Abstract
The present study identified that ω-3 long chain polyunsaturated fatty acids (ω-3 PUFAs), docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) demonstrate anti-proliferative effects in lung cancer A549 cells. MTS and cytotoxicity assays were conducted to confirm that ω-3 PUFAs induced cell death. Autophagy-associated gene and signaling pathways were also detected. Microtubule-associated protein light chain 3 (LC3) expression was found to be increased subsequent to treatment with DHA and EPA, and the expression of LC3-II was particularly increased. mRFP-GFP-LC3 fluorescence staining and p62 expression levels were used to detect autophagic flux. The present results indicate that DHA and EPA block autophagic flux, suggesting autophagosome accumulation. Subsequent to treatment with DHA and EPA, which interfered with autophagosomes, the expression of Beclin 1 was significantly decreased, while the expression of phosphorylated Akt and phosphorylated mammalian target of rapamycin was significantly increased. Therefore, DHA and EPA exert anti-proliferative effects by inhibiting autophagy in A549 cells, which highlights the potential of DHA and EPA for use in the prevention or treatment of lung cancer.
Keywords: ω-3 long chain polyunsaturated fatty acids, autophagy, Akt/mammalian target of rapamycin
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide (1), and the five-year survival rate remains extremely poor (2). Overall, ~75–85% of lung cancers cases are non-small cell lung cancer (NSCLC), which includes squamous cell carcinoma, adenocarcinoma and large cell carcinoma. The treatment of lung cancer involves the use of medical therapies such as surgery, radiation, chemotherapy and palliative care (3) in an attempt to successfully treat or reduce the adverse impact of malignant neoplasms originating in lung tissue. Chemotherapeutic agents are the main treatment measures for NSCLC, but the side-effects are usually difficult to tolerate (4).
Fish oils are an excellent source of long-chain ω-3 polyunsaturated fatty acids (ω-3 PUFAs), including eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). Fish oil supplements are increasingly recognized by clinical studies to be useful for the treatment of a variety of human afflictions, including cancer (5). Numerous studies reveal evidence for the capability of ω-3 PUFAs to decrease proliferation, exert a pro-apoptotic effect and inhibit angiogenesis in several in vitro models of colon cancer (6–9).
A previous study indicated that DHA and EPA inhibit the proliferation of A549 cells and induce apoptosis, with autophagy also being observed under transmission electron microscopy (10). Autophagy is a type of programmed cell death and it is an important process that is involved in various human pathologies. Previous studies suggest that autophagy is important in the regulation of cancer development and progression and also in determining the response of tumor cells to anticancer therapy (11–14). Several cell signaling pathways are implicated in regulating autophagy, including the phosphatidyl inositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway (15,16). The present study aimed to investigate the role of autophagy on DHA- and EPA-induced cell death in A549 cells, and the associated Akt/mTOR signaling pathway.
Materials and methods
Cell and reagents
The human non-small cell lung carcinoma A549 cell line was obtained from The Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were grown in modified RPMI-1640 medium (HyClone, Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a humidified incubator with a 5% CO2 atmosphere. DHA, EPA and 3-methyladenine (3-MA) were obtained from Sigma-Aldrich (St. Louis, MO, USA).
MTS was obtained from Promega (Madison, Wisconsin, USA). A lactate dehydrogenase (LDH) kit was obtained from Jiancheng Bioengineering Institute (Nanjing, China). Lipofectamine 2000 reagent and Opti-MEM I reduced serum medium were obtained from Life Technologies (Carlsbad, CA, USA). The pCDNA3.1-mRFP-GFP-LC3 plasmid was provided by the Biomedical Research Center of Sir Run Run Shaw Hospital (Hangzhou, Zhejiang, China). Rabbit monoclonal primary antibodies against human mTOR (catalog no. 2983), phosphorylated-mTOR [p-mTOR (Ser2448); catalog no. 5536], Akt (catalog no. 4691), p-Akt (Ser473; catalog no. 4060), Beclin-1 (catalog no. 3495), human microtubule-associated protein 1 light chain 3 isoform B (LC3B; catalog no. 3868), p62 (catalog no. 8025) and β-actin (catalog no. 8457) were obtained from Cell Signaling Technology (Danvers, MA, USA). A bicinchoninic acid (BCA) protein assay kit was obtained from Beyotime Institute of Biotechnology (Haimen, Jiangsu, China), and a protein extraction kit was obtained from KeyGEN (Nanjing, China).
MTS assay for cell viability
The cells were seeded at a density of 2.5×105 cells/well in six-well plates and incubated for 24 h. DHA (50 µg/ml) or EPA (60 µg/ml) was added to the wells for 24 h, while the control group was administered with complete medium, as previously described (17). MTS solution was added to each well and incubated at 37°C for 0.5–2 h. Absorbance values were detected at 490 nm using a microplate reader (SynergyHT2; BioTek, Winooski, VT, USA). Cell viability was calculated based on the following formula:
Cell viability (%) = A490 experimental / A490 control × 100
Each experiment was repeated three times.
LDH detected by enzyme-labeling
The cells were seeded at a density of 2.5×105 cells/well in six-well plates and were incubated with 50 µg/ml DHA or 60 µg/ml EPA for 24 h, then the culture supernatant was collected and added to a 96-well plate. The LDH cytotoxicity kit was used according to the manufacturer's instructions. Briefly, the standard liquid, matrix buffer, coenzyme I, dinitrophenylhydrazine and sodium hydroxide solution were combined and incubated at 37°C. Absorbance values were detected at 450 nm by the microplate reader. The activity of LDH was calculated in U/l based on the following formula, where 0.2 mmol/l represents the concentration of the standard liquid:
Each experiment was repeated three times.
Plasmid transfection and morphological analysis
To analyze autophagic flux, the A549 cells were transfected with an mRFP-GFP-LC3 plasmid. The A549 cells were seeded at a density of 2×105 cells/well in six-well plates. At 90–95% confluency, the cells were transfected using Lipofectamine 2000, Opti-MEM I reduced serum medium and the pCDNA3.1-mRFP-GFP-LC3 plasmid, according to the manufacturer's instructions, for 42 h. Following the addition of 50 µg/ml DHA or 60 µg/ml EPA, the cells were cultured for 24 h. Subsequent to was hing twice with phosphate buffered saline, the cells were examined by fluorescence microscopy (BX51, Olympus, Tokyo, Japan).
Western blot analysis
Following treatment with 50 µg/ml DHA, 60 µg/ml EPA and 5 mM 3-MA, the cells were lysed in cell lysis buffer with phenylmethylsulfonyl fluoride, phosphatase inhibitor and protease inhibitor, according to the instructions of the manufacturer of the protein extraction kit. The protein concentration in the lysate was quantified using a BCA protein assay kit. Equal amounts of protein for each sample were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). Subsequent to blocking with 5% non-fat milk for 1 h, the membranes were incubated with monoclonal primary antibodies against mTOR, p-mTOR (Ser2448), Akt, p-Akt (Ser473), Beclin-1, LC3B, p62 and β-actin overnight at 4°C. The membranes were then incubated with a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Cell Signaling Technology; catalog no. 7074) for 2 h. The bands were detected using ECL (Amresco LLC, Solon, OH, USA). The protein levels were quantitated by densitometry using Gel-Pro Analyzer software (Media Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
All experiments were repeated at least three times. Statistical analyses were performed using SPSS software, version 17.0 (SPSS, Inc., Chicago, IL, USA). The data are expressed as the mean ± standard deviation and were analyzed using one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
ω-3 PUFAs inhibit the proliferation of A549 cells
A549 cells were treated with 50 µg/ml DHA or 60 µg/ml EPA for 24 h. The MTS assay was used to examine the anti-proliferative effect of DHA/EPA on A549 cells. As shown in Fig. 1A, the experimental groups demonstrated significantly decreased cell proliferation compared with the control group. The cell viability rates in the DHA and EPA groups were 26.1±1.3 and 30.72±1.66%, respectively, when treated for 24 h (P<0.01 vs. control).
Figure 1.
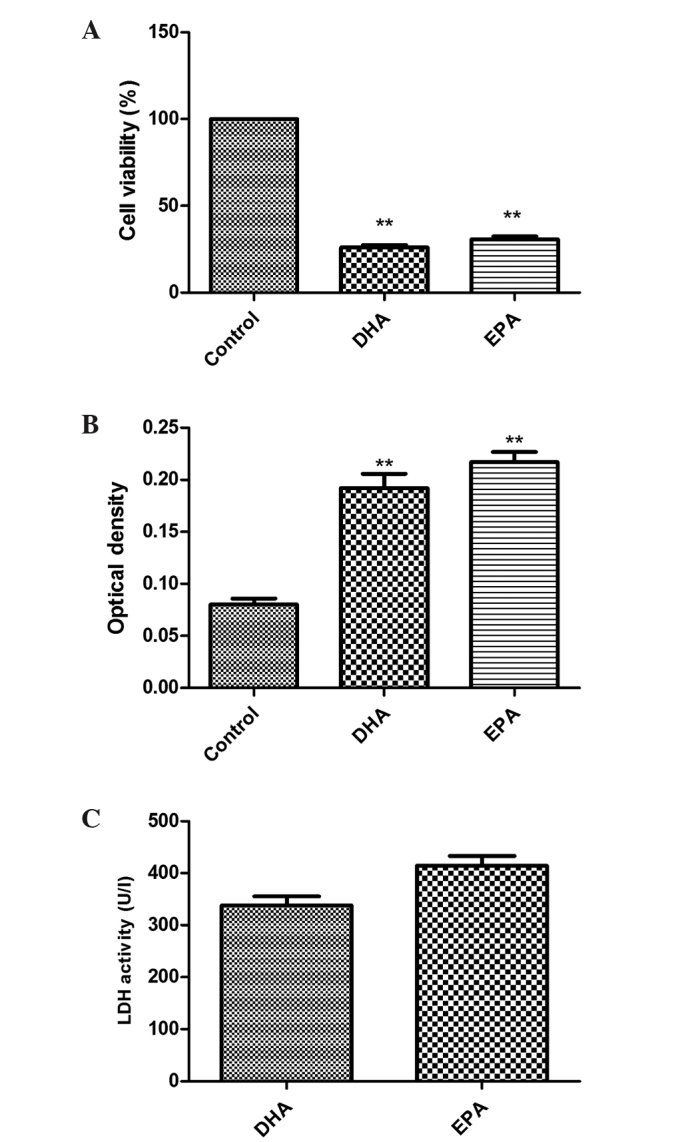
Effects of DHA and EPA on the growth of lung cancer A549 cells. (A) Cell viability (%). DHA (50 µg/ml) and EPA (60 µg/ml) were administered to A549 cells for 24 h and each agent significantly suppressed the proliferation of the A549 cells compared with the control cells (**P<0.01 vs. control). (B) LDH optical density. All experimental groups treated with 50 µg/ml DHA or 60 µg/ml EPA possessed a significantly higher optical density compared with the control group (**P<0.01 vs. control). (C) LDH activity (U/l). Compared with the control cells, LDH activity was evidently higher in the experimental groups. DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; LDH, lactate dehydrogenase; U/l, units per liter.
ω-3 PUFAs induce cytotoxicity in A549 cells
LDH is located in the cytoplasm and is released when the membrane is damaged. Therefore, the degree of adherent cell damage may be evaluated through the LDH levels in the supernatant. The higher the optical density (OD) was, the more LDH was released, and therefore there was elevated cytotoxicity. As shown in Fig. 1B, the LDH OD values subsequent to treatment were 0.08±0.006, 0.192±0.014 and 0.217±0.01 for the control, DHA and EPA groups, respectively; the values for the DHA and EPA groups differed significantly from that of the control group (both P<0.01). Therefore, DHA and EPA significantly inhibited the growth of A549 cells. LDH activity was calculated and the results are shown in Fig. 1C. Compared with the control group, the LDH activity of the treated groups was significantly increased (P<0.01).
Maturation of autophagosomes is inhibited by ω-3 PUFAs
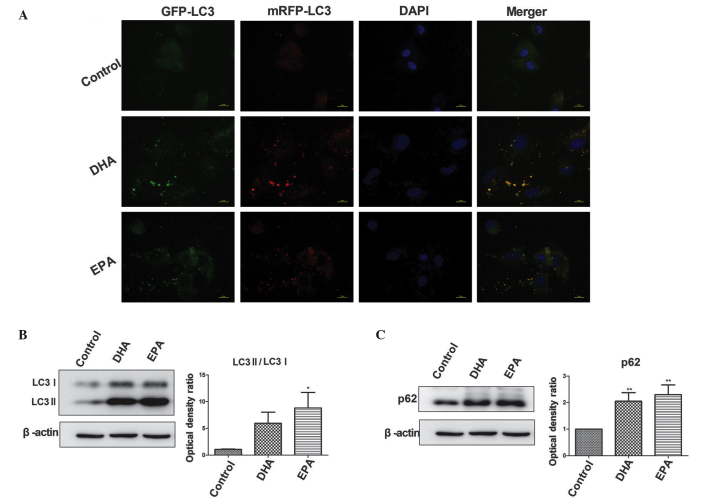
The pCDNA3.1-mRFP-GFP-LC3 plasmid was transfected into A549 cells. Cells undergoing autophagy demonstrated a notable quantity of punctate green fluorescent protein (GFP) and monomeric red fluorescent protein (mRFP) signals, while normal cells demonstrated a primarily diffuse GFP/mRFP signal. The fusion of the autophagosomes and lysosomes indicated the formation of autophagolysosomes, which are acidic vesicular organelles (18). In an acidic environment, the GFP signal is quenched while the mRFP signal is stable (19). As shown in Fig. 2A, the control group exhibited a diffuse GFP/mRFP-LC3 signal while the DHA and EPA groups exhibited evident punctate GFP/mRFP-LC3 signals. In the merged view, the LC3 signal in the experimental groups was yellow, indicating that GFP-LC3 was not suppressed. This result indicates a lack of formation of autophagolysosomes.
Figure 2.
Detection of autophagic flux. (A) Fluorescence staining revealed diffuse GFP/mRFP-LC3 signals in the control group but evident punctate GFP/mRFP-LC3 signals were present in the DHA and EPA groups, indicating that the addition of ω-3 long chain polyunsaturated fatty acids increased LC3 expression levels. The merged image of the GFP and mRFP signals is yellow, suggesting a blockage of autophagic degradation. (B) The results from the western blot analysis revealed increased levels of LC3, particularly LC3-II, subsequent to treatment with DHA and EPA, leading to a higher ratio of LC3-II/LC3-I compared with the control group (*P<0.05 vs. control). (C) The p62 expression levels were significantly higher in the cells treated with DHA and EPA compared with the control cells (**P<0.01 vs. contol). The results presented in (A–C) indicate that DHA and EPA blocked autophagic flux. DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; LC3, light chain 3; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
Western blot analysis
The processing of the mammalian homologue of the yeast Atg8 protein, LC3-I, was analyzed by western blot analysis. During autophagy, LC3-I undergoes cleavage and lipidation to yield LC3-II, and quantitation of the levels of LC3-I and LC3-II in culture provides a good measurement of the degree of autophagy (20). The LC3-II and LC3-I expression level ratio in the DHA and EPA groups was increased compared with the control group. In the DHA group, the ratio tended towards an elevated value, although this difference was not significant; however the ratio was significantly increased in the EPA group (P<0.05; Fig. 2B).
The level of p62 expression was significantly increased in the DHA and EPA groups compared with the control group (P<0.01; Fig. 2C), indicating an accumulation of p62, which is considered to be indicative of autophagy abnormality.
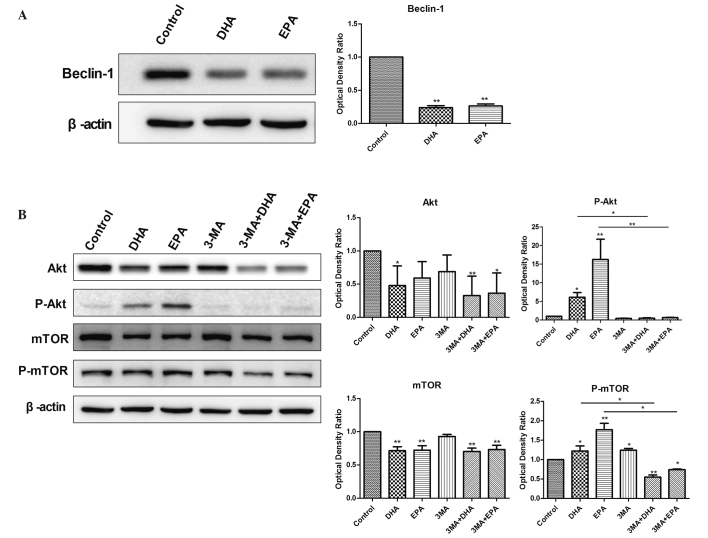
The present study also analyzed the levels of Beclin 1, an ATG gene product that is essential for autophagy. Compared with the control group, the expression of Beclin 1 was significantly decreased in the DHA and EPA treatment groups (P<0.01; Fig. 3A), indicating that treatment with either DHA or EPA significantly reduced Beclin 1 levels.
Figure 3.
Western blot analysis of the protein-level expression of Beclin-1 and the Akt/mTOR signaling pathway. (A) Beclin-1 expression levels in the DHA and EPA groups were significantly increased compared with the control group (**P<0.01 vs. control). (B) Compared with the control group, the expression levels of p-Akt and p-mTOR in the DHA and EPA groups were significantly increased (*P<0.05 vs. control), suggesting the activity of Akt and its downstream target mTOR. The administration of 3-MA to the groups treated with ω-PUFAs significantly decreased the p-Akt and p-mTOR levels compared with the administration of ω-PUFAs alone (*P<0.05 vs. control). Total Akt and mTOR levels were decreased in the group treated with ω-PUFAs alone compared with the groups also administered with 3-MA. DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; ω-PUFAs, ω-3 long chain polyunsaturated fatty acids; 3-MA, 3-methyladenine; mTOR, mammalian target of rapamycin; p-mTOR, phosphorylated mTOR; p-Akt, phosphorylated Akt.
Autophagy is inhibited by 3-MA through the blocking of autophagosome formation by the inhibition of type III PI3K. Compared with the control group, the expression of p-Akt and p-mTOR was significantly increased in the DHA and EPA groups (P<0.05; Fig. 3B). No significant difference in p-Akt expression was identified between the groups treated with 5 mM 3-MA, 3-MA combined with DHA, and 3-MA combined with EPA (P>0.05 vs. control), but p-mTOR expression was significantly reduced in the groups treated with 3-MA and DHA, and 3-MA and EPA (P<0.05). Therefore, treatment with DHA or EPA was able to significantly increase the levels of p-Akt and p-mTOR.
The total expression levels of the Akt protein were significantly decreased in the DHA group (P<0.05; Fig. 3B) and slightly (non-significantly) decreased in the EPA group compared with the control. The total expression levels of the mTOR protein were significantly decreased in the DHA and EPA treatment groups (P<0.01). The total Akt and mTOR levels were significantly decreased in the groups treated with 3-MA and DHA and 3-MA and EPA (P<0.05) and slightly, but not significantly, decreased in the 3-MA group. Therefore, treatment with DHA, EPA and 3-MA decreased the total expression level of Akt and mTOR.
Discussion
The major bioactive components of fish oil are the long-chain ω-3 PUFAs, which contain EPA and DHA. Consumption of EPA and DHA is associated with a decreased risk of breast (6,21,22), colon (8), prostate, and kidney (23) cancers. However, there have been few studies into the association between ω-3 PUFAs and lung cancer. As lung cancer is the leading malignant tumor in humans, with 75–85% of cases involving NSCLC (24), the A549 cell line was chosen for the present study.
The MTS assay performed to assess the cell viability indicated that DHA and EPA demonstrated antiproliferative effects in the A549 cells. In addition, the LDH activity of the culture supernatant was increased subsequent to treatment with DHA or EPA, due to cell membrane damage. Overall, these results are consistent with and confirm the results of a previous study (10).
Autophagic vacuoles have previously been observed under transmission electron microscopy (10). During the formation of autophagosomes, the LC3-I cytoplasmic form is cleaved and lipidated to generate the membranous LC3-II form (25,26). To determine the form of LC3 that is affected by the presence of DHA and EPA, western blot analysis was used to detect LC3-I and LC3-II levels. The results revealed higher levels of LC3, particularly LC3-II, leading to a significantly increased LC3-II to LC3-I ratio subsequent to treatment with ω-3 PUFAs compared with the control group (P<0.05). An increased LC3-II to LC3-I ratio indicates an increased quantity of autophagosomes. Autophagosome accumulation may result from either the increased formation of autophagosomes or from the blockage of autophagic degradation (27).
Beclin 1 is the mammalian ortholog of the yeast Vps30/Apg6 gene, which is required for autophagosome formation, and is monoallelically deleted in a high percentage of human carcinomas (28–30). Previous studies have revealed that the promotion of the expression of Beclin 1 through reduced autophagy demonstrates anticancer effects (25,26,31). The present study indicated that Beclin 1 expression levels were significantly decreased subsequent to treatment with DHA and EPA, which suggests that ω-3 PUFAs inhibit autophagosome formation in A549 cells.
There is conclusive evidence that the PI3K/Akt/mTOR pathway represents the major regulatory mechanism of autophagy (32). Numerous studies have revealed that the inhibition of the PI3K/Akt/mTOR signaling pathway activates autophagy and induces cell death (33,34). Selective inhibition of the Akt/mTOR signaling pathway reduces the level of macrophages and stabilizes vulnerable atherosclerotic plaques by promoting macrophage autophagy (35). The addition of a PI3K inhibitor overcomes cellular resistance to mTORC1 inhibitors, regardless of PTEN status, and therefore substantially expands the molecular phenotype of tumors likely to respond (16). Coordinated inhibition of the mTOR and autophagy pathways promotes apoptosis and may be a novel therapeutic paradigm for the treatment of melanoma (36). Targeted therapy using the PI3K/mTOR inhibitor NVP-BEZ235 significantly enhances doxorubicin-induced apoptosis in neuroblastoma cells (37).
However, certain studies report that activation of the PI3K/Akt/mTOR signaling pathway inhibits autophagy and induces cell death (38,39). mTOR is a highly conserved serine/threonine kinase that is involved in the regulation of cell responses to altered nutrition conditions, and is also involved in a number of energy-associated regulatory pathways (40). Unc-51 like autophagy activating kinase 1 (ULK1) and mammalian autophagy related 13 (mAtg13) are downstream signaling targets of mTOR. When the activity of mTOR is inhibited, the ability of mTOR to inhibit the phosphorylation of ULK1 and mAtg13 is decreased. Therefore, ULK1, mAtg13 and focal adhesion kinase family interacting protein of 200 kD are phosphorylated, which leads to autophagosome formation (41). By contrast, the activation of mTOR has been found to inhibit autophagosome formation. Inhibition of autophagy may enhance matrine-induced apoptosis in human hepatoma cells (42). The human papillomavirus-host cell interaction stimulates the PI3K/Akt/mTOR pathway and inhibits autophagy, which benefits the viral infection (43). In the present study, western blot analysis revealed significantly increased levels of p-Akt and p-mTOR subsequent to treatment with DHA and EPA, indicating activation of this signaling pathway and a decrease in autophagy signal in the early stage. The inhibitor of PI3 K 3-MA inhibits the upstream Akt/mTOR pathway, leading to a reduction in p-Akt and p-mTOR expression levels. The addition of DHA and EPA also decreased the total expression levels of Akt and mTOR, as demonstrated by western blot analysis.
During autophagolysosome formation, an increased quantity of autophagosomes may indicate accumulation and interfere with the formation of autophagolysosomes (44). mRFP-GFP-LC3 is used to distinguish early and late autophagosomes. The GFP tag is rapidly suppressed in the acidic environment of the autophagolysosome, leaving only the mRFP tag detectable (45,46). Fig. 2B shows the appearance of yellow fluorescence in DHA- and EPA-treated A549 cells, indicating decreased autophagolysosome formation and diminished autophagic flux.
Autophagic flux can also be measured by decreased p62 protein level, since p62 acts as an autophagosomal cargo receptor for ubiquitinated proteins, which is degraded in the autophagolysosome (47). The addition of DHA and EPA to A549 cells for 24 h increased p62 levels, suggesting a block in the autophagic flux.
In conclusion, the present study has revealed that DHA and EPA exert anti-proliferative and cytotoxic effects on A549 cells. DHA and EPA interfere with autophagosome formation by activating the Akt/mTOR signaling pathway, thereby reducing the autophagy signal prematurely and decreasing the Beclin 1 expression level. DHA and EPA also block the maturation of autophagy and diminish autophagic flux. Inhibition of autophagy promotes cell death in A549 lung cancer cells. The present data identify a plausible mechanism by which DHA and EPA exert potent anticancer activity, and suggest the potential application of ω-3 PUFAs as a tool or viable drug in anticancer therapies.
Acknowledgements
The present study was supported by a grant from the Zhejiang Provincial Natural Science Foundation of China (grant no., 64212006). The authors thank the Zhejiang Provincial Key Laboratory of Gastroenterology and Biomedical Research Center of Sir Run Run Shaw Hospital for providing experimental facilities, instruments and guidance.
References
- 1.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD, Jacobsen J, et al. Early palliative care for patients with metastatic non-small-cell lung cancer. N Engl J Med. 2010;363:733–742. doi: 10.1056/NEJMoa1000678. [DOI] [PubMed] [Google Scholar]
- 4.Andrews J, Yeh P, Pao W, Horn L. Molecular predictors of response to chemotherapy in non-small cell lung cancer. Cancer J. 2011;17:104–113. doi: 10.1097/PPO.0b013e318213f3cf. [DOI] [PubMed] [Google Scholar]
- 5.Fetterman JW, Jr, Zdanowicz MM. Therapeutic potential of n-3 polyunsaturated fatty acids in disease. Am J Health Syst Pharm. 2009;66:1169–1179. doi: 10.2146/ajhp080411. [DOI] [PubMed] [Google Scholar]
- 6.Zheng JS, Hu XJ, Zhao YM, Yang J, Li D. Intake of fish and marine n-3 polyunsaturated fatty acids and risk of breast cancer: Meta-analysis of data from 21 independent prospective cohort studies. BMJ. 2013;346:f3706. doi: 10.1136/bmj.f3706. [DOI] [PubMed] [Google Scholar]
- 7.De Carlo F, Witte TR, Hardman WE, Claudio PP. Omega-3 eicosapentaenoic acid decreases CD133 colon cancer stem-like cell marker expression while increasing sensitivity to chemotherapy. PLoS One. 2013;8:e69760. doi: 10.1371/journal.pone.0069760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berstad P, Thiis-Evensen E, Vatn MH, Almendingen K. Fatty acids in habitual diet, plasma phospholipids, and tumour and normal colonic biopsies in young colorectal cancer patients. J Oncol. 2012;2012:254801. doi: 10.1155/2012/254801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards IJ, Berquin IM, Chen YQ, O'Flaherty JT. ω-3 PUFAs, breast and prostate cancer: experimental studies. In: Calviello G, Serini S, editors. Dietary Omega-3 Polyunsaturated Fatty Acids and Cancer. Springer; New York, NY: 2010. pp. 167–188. [DOI] [Google Scholar]
- 10.Yao QH, Zhang XC, Fu T, Gu JZ, Wang L, Wang Y, Lai YB, Wang YQ, Guo Y. ω-3 polyunsaturated fatty acids inhibit the proliferation of the lung adenocarcinoma cell line A549 in vitro. Mol Med Rep. 2014;9:401–406. doi: 10.3892/mmr.2013.1829. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XQ, Huang XF, Hu XB, Zhan YH, An QX, Yang SM, Xia AJ, Yi J, Chen R, et al. Apogossypolone, a novel inhibitor of antiapoptotic Bcl-2 family proteins, induces autophagy of PC-3 and LNCaP prostate cancer cells in vitro. Asian J Androl. 2010;12:697–708. doi: 10.1038/aja.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie BS, Zhao HC, Yao SK, Zhuo DX, Jin B, Lv DC, Wu CL, Ma DL, Gao C, et al. Autophagy inhibition enhances etoposide-induced cell death in human hepatoma G2 cells. Int J Mol Med. 2011;27:599–606. doi: 10.3892/ijmm.2011.607. [DOI] [PubMed] [Google Scholar]
- 13.Trejo-Solis C, Jimenez-Farfan D, Rodriguez-Enriquez S, Fernandez-Valverde F, Cruz-Salgado A, Ruiz-Azuara L, Sotelo J. Copper compound induces autophagy and apoptosis of glioma cells by reactive oxygen species and JNK activation. BMC Cancer. 2012;12:156. doi: 10.1186/1471-2407-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeisser H, Fey SB, Horowitz J, Fischer ER, Balinsky CA, Miyake K, Bekisz J, Snow AL, Zoon KC. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy. 2013;9:683–696. doi: 10.4161/auto.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viola G, Bortolozzi R, Hamel E, Moro S, Brun P, Castagliuolo I, Ferlin MG, Basso G. MG-2477, a new tubulin inhibitor, induces autophagy through inhibition of the Akt/mTOR pathway and delayed apoptosis in A549 cells. Biochem Pharmacol. 2012;83:16–26. doi: 10.1016/j.bcp.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S, Xiao X, Meng X, Leslie KK. A mechanism for synergy with combined mTOR and PI3 kinase inhibitors. PLoS One. 2011;6:e26343. doi: 10.1371/journal.pone.0026343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao QH, Zhang XC, Fu T, Gu JZ, Wang L, Wang Y, Lai YB, Wang YQ, Guo Y. ω-3 polyunsaturated fatty acids inhibit the proliferation of the lung adenocarcinoma cell line A549 in vitro. Mol Med Rep. 2014;9:401–406. doi: 10.3892/mmr.2013.1829. [DOI] [PubMed] [Google Scholar]
- 18.Shin SW, Kim SY, Park JW. Autophagy inhibition enhances ursolic acid-induced apoptosis in PC3 cells. Biochim Biophys Acta. 2012;1823:451–457. doi: 10.1016/j.bbamcr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, Pan X, Li F, Zhang Y, Lu X. Expression of Beclin 1 and LC3 in FIGO stage I–II cervical squamous cell carcinoma and relationship to survival. Tumour Biol. 2012;33:1653–1659. doi: 10.1007/s13277-012-0421-4. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, Kolonel LN, Henderson BE, Wilkens LR. Dietary fat and breast cancer in postmenopausal women according to ethnicity and hormone receptor status: The Multiethnic Cohort Study. Cancer Prev Res (Phila) 2012;5:216–228. doi: 10.1158/1940-6207.CAPR-11-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan SF, Cantwell MM, Cardwell CR, Velentzis LS, Woodside JV. Dietary patterns and breast cancer risk: A systematic review and meta-analysis. Am J Clin Nutr. 2010;91:1294–1302. doi: 10.3945/ajcn.2009.28796. [DOI] [PubMed] [Google Scholar]
- 23.MacLean CH, Newberry SJ, Mojica WA, Khanna P, Issa AM, Suttorp MJ, Lim YW, Traina SB, Hilton L, Garland R, et al. Effects of omega-3 fatty acids on cancer risk: A systematic review. JAMA. 2006;295:403–415. doi: 10.1001/jama.295.4.403. [DOI] [PubMed] [Google Scholar]
- 24.Hansakul P, Aree K, Tanuchit S, Itharat A. Growth arrest and apoptosis via caspase activation of dioscoreanone in human non-small-cell lung cancer A549 cells. BMC Complement Altern Med. 2014;14:413. doi: 10.1186/1472-6882-14-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye LH, Li WJ, Jiang XQ, Chen YL, Tao SX, Qian WL, He JS. Study on the autophagy of prostate cancer PC-3 cells induced by oridonin. Anat Rec (Hoboken) 2012;295:417–422. doi: 10.1002/ar.21528. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez CG, Penfornis P, Oskowitz AZ, Boonjindasup AG, Cai DZ, Dhule SS, Rowan BG, Kelekar A, Krause DS, Pochampally RR. Activation of autophagy in mesenchymal stem cells provides tumor stromal support. Carcinogenesis. 2011;32:964–972. doi: 10.1093/carcin/bgr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 28.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DM, Patel S, Raffoul F, Haller E, Mills GB, Nanjundan M. Arsenic trioxide induces a beclin-1-independent autophagic pathway via modulation of SnoN/SkiL expression in ovarian carcinoma cells. Cell Death Differ. 2010;17:1867–1881. doi: 10.1038/cdd.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicotra G, Mercalli F, Peracchio C, Castino R, Follo C, Valente G, Isidoro C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol. 2010;23:937–950. doi: 10.1038/modpathol.2010.80. [DOI] [PubMed] [Google Scholar]
- 31.Smuder AJ, Kavazis AN, Min K, Powers SK. Exercise protects against doxorubicin-induced markers of autophagy signaling in skeletal muscle. J Appl Physiol 1985. 2011;111:1190–1198. doi: 10.1152/japplphysiol.00429.2011. [DOI] [PubMed] [Google Scholar]
- 32.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Gao C, Yao S, Xie B. Blocking autophagic flux enhances matrine-induced apoptosis in human hepatoma cells. Int J Mol Sci. 2013;14:23212–23230. doi: 10.3390/ijms141223212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhai C, Cheng J, Mujahid H, Wang H, Kong J, Yin Y, Li J, Zhang Y, Ji X, Chen W. Selective inhibition of PI3K/Akt/mTOR signaling pathway regulates autophagy of macrophage and vulnerability of atherosclerotic plaque. PLoS One. 2014;9:e90563. doi: 10.1371/journal.pone.0090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westhoff MA, Faham N, Marx D, Nonnenmacher L, Jennewein C, Enzenmuller S, Gonzalez P, Fulda S, Debatin KM. Sequential dosing in chemosensitization: Targeting the PI3K/Akt/mTOR pathway in neuroblastoma. PLoS One. 2013;8:e83128. doi: 10.1371/journal.pone.0083128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surviladze Z, Sterk RT, DeHaro SA, Ozbun MA. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J Virol. 2013;87:2508–2517. doi: 10.1128/JVI.02319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Khoury V, Pierson S, Szwarcbart E, Brons NH, et al. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia. 2014;28:1636–1646. doi: 10.1038/leu.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiao H, Zhang Z, Ma Q, Fu W, Liu Z. Mechanism underlying the inhibitory effect of Apelin-13 on glucose deprivation-induced autophagy in rat cardiomyocytes. Exp Ther Med. 2013;5:797–802. doi: 10.3892/etm.2013.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Gao C, Yao S, Xie B. Blocking autophagic flux enhances matrine-induced apoptosis in human hepatoma cells. Int J Mol Sci. 2013;14:23212–23230. doi: 10.3390/ijms141223212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surviladze Z, Sterk RT, DeHaro SA, Ozbun MA. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J Virol. 2013;87:2508–2517. doi: 10.1128/JVI.02319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J, Chung S, An H, et al. Haloperidol and clozapine block formation of autophagolysosomes in rat primary neurons. Neuroscience. 2012;209:64–73. doi: 10.1016/j.neuroscience.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 45.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 46.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by sirt1 plays an essential role in mediating starvation induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]