Abstract
Objectives
To determine viral and immune factors involved in transmission and control of HIV-1 infection in persons without functional CCR5
Design
Understanding transmission and control of HIV-1 in persons homozygous for CCR5Δ32 is important given efforts to develop HIV-1 curative therapies aimed at modifying or disrupting CCR5 expression.
Methods
We identified two HIV-infected CCR5Δ32/Δ32 individuals among a cohort of patients with spontaneous control of HIV-1 infection without antiretroviral therapy and determined co-receptor usage of the infecting viruses. We assessed genetic evolution of full-length HIV-1 envelope sequences by single-genome analysis from one participant and his sexual partner, and explored HIV-1 immune responses and HIV-1 mutations following virologic escape and disease progression.
Results
Both participants experienced viremia of less than 4,000 RNA copies/ml with preserved CD4+ T cell counts off ART for at least 3.3 and 4.6 years after diagnosis, respectively. One participant had phenotypic evidence of X4 virus, had no known favorable HLA alleles, and appeared to be infected by minority X4 virus from a pool that predominately used CCR5 for entry. The second participant had virus that was unable to use CXCR4 for entry in phenotypic assay but was able to engage alternative viral coreceptors (e.g. CXCR6) in vitro.
Conclusions
Our study demonstrates that individuals may be infected by minority X4 viruses from a population that predominately uses CCR5 for entry, and that viruses may bypass traditional HIV-1 coreceptors (CCR5 and CXCR4) completely by engaging alternative coreceptors to establish and propagate HIV-1 infection.
Keywords: HIV-1, CCR5, CCR5 Δ32 mutation, HIV tropism, HIV controller, viral coreceptor
INTRODUCTION
HIV-1 that uses CCR5 for entry (R5 virus) plays an important role in early infection. A 32 base-pair deletion in CCR5 (Δ32) alters its structure and expression making it unusable by R5 virus. The prevalence of CCR5 Δ32 homozygosity (CCR5Δ32/Δ32) is approximately 1% in the Northern European population and rare in other ethnic groups [1]. CCR5 Δ32 homozygosity confers a high degree of resistance against infection by HIV-1, and, as a result, only a handful of CCR5Δ32/Δ32 HIV-infected individuals have been identified. All CCR5Δ32/Δ32 patients reported to date have been infected with viruses that are able to use CXCR4 for entry [X4 virus or dual/mixed tropic (D/M) virus]. A majority of these individuals experience rapid loss of CD4+ T cells [2–16], a phenomenon that also has been described in cohorts of CCR5 wild-type patients infected with X4 virus [7, 17–24].
Understanding transmission and control of HIV-1 in persons homozygous for CCR5Δ32 is important given efforts to develop HIV-1 curative therapies aimed at modifying or disrupting CCR5 expression [25–27]. Emergence of X4-D/M virus in many patients over time may be due to changes in the availability of CCR5-expressing target cells [28], an observation supported by the report of viral rebound and R5 to X4 coreceptor-usage switching in an individual following allogeneic stem cell transplantation with CCR5Δ32/Δ32 cells [29]. The study of HIV-1-infected CCR5Δ32/Δ32 individuals has the potential to provide valuable insights into mechanisms of HIV-1 transmission, disease progression and the evolution of coreceptor usage in the setting of CCR5 modification.
Although HIV-1 predominately uses CCR5 and/or CXCR4 for cell entry, a proportion of HIVand SIV strains have the capacity to use alternative coreceptors other than CCR5 or CXCR4 [5, 6, 8, 30–35]. Use of alterative coreceptors by HIV-1 from an individual heterozygous for CCR5 Δ32, and exclusive use of GPR15 by HIV-1 during primary infection in one CCR5 wild-type patient have previously been observed [35, 36]. These findings suggest that alternative coreceptors may play a role in HIV-1 transmission and infection in patients with reduced CCR5 expression and function, but the role of HIV-1 alternative coreceptor use in CCR5Δ32/Δ32 individuals is unknown.
We identified two CCR5Δ32/Δ32 individuals with persistent, low-level plasma HIV-1 loads in the absence of antiretroviral therapy (ART) and determined the co-receptor usage of their viruses. We also assessed evolution of HIV-1 env by single-genome analysis of full-length env sequences from one participant and his sexual partner, and explored HIV-1 immune responses and escape mutations associated with increased viremia (virologic escape) and disease progression.
METHODS
Patient Samples and CCR5 genotyping
CCR5Δ32/Δ32 patients were identified from a genome-wide association study of HIV-1 disease control, including nucleotide polymorphisms in viral coreceptor genes [37]. Subsequent PCR screening of PBMC DNA for the presence of two mutant CCR5 copies was then performed as described [38]. Bidirectional sequencing of CCR5 was performed to verify the findings from PCR screening. Plasma, cryopreserved peripheral blood mononuclear cells (PBMCs), and clinical laboratory information were obtained from the International HIV Controllers Study (http://ragoninstitute.org/hivcontrollers). Plasma from a sexual partner of one participant thought to be the source of HIV-1 transmission was also obtained. The Partners Healthcare Institutional Review Board approved this study.
Phenotypic viral coreceptor usage
Virus was concentrated by centrifuging 500 to 1000 μl of plasma at 17,000×g for 1.5 hours at 4°C prior to RNA extraction using the QIAamp Viral RNA Mini Kit (Qiagen). Envelope genes were then amplified using nested PCR as described [39, 40]. Full-length env sequences were amplified and sequenced from PBMC DNA extracted using the QIAamp DNA Mini Kit (Qiagen). PCRs were performed in triplicate wells and combined prior to further processing. Bidirectional sequencing of full-length envelope was performed using published primers [41]. Population or single genome sequences of the third variable loop (V3) of HIV-1 envelope were applied to theGeno2Pheno coreceptor usage prediction algorithm [42].
An in-house phenotypic assay able to detect minority X4 or D/M virus present at 1% or greater of the virus population using pseudoviruses incorporating a luciferase reporter gene and full-length env amplicons from population or single genome viral RNA was performed as described [40]. Phenotyping was repeated if the luciferase signal on indicator cell lines was less than 20-fold higher than the signal generated from envelope-deleted pseudoviral vectors alone.
Plasma virus recovery and expansion
If RNA extraction and subsequent PCR amplification failed to yield envelope sequences, nested primers for HIV-1 subtypes A throughF were used to attempt HIV-1 env, gag and nef gene amplification as described [41]. A viral expansion protocol was a implemented in which 500 μL ml of plasma was added to 2 million uninfected CD4+ T cell blastsstimulated with αCD3/CD28 antibodies (100 ng/ml) and IL-2 (100 units/ml); MOLT4-CCR5 feeder cells were added on day 2. Viral RNA was extracted from culture supernatants concentrated by centrifugation on day 7 and subjected to reverse transcription and real-time PCR quantification as above.
Phenotypic determination of alternative viral coreceptor usage
We performed infectivity assays using GHOST [3] cell lines obtained from the NIH AIDS Research & Reference Reagent Program [30]. Parental cell lines that express CD4 but no other viral coreceptors in addition to lines expressing CCR1, CCR2b, CCR3, CCR4, CXCR4, CCR5, CCR8, BOB/GPR15, CXCR6 (BONZO/STRL33) and V28/CX3CR1 were used in the assay. 15,000 GHOST cells were added to each well of a 96-well plate in high glucose DMEM +10% fetal bovine serum (FBS) and incubated for 24 hours at 37°C and 5% CO2. The parental cell line was incubated in medium supplemented with 500 μg/ml of G418, 100 μg/ml hygromycin, and penicillin/streptomycin. The same medium with the addition of 1μg/ml of puromycin was used for cells with coreceptor transfectants. 75 uL of pseudovirus incorporating a luciferase reporter gene and full-length env amplicons from population viral RNA as above in addition to a HIV-1 pseudoviral control backbone without envelope and a pNL4-3 positive control psueudovirus were added to wells in a total volume of 150 uL. Plates were incubated for 48–72 hours followed by washing, cell lysis and the quantification of luciferase relative light units (RLUs) using a Fluostar Optima fluorescent plate reader (BMG Labtech). All assays were run in triplicate wells and repeated at least twice.
To test whether or not entry into alternative viral coreceptors can be inhibited in vitro, the experiment was repeated using GHOST CXCR6 cells with or without the addition of a rabbit anti-CXCR6 polyclonal antibody (Sigma Aldrich SAB2100513) at concentrations ranging from 50 ng/ml to 1 μg/ml incubated one hour prior to addition of pseudovirus and pseudoviral backbone control.
Full-length HIV-1 envelope single-genome analysis
Single genome amplification and sequencing of full-length envelope sequences from longitudinal time points were performed based on an existing protocol [43]. Briefly, viral RNA was reversed transcribed followed by nested PCR of various RNA dilutions to achieve 1 or fewer HIV-1 copies per reaction well. Sequences were kept if <30% of PCR wells were positive and there were no ambiguous peaks from sequencing chromatograms. Maximum likelihood trees of near full-length envelope sequences were constructed to compare sequence divergence using the PhyML/PAUP plugins for Geneious Pro (Biomatters) after statistical selection of best-fit models of nucleotide substitution (jModelTest). Large gaps and sequence segments with ambiguous nucleotide alignments were stripped prior to model selection and phylogenetic analysis.
HIV-1 gag and nef sequencing, and HLA-associated immune escape mutations
Full length population gag and nef gene amplification and sequencing from plasma RNA and PBMC DNA were performed as described [44]. Ambiguous bases were called if minority sequencing chromatogram peaks were 25% or higher of the peak height from bidirectional strands. Known cytotoxic T-lymphocyte (CTL) epitopes and probable epitopes based on HLA-specific anchor motifs in the gag and nef protein sequences were identified using the epitope location finder tool from the Los Alamos HIV database.
Assessment of HIV-specific T cell responses
IFN-γ enzyme-linked immunospot (Elispot) assays were performed on PBMCs using comprehensive HLA-specific peptide panel as described involving 50,000 to 100,000 cells per reaction well [45]. The number of antigen-specific cells defined by the number of spot-forming cells (SFC) per million PMBC was calculated by subtracting the mean negative-control values. An antigen-specific PBMC response was considered positive only if it was ≥55 SFC/106 PBMC, at least 4 times the mean background, and also, ≥4 times the standard deviation of the number of SFC/106 PBMC within the negative controls.
Data analysis
Phylogenic and statistical analysis were performed using Geneious v. R7 (Biomatters) and Prism 5.0 (GraphPad Software).
RESULTS
Clinical course and host genetics
We identified two HIV-1 infected men with a CCR5Δ32/Δ32 genotype from a large genome-wide association study of the International HIV Controller Cohort. PCR screening and subsequent full-length CCR5 sequencing confirmed that both CCR5 alleles in these two individuals carried the Δ32 mutation. Screening revealed that participant A’s presumed source partner was CCR5 wild-type. Figure 1 shows longitudinal results from clinical viral load monitoring and CD4+ T lymphocyte counts for both participants. Participants A and B had sexually acquired HIV-1 infection and experienced asymptomatic viremia of less than 4,000 HIV-1 RNA copies/ml with preserved CD4+ T cell counts off ART for at least 3.3 and 4.6 years, respectively. Both participants eventually experienced increases in HIV-1 RNA to more than 10,000 copies/ml and decreasing CD4+ T cell counts; participant B started ART in January, 2012. Participant A had no known favorable HLA A or B alleles whereas Participant B had one copy of the B*5201 allele, which has been associated with slower HIV-1 disease progression [46].
Fig. 1.
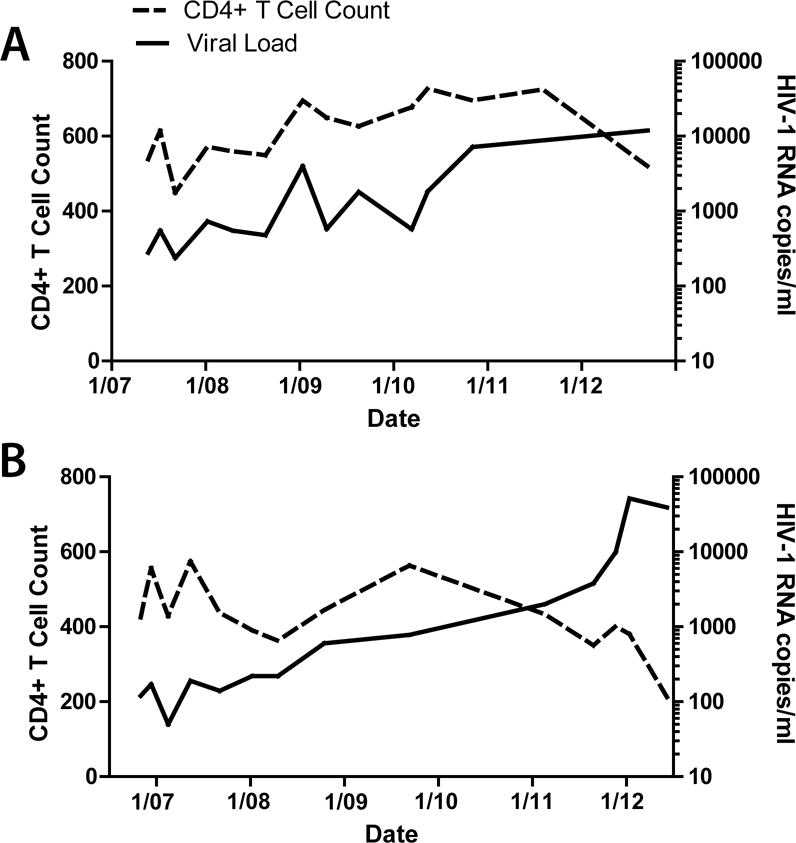
HIV-1 viral load and CD4+ T cell counts for participants A and B (top and bottom panels, respectively). Both participants experienced slow disease progression with persistent, but low-level viremia for 3–4 years prior to rising viral loads (virologic escape) and declining CD4+ T cell counts. Participant B initiated ART in January of 2012.
Participant A coreceptor usage
Coreceptor usage by HIV-1 obtained from Participant A and the presumed source partner was determined by env genotype and by phenotypic assay. The geno2pheno algorithm applied to V3 loop sequences from time-points during virologic control off ART (April 2009) and after virologic escape (September 2012) predicted that virus was only able to use CXCR4 for entry (false positive rates of classifying an R5-virus falsely as X4, or FPR, of 0.7 and 1.3, respectively). Phenotypic assays performed at both time points demonstrated the ability to use only CXCR4 for entry (Figure 2). However, the geno2pheno algorithm predicted CCR5 usage from the suspected infecting partner 2 years prior to the first sample obtained from participant A, and CCR5 usage was also observed in phenotypic assays. In addition, a faint, but reproducible signal in CXCR4-expressing cells was observed in assays using viruses pseudotyped with the envelope from virus of participant A’s partner, which was completely inhibited by the small-molecule CXCR4 inhibitor AMD3100 (Figure 2).
Fig. 2.
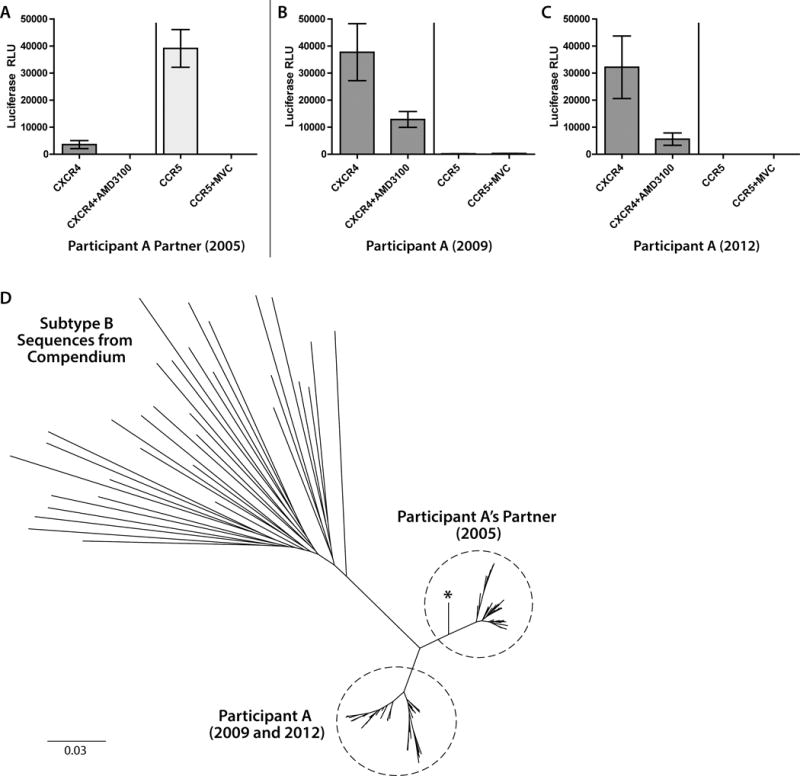
Coreceptor usage determined by phenotypic assay for participant A and his suspected infecting sexual partner. Pseudoviruses incorporating full-length env amplicons from population viral RNA from participant A’s partner used CCR5-expressing U87 cells for entry and led to a faint, but reproducible signal in CXCR4-expressing cells which was inhibited with the addition of the small-molecule CXCR4 inhibitor AMD3100 (A). Pseudoviruses incorporating envelope amplicons from participants A used only CXCR4 for entry from pre- and post-virologic escape time-points (panels B and C, respectively). Error bars represent standard error measurement from triplicate wells of each phenotypic assay. Panel D shows unrooted phylogenetic tree of near full-length HIV-1 env sequences from plasma RNA single genome amplification from participant A, participant A’s suspected infecting partner and an outgroup of compendium of subtype B sequences. The maximum likelihood tree was generated using the best fit selected General Time Reversible model. Sequences from participant A and his partner form distinct clusters as shown in the dashed circles, but share a common branch point from other subtype B compendium sequences. One sequence from participant A’s partner (denoted by an asterisk) had a V3 loop with closer homology to V3 sequences from participant A despite an overall higher homology to his own full-length env sequences.
Participant A single genome analysis
In order to assess viral sequence changes between isolates obtained from participant A’s sexual partner and from participant A before and after virologic escape, single genome sequencing and phylogenetic analysis of full-length envelope sequences from plasma RNA were performed. We obtained 49 sequences from plasma collected from participant A’s partner in December of 2005, 11 sequences prior to loss of virologic control from participant A (collected in April 2009) and 41 sequences obtained after virologic escape in September 2012. All sequences were determined to be subtype B. Env sequences from participant A and his partner formed separate clusters, but shared a common phylogenetic branch point and were more closely related to each other than to 33 unique subtype B sequences obtained from the Los Alamos HIV-1 sequence compendium (Figure 2). All but one of the viral sequences from participant A’s partner were predicted by geno2pheno to use CCR5 alone; phenotypic assay of peudoviruses incorporating env amplicons from two representative envs sequences confirmed the R5 phenotype of these envelopes (Figure 3). The one V3 loop sequence predicted to use CXCR4 and subsequent phenotypic determination of pseudoviruses incorporating this full-length env amplicon revealed pure X4virus. This V3 loop sequence from this virus was more highly related to those of the X4 sequences from participant A, but the non-V3 segment shared greater homology to his other R5 variants (Figure 3).
Fig. 3.
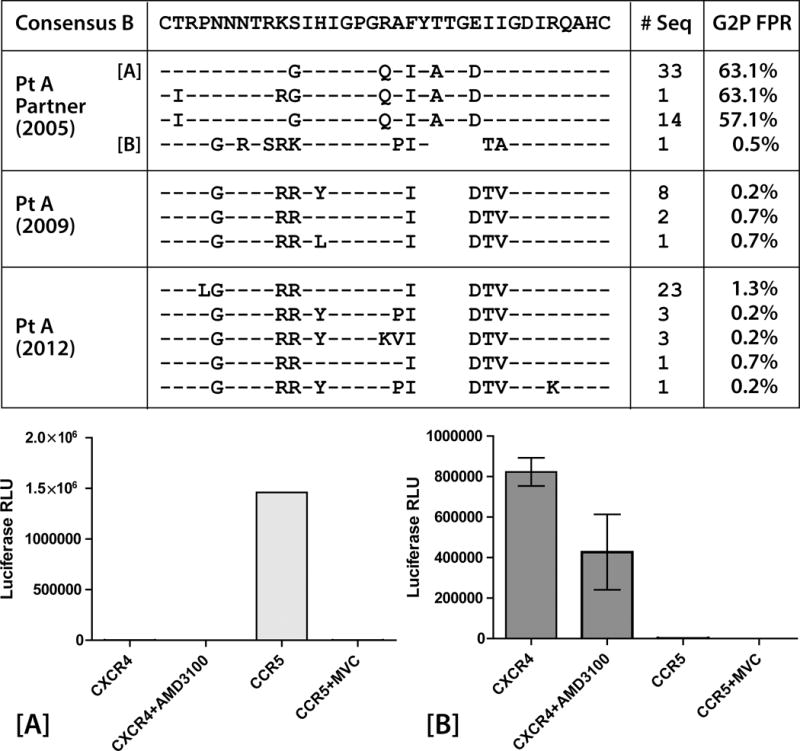
Conservation alignment of HIV-1 env V3 loop amino acid sequences from single genome PCR amplification and sequencing from participant A before and after virologic escape (2009 and 2012 respectively) and his sexual partner in 2005. One V3 loop sequence from participant A’s partner was more similar to V3 loop sequences from participant A, with a GPGR V3 crown motif in lieu of GPGQ. A positively charged amino acid in V3 loop position 11 (arginine) in all of participant A’s sequences and the related V3 loop sequence from his partner (lysine) was observed, and the geno2pheno (G2P) algorithm predict X4-D/M virus (FPR = false positive rate of classifying an R5-virus as X4) from these sequences. Results from phenotypic coreceptor usage assays using pseudoviruses incorporating single genome envelope sequences from participants A’s partner are shown in the bottom frame. Pseudovirus incorporating a representative sequence from participantA partner’s majority V3 loop sequences [A] used only CCR5 for entry. Virus incorporating the partner envelope sequence predicted to be X4-D/M [B] used only CXCR4. Error bars represent standard error measurement from triplicate wells of each phenotypic assay.
Participant B coreceptor usage
The geno2pheno algorithm of V3 population sequences from Participant B 2008 plasma RNA and 2011 and 2012 cell-associated HIV-1 DNA predicted the ability for the virus to use CXCR4 for entry, with FPRs all less than or equal to 1.5% as shown in Figure 4. However, pseudoviruses derived from 2008 plasma RNA amplicons were only able to use CCR5 for entry in the phenotypic assay, with no significant luciferase signal in cells expressing CXCR4 despite repetition of the experiment with amplicons from two plasma aliquots (Figure 4). Pseudoviruses incorporating population cell-associated HIV-1 DNA envelope sequences from 2008 and 2011 were unable to infect U87 cells expressing either CCR5 or CXCR4, but virus incorporating DNA from the 2012 post-virologic escape PBMCs was able to use both CXCR4 and CCR5, although AMD3100 either incompletely suppressed or potentiated entry in CXCR4-expressing U87 cell (Figure 4).
Fig. 4.

Participant B population HIV-1 env V3 loop amino acid sequences and results of the phenotypic coreceptor usage assay. Envelope sequences were obtained from plasma RNA prior to virologic escape (2008) and from cell-associated DNA from 2011 and 2012, after rising viral loads. The geno2pheno (G2P) algorithm predicted that all virus or proviruses would use CXCR4 given low FPRs and positively charged amino acids in V3 position 11 (arginine). However, pseudoviruses incorporating population envelope sequences from 2008 RNA used only CCR5 for entry. HIV-1 RNA was unable to be recovered or amplified from post-virologic escape time points but pseudoviruses incorporating cell-associated DNA envelope sequences from 2012 were able to use CXCR4 and CCR5 (dual/mixed tropic), but at lower levels than viruses derived from RNA. AMD3100 either incompletely suppressed or potentiated entry in CXCR4-expressing U87 cells. Pseudovirsues derived from DNA in 2008 and 2011 were unable to enter U87 cells expressing either CXCR4 or CCR5.
Amplification and sequencing of HIV-1 env, gag or pol from plasma RNA after virologic escape was unsuccessful despite repeated attempts with primers spanning various HIV-1 subtypes. Amplification was also unsuccessful following attempts to amplify virus by co-culture in vitro.
Alternative coreceptor usage
Given the lack of CXCR4 usage determined by phenotypic assay from Participant B’s virus, pseudoviruses incorporating plasma RNA population env amplicons from 2008 were tested on GOHST cells expressing CD4 alone, or alternative viral coreceptors as shown in Figure 5. Low but reproducible levels of entry determined by luciferase activity were observed in cell lines expressing CXCR6, and to a lesser extent GPR15 and CCR1. No entry was observed in cells expressing CXCR4, but a strong signal was observed in cells expressing CCR5. Addition of 0.05 to 1 μg/mL of anti-CXCR6 antibody to the assay incorporating CXCR6-transfected GHOST cells resulted in a 27 % to 41% reduction in viral entry as measured by luciferase RLUs. An NL4-3 control virus was able to use CXCR4-expressing GHOST, and to a very low degree, the CD4-only parental cell line cells in parallel experiments. Although luciferase levels were too low to make definitive statements about the presence or absence of alternative coreceptor usage in experiments involving pseudoviruses from participant A, virus from his sexual partners was only able to use CCR5.
Fig. 5.
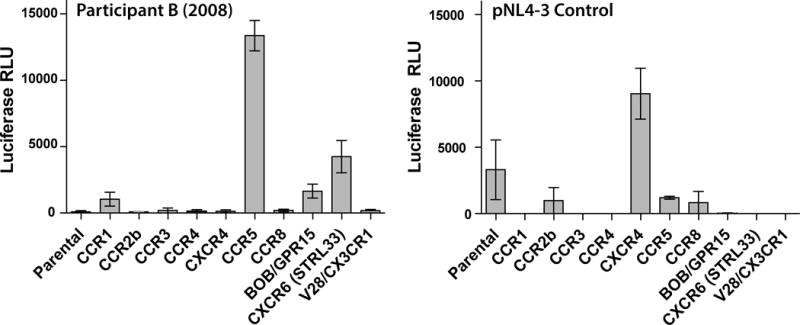
Phenotypic coreceptor usage experiments for participant B using GHOST cells expressing a variety of alternative HIV-1 coreceptors. Cells were infected with pseudoviruses incorporating 2008 plasma RNA full-length population envelope sequences. Low but reproducible levels of entry as determined by luciferase activity were observed in cell lines expressing CXCR6, and to a lesser extent GPR15 and CCR1. No entry was observed in cells expressing CXCR4 as demonstrated in the prior coreceptor usage experiments, but a strong signal was observed in cells expressing CCR5. Pseudoviruses incorporating envelope from a known X4 viral clone (pNL4-3) was used as a control in parallel experiments, and was able to use CXCR4 for entry as shown in the right panel. Error bars represent standard error measurement from duplicate experiments. Luciferase activity from X4 control pseudoviruses in the CD4 parental cell line was intermittently detectable at low levels, suggesting the possibility of endogenous coreceptor expression in the parental cell line.
Longitudinal sequencing of HIV-1 genes
Amplification and population sequencing of near full-length HIV-1 env, nef, and gag was performed on plasma RNA and cell-associated DNA from PBMCs collected from various time-points off ART (RNA from 4/2009 and 9/2012 for Participant A; RNA from 10/2008, and DNA from 2/2011 and 6/2012 for participant B). One novel mutation in a known HLA B*4001-restricted nef epitope from participant BPBMC DNA previously associated with immune escape [47] was observed after virologic escape compared with prior RNA or DNA sequences. No amino acid changes or known escape mutations in any other epitopes were observed in samples from all other time points from participants A and B. Viral RNA was unable to be amplified or detected from post-escape plasma samples from participant B as above.
HIV-1 specific immune responses following virologic escape
Participant A’s PBMCs from 2012 demonstrated a strong HIV-1-specific T cell immune response to the B*1501-GY9(p24) restricted epitope by INF-γ ELISpot assay (1730 SFC/106 PBMCs), and a weaker response to A*11-SK9 (env; 60 SFC/106 PBMCs); no significant responses were detected to other peptides spanning various HIV-1 regions from post-viral escape samples. Participant B PBMCs from 2012 demonstrated activation upon stimulation by several HLA-restricted epitopes spanning env, p24, nef, reverse transcriptase, p17, and gp 41 (Supplementary Table 1). A particular strong response was demonstrated for the B*4001-KL9 (nef) epitope (1000 SFC/106 PBMCs) post-virologic escape, the epitope described above with a novel amino acid mutation in proviral DNA following virologic escape.
DISCUSSION
We studied two CCR5Δ32/Δ32 HIV-1-infected participants that experienced virologic control without ART for approximately 3 to 4 years after diagnosis. Investigation of viral evolution, coreceptor usage, and immune control yielded several important and novel findings, including the presence of HIV-1 that appears to engage alternative corececptors rather than CXCR4 for entry, and the ability for an individual without functional CCR5 to become infected with a minority X4 viral population. A majority of CCR5Δ32/Δ32 individuals experienced rapid disease progression [2–16], an observation previously attributed to the fact that all described CCR5Δ32/Δ32 participants were infected with X4-D/M virus. However, participant A experienced virologic control off ART with plasma HIV-1 RNA levels under 4,000 copies/ml years after diagnosis with preserved CD4+ T cell counts greater than 400 cells/μl with X4 virus and no known favorable HLA alleles. He had a relatively narrow HIV-1-specific post-virologic escape PBMC response to HLA-restricted peptides and no known CTL escape mutations in gag or nef genes. Taken together this data suggest that rapid disease progression cannot be attributed to the presence of X4 virus alone.
Participant A appears to have been infected from a pool of HIV-1 viruses that predominately uses CCR5 for entry, and our findings suggest that minority X4 viruses are able to establish infection in hosts without functional CCR5. Although no direct link could be demonstrated between viruses from participant A and his suspected infecting partner, full-length env sequences from both individuals shared a common phylogenic branch point and were distinct from a subtype B compendium envelope sequence outgroup. Furthermore, the V3 loop sequence of participant A’s partner X4 viral strain was more highly related to participant A than to his own R5 viral sequences. These env sequences shared a 4 amino-deletion in a V3 loop region previously associated with the ability for an R5 HIV-1 strain to use CXCR4 after weeks of in vivo replication and appearance of the deletion [48]. Envelope V3 loop sequences from both participant A and B differed from the HIV-1 clade B consensus sequence by 5–10 amino acid positions, but further study of a larger number of patients will be needed in order to understand the relationship between V3-loop diversity and viral entry in cells lacking functional CCR5.
The lack of matching full-length envelope sequences between participant A and his partner may have been due to several factors, including the limited number of sequences able to be obtained by single-genome analysis, and years of viral replication between sample time points. RNA extraction and single genome analysis of the participant and partner envelope sequences were performed months apart using different reagents, minimizing the potential for cross-sample PCR or sequencing contamination.
Viremic control in participant B may be related to several unique virologic and immunologic findings. For example, participant B was found to have one copy of a HLA B*52 allele previously associated with slower HIV-1 progression [37, 46]. The participant also developed a novel amino acid mutation in a known HLA-B*4001-restricted epitope identified in HIV-1 DNA following rapid increase in viral loads [47]. However, a HIV-1 specific PBMC response to this epitope suggests immune adaptation in the setting of ongoing viral replication, and does not necessarily explain years of virologic control off ART.
Participant B also had evidence of plasma-derived HIV-1 that was unable to use CXCR4 for entry in phenotypic experiments despite genotypic prediction to do so. However, pseudoviruses derived from Participant B’s pre-virologic escape plasma were able to engage one or more alternative co-receptor for entry (e.g. CXCR6). HIV-1 normally prefers to use CXCR4 or CCR5 for entry [31], and the exclusive use of other coreceptors may explain in part, observed low-level viremia and stable CD4+ T cell counts off ART for over 4 years. It is also possible that the virus used other, unidentified mechanisms for cell entry or cell-to-cell transmission. Interestingly, the exclusive use of the coreceptor GPR15 has been reported in one CCR5 wild-type individual in the setting of primary infection with a very high peak viral load peak (87.7 million RNA copies/ml), suggesting efficient use of this coreceptor during early stages of infection [35]. HIV-1 able to use a broad range of alternative coreceptors has also been observed in a CCR5Δ32/WT individual, but unlike our participant, virus was able to use CXCR4 for entry [36]. Of note, small-molecule CCR5 inhibition has not been shown to lead to selection of virus able to use alternative coreceptors for entry in vivo, in which a majority of drug failures are due to selection of pre-existing X4 minority variants [49].
Pseudoviruses derived from participant B’s post viral escape population DNA env sequences were able to use low-levels of CXCR4 in addition to CCR5 in the phenotypic assays. It is not known, however, whether or not plasma-derived viruses from post-virologic escape time points had the capacity to use CXCR4 in vivo. It is possible that the virus evolved to use CXCR4 or alternative coreceptors more efficiently over time leading to virologic escape. Unfortunately, repeated attempts to amplify HIV-1 RNA from sample time points after virologic escape were unsuccessful; sample integrity may have contributed to the lack of amplifiable RNA. In summary, we report the two cases of HIV-1control off ART in the setting of CCR5Δ32 homozygosity, although viremic control was not durable. Our study demonstrates that individuals may be infected by minority X4 viruses from a population that predominately uses CCR5 for entry, and that viruses may bypass traditional HIV-1 coreceptors (CCR5 and CXCR4) completely by engaging alternative coreceptors to establish and propagate HIV-1 infection in individuals lacking functional CCR5.
Supplementary Material
Acknowledgments
Sources of Funding: This work was supported by the NIAID (R37 AI55357; K23AI098480; K24 AI069994), the Delaney AIDS Research Enterprise (DARE; AI096109), The Foundation for AIDS Research (amfAR ARCHE grant), the Bill and Melinda Gates Foundation, and the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763).
Footnotes
Conflicts of Interests: No conflicts of interest reported.
Author Contributions: TJH designed the study, performed experiments and wrote the manuscript; EH and ZH performed experiments and helped analyze data; HJS, CN, JNM and SGD provided samples, clinical data and contributed to manuscript preparation; DRK provided study design oversight and contributed to manuscript preparation; FFP helped design the study and/or performed experiments and contributed to manuscript preparation.
References
- 1.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 2.Balotta C, Bagnarelli P, Violin M, Ridolfo AL, Zhou D, Berlusconi A, et al. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. Aids. 1997;11:F67–71. doi: 10.1097/00002030-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Gray L, Churchill MJ, Keane N, Sterjovski J, Ellett AM, Purcell DF, et al. Genetic and functional analysis of R5X4 human immunodeficiency virus type 1 envelope glycoproteins derived from two individuals homozygous for the CCR5delta32 allele. J Virol. 2006;80:3684–3691. doi: 10.1128/JVI.80.7.3684-3691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien TR, Winkler C, Dean M, Nelson JA, Carrington M, Michael NL, et al. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 5.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, et al. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog. 2010;6:e1001064. doi: 10.1371/journal.ppat.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rucker J, Edinger AL, Sharron M, Samson M, Lee B, Berson JF, et al. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard HW, Celum C, Michael NL, O’Brien S, Dean M, Carrington M, et al. HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion. J Acquir Immune Defic Syndr. 2002;29:307–313. doi: 10.1097/00126334-200203010-00013. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu N, Tanaka A, Oue A, Mori T, Ohtsuki T, Apichartpiyakul C, et al. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. Aids. 2009;23:761–769. doi: 10.1097/QAD.0b013e328326cc0d. [DOI] [PubMed] [Google Scholar]
- 9.Steinfelder S, Floess S, Engelbert D, Haeringer B, Baron U, Rivino L, et al. Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood. 2011;117:2839–2846. doi: 10.1182/blood-2010-06-293027. [DOI] [PubMed] [Google Scholar]
- 10.Gorry PR, Zhang C, Wu S, Kunstman K, Trachtenberg E, Phair J, et al. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5 Delta 32 allele. Lancet. 2002;359:1832–1834. doi: 10.1016/S0140-6736(02)08681-6. [DOI] [PubMed] [Google Scholar]
- 11.Oh DY, Jessen H, Kucherer C, Neumann K, Oh N, Poggensee G, et al. CCR5Delta32 genotypes in a German HIV-1 seroconverter cohort and report of HIV-1 infection in a CCR5Delta32 homozygous individual. PLoS One. 2008;3:e2747. doi: 10.1371/journal.pone.0002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iversen AK, Christiansen CB, Attermann J, Eugen-Olsen J, Schulman S, Berntorp E, et al. Limited protective effect of the CCR5Delta32/CCR5Delta32 genotype on human immunodeficiency virus infection incidence in a cohort of patients with hemophilia and selection for genotypic X4 virus. J Infect Dis. 2003;187:215–225. doi: 10.1086/345881. [DOI] [PubMed] [Google Scholar]
- 13.Heiken H, Becker S, Bastisch I, Schmidt RE. HIV-1 infection in a heterosexual man homozygous for CCR-5 delta32. Aids. 1999;13:529–530. doi: 10.1097/00002030-199903110-00017. [DOI] [PubMed] [Google Scholar]
- 14.Kuipers H, Workman C, Dyer W, Geczy A, Sullivan J, Oelrichs R. An HIV-1-infected individual homozygous for the CCR-5 delta32 allele and the SDF-1 3’A allele. Aids. 1999;13:433–434. doi: 10.1097/00002030-199902250-00025. [DOI] [PubMed] [Google Scholar]
- 15.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 16.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR5 delta 32. Seroco Study Group. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 17.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daar ES, Kesler KL, Petropoulos CJ, Huang W, Bates M, Lail AE, et al. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis. 2007;45:643–649. doi: 10.1086/520650. [DOI] [PubMed] [Google Scholar]
- 19.Delobel P, Sandres-Saune K, Cazabat M, Pasquier C, Marchou B, Massip P, et al. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2005;38:382–392. doi: 10.1097/01.qai.0000152835.17747.47. [DOI] [PubMed] [Google Scholar]
- 20.Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 21.Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 22.Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 24.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors–central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 25.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 26.Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5{Delta}32/{Delta}32 stem cell transplantation. Blood. 2010 doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 27.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:90–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosier DE. How HIV changes its tropism: evolution and adaptation? Curr Opin HIV AIDS. 2009;4:125–130. doi: 10.1097/COH.0b013e3283223d61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kordelas L, Verheyen J, Beelen DW, Horn PA, Heinold A, Kaiser R, et al. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 30.Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cashin K, Jakobsen MR, Sterjovski J, Roche M, Ellett A, Flynn JK, et al. Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection. Retrovirology. 2013;10:98. doi: 10.1186/1742-4690-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Islam S, Shimizu N, Hoque SA, Jinno-Oue A, Tanaka A, Hoshino H. CCR6 functions as a new coreceptor for limited primary human and simian immunodeficiency viruses. PLoS One. 2013;8:e73116. doi: 10.1371/journal.pone.0073116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedellec R, Coetzer M, Shimizu N, Hoshino H, Polonis VR, Morris L, et al. Virus entry via the alternative coreceptors CCR3 and FPRL1 differs by human immunodeficiency virus type 1 subtype. J Virol. 2009;83:8353–8363. doi: 10.1128/JVI.00780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol. 2014;88:1858–1869. doi: 10.1128/JVI.02477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang C, Parrish NF, Wilen CB, Li H, Chen Y, Pavlicek JW, et al. Primary infection by a human immunodeficiency virus with atypical coreceptor tropism. J Virol. 2011;85:10669–10681. doi: 10.1128/JVI.05249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorry PR, Dunfee RL, Mefford ME, Kunstman K, Morgan T, Moore JP, et al. Changes in the V3 region of gp120 contribute to unusually broad coreceptor usage of an HIV-1 isolate from a CCR5 Delta32 heterozygote. Virology. 2007;362:163–178. doi: 10.1016/j.virol.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muxel SM, Borelli SD, Amarante MK, Voltarelli JC, Aoki MN, de Oliveira CE, et al. Association study of CCR5 delta 32 polymorphism among the HLA-DRB1 Caucasian population in Northern Parana, Brazil. J Clin Lab Anal. 2008;22:229–233. doi: 10.1002/jcla.20225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrich TJ, Tsibris AM, Lewine NR, Konstantinidis I, Leopold KE, Sagar M, et al. Evolution of CCR5 antagonist resistance in an HIV-1 subtype C clinical isolate. J Acquir Immune Defic Syndr. 2010;55:420–427. doi: 10.1097/QAI.0b013e3181f25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin NH, Negusse DM, Beroukhim R, Giguel F, Lockman S, Essex M, et al. The design and validation of a novel phenotypic assay to determine HIV-1 coreceptor usage of clinical isolates. J Virol Methods. 2010 doi: 10.1016/j.jviromet.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders-Buell E, Salminen MO, McCutchan FE. Sequencing Primers for HIV-1. The Human Retroviruses and AIDS 1995 Compendium: The HIV Sequence Database and Analysis Project. 1995:III.15–III.21. [Google Scholar]
- 42.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 43.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JZ, Brumme ZL, Brumme CJ, Wang H, Spritzler J, Robertson MN, et al. Factors associated with viral rebound in HIV-1-infected individuals enrolled in a therapeutic HIV-1 gag vaccine trial. J Infect Dis. 2011;203:976–983. doi: 10.1093/infdis/jiq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altfeld MA, Trocha A, Eldridge RL, Rosenberg ES, Phillips MN, Addo MM, et al. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/jvi.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira SL, de Sa NB, Campos DP, Coelho AB, Guimaraes ML, Leite TC, et al. Association of the HLA-B*52 allele with non-progression to AIDS in Brazilian HIV-1-infected individuals. Genes Immun. 2014 doi: 10.1038/gene.2014.14. [DOI] [PubMed] [Google Scholar]
- 47.Almeida CA, Bronke C, Roberts SG, McKinnon E, Keane NM, Chopra A, et al. Translation of HLA-HIV associations to the cellular level: HIV adapts to inflate CD8 T cell responses against Nef and HLA-adapted variant epitopes. J Immunol. 2011;187:2502–2513. doi: 10.4049/jimmunol.1100691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nedellec R, Coetzer M, Lederman MM, Offord RE, Hartley O, Mosier DE. Resistance to the CCR5 inhibitor 5P12-RANTES requires a difficult evolution from CCR5 to CXCR4 coreceptor use. PLoS One. 2011;6:e22020. doi: 10.1371/journal.pone.0022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


