genes in the 16 samples shown in Fig. 7E. As in the heat maps in Fig. 7A and B, two major clusters could be deduced that encompass genes coinduced in wt EBV-, K1 EBV-, and K15 EBV-infected cells but downregulated in ΔLMP2A EBV-infected cells and vice versa (see the top and bottom halves of the heat map in Fig. 7, respectively). In essence, this finding again underscores that the expression of LMP2A, K1, or K15 activates similar pathways and probably executes related functions in this infection model with primary B lymphocytes.
DISCUSSION
Wild-type EBV but not ΔLMP2A EBV rescued infected BCR− GC B cells from apoptosis (4), indicating that an active LMP2A signal is required to support the survival of these B cells at the initial stage of infection. It is likely that LMP2A provides early antiapoptotic functions in conjunction with the two viral bcl-2 homologues, BHRF1 and BALF1 (48), expressed in the prelatent phase of EBV infection (49). The presumed concerted action of several viral genes dedicated to cellular survival suggests that infected cells have to cope with different proapoptotic signals, e.g., DNA damage response signals (50) and interferon induction (32), which the virus must counteract for its success. LMP2A might complement the two redundant viral BCL-2 proteins (48) and induce cellular target genes (Fig. 7) that indirectly support B cell survival, as reported in transgenic mice (51) and Burkitt's lymphoma cell lines expressing LMP2A (52).
The KSHV proteins K1 and K15 have been reported to have multiple functions depending on the host cell (see reference 16 for a recent review). For example, K1 and K15 were found to interfere with BCR signaling (20, 21, 41), but cross-linking of BCR+ LCLs infected with K1 EBV or K15 EBV induced calcium influx and phosphorylation of Syk and PLCγ2, indicating that K1 or K15 do not block BCR-emanating signals in our model (Fig. 5). However, we cannot discard the possibility that the EBV recombinants expressed levels of K1 or K15 proteins sufficient to mimic a “trickling” BCR (53) but insufficient to antagonize BCR-dependent signaling, as observed after ectopic expression of K1 or K15 (20, 21, 41).
The functions of LMP2A, K1, and K15 were analyzed in two different approaches: we studied the survival and proliferation of infected BCR+ and BCR− B cells over time and obtained gene expression profiles of unsorted, mainly BCR+ B cells shortly after infection. With both approaches we found that K1 and K15 activities are indistinguishable (Fig. 4 and 5) and closely related to phenotypes and functions of LMP2A (Fig. 7). Microarrays have become standard tools to provide insights into gene regulation and the underlying biological processes. Their biomathematical analysis and interpretation, however, is not standardized. Deliberately, we used ICA, an unsupervised, hypothesis-free approach, to analyze the microarray data sets from B cells of four individual donors infected with the four virus stocks. We wanted to challenge our initial observation in which K1 and K15 expression evoked comparable, yet noncomplementing phenotypes in BCR− B cells, replacing LMP2A at least partially (Fig. 3). Reassuringly, ICA in conjunction with subsequent hierarchical clustering, another unsupervised technique of data analysis, confirmed our initial observation (Fig. 7). The microarray data nicely recapitulated our findings in BCR− B cells and identified two different groups: samples infected with wt EBV, K1 EBV, and K15 EBV and the group of four samples infected with ΔLMP2A EBV (Fig. 7). Dendrograms reflect the relationship of the different samples and indicated that K1 and K15 samples are almost indistinguishable in this experimental setting but related to LMP2A+ and yet very distinct from LMP2A− samples.
Interestingly, certain target genes of LMP2A, K1 and K15 shown in Fig. 7E underscore the functional contribution of this group of herpesviral oncogenes to B cell survival and activation. For example, CD80 is highly upregulated in wt EBV, moderately induced in K1 and K15 but downregulated in ΔLMP2A EBV-infected B cells. This finding is further supported by CD86 surface expression measured by FACS that parallels CD80 gene regulation. Both receptors are coinduced by the expression of LMP2A (54), K1, and K15, suggesting that the three encoded viral proteins uniformly contribute to B cell activation.
Similarly, PIK3R5, the regulatory subunit of phosphatidylinositol 3-kinase (PI3K), which itself is critically involved in LMP2A signaling (55, 56), is upregulated by LMP2A, K1, and K15 but downregulated if LMP2A is absent. This observation suggests that K1 and K15 indeed activate PI3K-dependent signaling pathways (57) as LMP2A does. B cell survival depends on PI3K-dependent signaling (58), which might explain the antiapoptotic effects of LMP2A, K1, and K15 proteins seen in our model.
CARD16 and CASP1 encoding COP1 and caspase 1, respectively, are critically involved in activating apoptosis signals and are upregulated in ΔLMP2A EBV-infected B cells but not in other samples, suggesting that LMP2A might prevent the initial upregulation of proapoptotic cellular regulators upon infection. The three selected examples only skim the data uncovered by our microarray analysis but suggest that our model is informative reflecting an important aspect of EBV's and KSHV's biology.
Models of B cell infection have been scarce and limit our knowledge of KSHV-induced B cell lymphomagenesis (see reference 24 for a recent review), although studies with activated primary cells from peripheral blood mononuclear cells or tonsils showed a 5 to 10% rate of KSHV infection (59–61). In this setting, KSHV supported the short-term proliferation of tonsillar B cells only, suggesting that this virus contributes to cellular survival (59). Both K1 and K15 are expressed in PELs and have been linked to the cellular transformation and/or maintenance of latent B cell infection with KSHV (16). It is not clear whether EBV or KSHV infect human BCR− B cells in vivo, but in vitro these cells only survive and proliferate, eventually, if infected with EBV strains expressing LMP2A, K1, or K15, thus supplying the proapoptotic B cells with a BCR-like survival signal.
Our findings suggest that K1 and K15 are instrumental in KSHV-associated B cell lymphoma and might explain the existence of BCR− PEL cells (14, 15). K1 and K15 might also contribute to other malignancies, including Kaposi's sarcoma or multicentric Castleman's disease, because K1 is regularly expressed in biopsy specimens of these entities (62).
ACKNOWLEDGMENTS
We thank Dietmar Martin and Kerstin Maier for critical and important support with the microarray hybridizations and data acquisition, Christine Göbel for help and expertise in preparing, purifying, and infecting primary B cells, and Dagmar Pich for experimental advice and assistance during cell sorting experiments.
M.G. is an Alexander-von-Humboldt Fellow. Our work was supported by funds from the Deutsche Forschungsgemeinschaft (SPP1230, SFB1054/TP B05, SFB1064/TP A13, and SFB-TR36/TP A04), the National Institutes of Health (grant CA70723), and Deutsche Krebshilfe (grants 107277 and 109661).
REFERENCES
- 1.Kuppers R. 2009. The biology of Hodgkin's lymphoma. Nat Rev Cancer 9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 2.Matolcsy A, Nador RG, Cesarman E, Knowles DM. 1998. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am J Pathol 153:1609–1614. doi: 10.1016/S0002-9440(10)65749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brauninger A, Schmitz R, Bechtel D, Renne C, Hansmann ML, Kuppers R. 2006. Molecular biology of Hodgkin's and Reed/Sternberg cells in Hodgkin's lymphoma. Int J Cancer 118:1853–1861. doi: 10.1002/ijc.21716. [DOI] [PubMed] [Google Scholar]
- 4.Mancao C, Hammerschmidt W. 2007. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 110:3715–3721. doi: 10.1182/blood-2007-05-090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaufils P, Choquet D, Mamoun RZ, Malissen B. 1993. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J 12:5105–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alber G, Kim KM, Weiser P, Riesterer C, Carsetti R, Reth M. 1993. Molecular mimicry of the antigen receptor signaling motif by transmembrane proteins of the Epstein-Barr virus and the bovine leukaemia virus. Curr Biol 3:333–339. doi: 10.1016/0960-9822(93)90196-U. [DOI] [PubMed] [Google Scholar]
- 7.Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K. 2004. B cell receptor signal strength determines B cell fate. Nat Immunol 5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 8.Kuppers R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat Rev Immunol 3:801–812. doi: 10.1038/nri1201. [DOI] [PubMed] [Google Scholar]
- 9.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 10.Horenstein MG, Nador RG, Chadburn A, Hyjek EM, Inghirami G, Knowles DM, Cesarman E. 1997. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood 90:1186–1191. [PubMed] [Google Scholar]
- 11.Jenner RG, Boshoff C. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim Biophys Acta 1602:1–22. [DOI] [PubMed] [Google Scholar]
- 12.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648–2654. [PubMed] [Google Scholar]
- 13.Arguello M, Sgarbanti M, Hernandez E, Mamane Y, Sharma S, Servant M, Lin R, Hiscott J. 2003. Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene 22:964–973. doi: 10.1038/sj.onc.1206270. [DOI] [PubMed] [Google Scholar]
- 14.Carbone A, Cilia AM, Gloghini A, Capello D, Todesco M, Quattrone S, Volpe R, Gaidano G. 1998. Establishment and characterization of EBV-positive and EBV-negative primary effusion lymphoma cell lines harbouring human herpesvirus type-8. Br J Haematol 102:1081–1089. doi: 10.1046/j.1365-2141.1998.00877.x. [DOI] [PubMed] [Google Scholar]
- 15.Said W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho SK, de Vos S, Cesarman E, Knowles DM, Koeffler HP. 1996. Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood 87:4937–4943. [PubMed] [Google Scholar]
- 16.Wen KW, Damania B. 2010. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett 289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkmann MM, Pietrek M, Dittrich-Breiholz O, Kracht M, Schulz TF. 2007. Modulation of host gene expression by the K15 protein of Kaposi's sarcoma-associated herpesvirus. J Virol 81:42–58. doi: 10.1128/JVI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers RC, Jung JU. 1998. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat Med 4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 19.Prakash O, Tang ZY, Peng X, Coleman R, Gill J, Farr G, Samaniego F. 2002. Tumorigenesis and aberrant signaling in transgenic mice expressing the human herpesvirus-8 K1 gene. J Natl Cancer Inst 94:926–935. doi: 10.1093/jnci/94.12.926. [DOI] [PubMed] [Google Scholar]
- 20.Lee BS, Alvarez X, Ishido S, Lackner AA, Jung JU. 2000. Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi's sarcoma-associated herpesvirus K1. J Exp Med 192:11–21. doi: 10.1084/jem.192.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietrek M, Brinkmann MM, Glowacka I, Enlund A, Havemeier A, Dittrich-Breiholz O, Kracht M, Lewitzky M, Saksela K, Feller SM, Schulz TF. 2010. Role of the Kaposi's sarcoma-associated herpesvirus K15 SH3 binding site in inflammatory signaling and B-cell activation. J Virol 84:8231–8240. doi: 10.1128/JVI.01696-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller CL, Longnecker R, Kieff E. 1993. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J Virol 67:3087–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones T, Ye F, Bedolla R, Huang Y, Meng J, Qian L, Pan H, Zhou F, Moody R, Wagner B, Arar M, Gao SJ. 2012. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J Clin Invest 122:1076–1081. doi: 10.1172/JCI58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boshoff C. 2011. Unraveling virus-induced lymphomagenesis. J Clin Invest 121:838–841. doi: 10.1172/JCI46499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog 6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulvertaft JV. 1964. Cytology of Burkitt's tumour (African lymphoma). Lancet i:238–240. [DOI] [PubMed] [Google Scholar]
- 27.Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J 21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancao C, Altmann M, Jungnickel B, Hammerschmidt W. 2005. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood 106:4339–4344. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jochum S, Moosmann A, Lang S, Hammerschmidt W, Zeidler R. 2012. The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog 8:e1002704. doi: 10.1371/journal.ppat.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jochum S, Ruiss R, Moosmann A, Hammerschmidt W, Zeidler R. 2012. RNAs in Epstein-Barr virions control early steps of infection. Proc Natl Acad Sci U S A 109:E1396–E1404. doi: 10.1073/pnas.1115906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalla M, Gobel C, Hammerschmidt W. 2012. The lytic phase of Epstein-Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J Virol 86:447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dirmeier U, Neuhierl B, Kilger E, Reisbach G, Sandberg ML, Hammerschmidt W. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res 63:2982–2989. [PubMed] [Google Scholar]
- 35.Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 36.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. 2005. Independence and reproducibility across microarray platforms. Nat Methods 2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 38.Cardoso JF, Souloumiac A. 1996. Jacobi angles for simultaneous diagonalization. SIAM J on Matrix Analysis and Applications 17:161–164. doi: 10.1137/S0895479893259546. [DOI] [Google Scholar]
- 39.Bechtel D, Kurth J, Unkel C, Kuppers R. 2005. Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood 106:4345–4350. doi: 10.1182/blood-2005-06-2342. [DOI] [PubMed] [Google Scholar]
- 40.Chaganti S, Bell AI, Pastor NB, Milner AE, Drayson M, Gordon J, Rickinson AB. 2005. Epstein-Barr virus infection in vitro can rescue germinal center B cells with inactivated immunoglobulin genes. Blood 106:4249–4252. doi: 10.1182/blood-2005-06-2327. [DOI] [PubMed] [Google Scholar]
- 41.Choi JK, Lee BS, Shim SN, Li M, Jung JU. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J Virol 74:436–446. doi: 10.1128/JVI.74.1.436-446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuhierl B, Feederle R, Hammerschmidt W, Delecluse HJ. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc Natl Acad Sci U S A 99:15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutter D, Langmann T, Ugocsai P, Moehle C, Seibold E, Splettstoesser WD, Gruber P, Lang EW, Schmitz G. 2009. Analyzing time-dependent microarray data using independent component analysis derived expression modes from human macrophages infected with F. tularensis holartica. J Biomed Inform 42:605–611. doi: 10.1016/j.jbi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Schachtner R, Lutter D, Knollmuller P, Tome AM, Theis FJ, Schmitz G, Stetter M, Vilda PG, Lang EW. 2008. Knowledge-based gene expression classification via matrix factorization. Bioinformatics 24:1688–1697. doi: 10.1093/bioinformatics/btn245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SI, Batzoglou S. 2003. Application of independent component analysis to microarrays. Genome Biol 4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutter D, Ugocsai P, Grandl M, Orso E, Theis F, Lang EW, Schmitz G. 2008. Analyzing M-CSF-dependent monocyte/macrophage differentiation: expression modes and meta-modes derived from an independent component analysis. BMC Bioinformatics 9:100. doi: 10.1186/1471-2105-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebermeister W. 2002. Linear modes of gene expression determined by independent component analysis. Bioinformatics 18:51–60. doi: 10.1093/bioinformatics/18.1.51. [DOI] [PubMed] [Google Scholar]
- 48.Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalla M, Hammerschmidt W. 2012. Human B cells on their route to latent infection: early but transient expression of lytic genes of Epstein-Barr virus. Eur J Cell Biol 91:65–69. doi: 10.1016/j.ejcb.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Portis T, Longnecker R. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619–8628. doi: 10.1038/sj.onc.1207905. [DOI] [PubMed] [Google Scholar]
- 52.Fukuda M, Longnecker R. 2005. Epstein-Barr virus (EBV) latent membrane protein 2A regulates B-cell receptor-induced apoptosis and EBV reactivation through tyrosine phosphorylation. J Virol 79:8655–8660. doi: 10.1128/JVI.79.13.8655-8660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature 381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 54.Portis T, Dyck P, Longnecker R. 2003. Epstein-Barr Virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood 102:4166–4178. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- 55.Miller CL, Burkhardt AL, Lee JH, Stealey B, Longnecker R, Bolen JB, Kieff E. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant-negative effects on protein-tyrosine kinases. Immunity 2:155–166. doi: 10.1016/S1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 56.Engels N, Merchant M, Pappu R, Chan AC, Longnecker R, Wienands J. 2001. Epstein-Barr virus latent membrane protein 2A (LMP2A) employs the SLP-65 signaling module. J Exp Med 194:255–264. doi: 10.1084/jem.194.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomlinson CC, Damania B. 2004. The K1 protein of Kaposi's sarcoma-associated herpesvirus activates the Akt signaling pathway. J Virol 78:1918–1927. doi: 10.1128/JVI.78.4.1918-1927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. 2009. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassman LM, Ellison TJ, Kedes DH. 2011. KSHV infects a subset of human tonsillar B cells, driving proliferation and plasmablast differentiation. J Clin Invest 121:752–768. doi: 10.1172/JCI44185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myoung J, Ganem D. 2011. Infection of primary human tonsillar lymphoid cells by KSHV reveals frequent but abortive infection of T cells. Virology 413:1–11. doi: 10.1016/j.virol.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rappocciolo G, Hensler HR, Jais M, Reinhart TA, Pegu A, Jenkins FJ, Rinaldo CR. 2008. Human herpesvirus 8 infects and replicates in primary cultures of activated B lymphocytes through DC-SIGN. J Virol 82:4793–4806. doi: 10.1128/JVI.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damania B. 2007. DNA tumor viruses and human cancer. Trends Microbiol 15:38–44. doi: 10.1016/j.tim.2006.11.002. [DOI] [PubMed] [Google Scholar]