Abstract
Ocean Sampling Day was initiated by the EU-funded Micro B3 (Marine Microbial Biodiversity, Bioinformatics, Biotechnology) project to obtain a snapshot of the marine microbial biodiversity and function of the world’s oceans. It is a simultaneous global mega-sequencing campaign aiming to generate the largest standardized microbial data set in a single day. This will be achievable only through the coordinated efforts of an Ocean Sampling Day Consortium, supportive partnerships and networks between sites. This commentary outlines the establishment, function and aims of the Consortium and describes our vision for a sustainable study of marine microbial communities and their embedded functional traits.
Keywords: Ocean sampling day, OSD, Biodiversity, Genomics, Health Index, Bacteria, Microorganism, Metagenomics, Marine, Micro B3, Standards
Background
Marine microbes inhabit all marine habitats, are the engines of the ocean’s major biogeochemical cycles, and form the basis of the marine food web [1]. Over the past decades scientists have aimed to understand marine microorganisms, but technical and computational limitations have restricted studies to a local scale. Fortunately, with technological advancements and decreasing sequencing costs, genomic studies have become feasible on a global scale. The first landmark marine metagenome studies were published by the J Craig Venter Institute, beginning with a pilot sampling project in the Sargasso Sea followed by the Global Ocean Sampling (GOS) expedition [2]. The Tara Ocean project expanded this further by integrating the marine genetic, morphological, and functional biodiversity in its environmental context at global ocean scale and at multiple depths [3]. The Micro B3 (Marine Microbial Biodiversity, Bioinformatics, Biotechnology) project now aims to investigate global marine microbial biodiversity and has pioneered the idea to do this on a single orchestrated Ocean Sampling Day (OSD).
Main text
Ocean Sampling Day
OSD is a simultaneous, collaborative, global mega-sequencing campaign to analyze marine microbial community composition and functional traits on a single day. On June 21st 2014 – the world’s first major OSD event – scientists around the world collected 155 16S/18S rRNA amplicon data sets, 150 metagenomes, and a rich set of environmental metadata. Standardized procedures, including a centralized hub for laboratory work and data processing via the Micro B3 Information System (Micro B3-IS), assured a high level of consistency and data interoperability [4]. Application of the Marine Microbial Biodiversity, Bioinformatics and Biotechnology (M2B3) standards ensures sustainable data storage and retrieval in respective domain-specific data archives [4]. OSD generated the largest standardized data set on marine microbes taken on a single day, which we consider complementary to other large-scale sequencing projects.
The solstice was chosen to test the hypothesis that diversity negatively correlates with day-length [5]. Data analysis will target three main areas: biodiversity, gene functions, and ecological models. OSD sampling sites are typically located in coastal regions within exclusive economic zones (EEZ). Therefore, the OSD data set provides a unique opportunity to test anthropogenic influences on microbial population ecology. We will perform a multi-level assessment of the human impact on microbial mediated biogeochemical cycles. Questions we would like to answer are: (i) what are the important factors (physical-chemical and biological) in structuring biodiversity patterns and range margins, and (ii) are functions associated with heavy metals, antibiotics or fecal indicators correlated with OSD sites exposed to higher human impact? We are confident that the simultaneous collection of samples will result in the discovery of new ecological patterns providing key information towards understanding environmental vulnerability and resilience.
Open access strategy and sharing of data
All OSD data are archived and immediately made openly accessible without an embargo period, following the Fort Lauderdale rules for sharing data [6]. Sequence and contextual data are publicly available via the International Nucleotide Sequence Database Collaboration (INSDC) umbrella study PRJEB5129 and at PANGAEA. A model agreement and OSD Data Policy [4] was developed in compliance with the Convention on Biological Diversity and the Nagoya Protocol on Access and Benefit Sharing (ABS) for the utilization on genetic resources in a fair and equitable way. An ABS Helpdesk exists to support OSD participants’ legal questions. Furthermore, the Mediterranean Science Commission (CIESM) developed the CIESM Charter on ABS, which has been endorsed by 391 scientists from 49 countries (as of April 2015).
The OSD Consortium
At the 16th Genomics Standards Consortium (GSC) meeting in 2014, the OSD community agreed to form the OSD Consortium. Led by the five OSD Coordinators and comprising of up to 130 OSD Site Coordinators and their teams, the OSD Consortium installed the infrastructure and expertise allowing coordinated OSD events to take place. Furthermore, the OSD Consortium aims to foster collaborations and share expertise among and beyond the OSD network, and to connect scientists in a worldwide environmental movement.
Membership and governance
OSD membership is open to anyone and is earned by participation. Registered participants are provided with privileged access to the OSD network of sites, as well as training activities. OSD samples are prioritized for all types of data generation (as funds and resources allow). In return, participants agree to provide samples according to OSD’s standardized procedures and to work under the umbrella of the OSD Data Policy, which requires open sharing of data and to respect the national legal sampling framework.
The OSD network of sites
Participants from 191 sampling sites signed up for the main OSD event; these sites range from tropical waters to polar environments (Fig. 1). All major oceanic divisions (Pacific, Atlantic, Indian, Antarctic and Arctic Ocean) and continents are covered with 81 and 37 sites in Europe and North America, respectively. The majority of sites are located in the Northern Hemisphere (172), including 36 sites in the Mediterranean and three sites in the Black Sea.
Fig. 1.
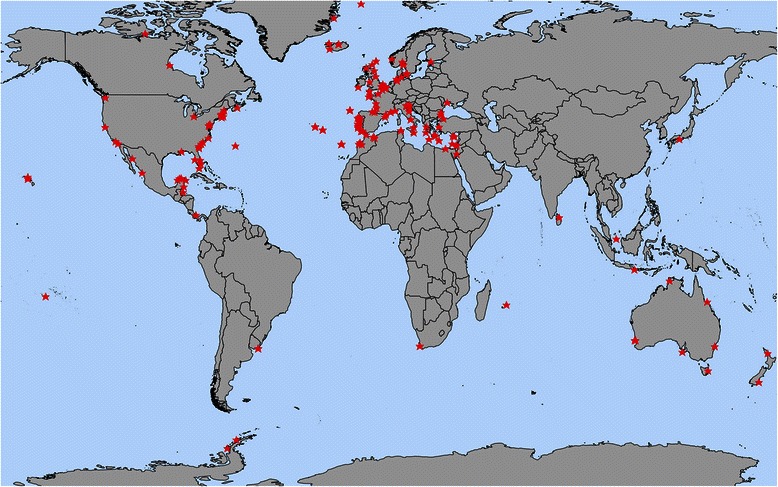
Map of registered sites for OSD 2014
OSD partnerships
Endorsement of the community and fruitful partnerships are essential. Supported by the Argonne National Laboratory, the generous cooperation with the Earth Microbiome Project (EMP) [7] enabled us to perform amplicon sequencing for OSD pilot events; these were conducted on each of the solstices in 2012 and 2013. In return, OSD data is EMP compliant and contributes towards construction of a global catalog of microbial diversity [7]. Cooperation with the LifeWatch project secured additional 18S rRNA gene sequencing, while Pacific Bioscience contributed sequencing of near-full-length 16S rRNA gene amplicons and metagenomes from selected OSD sampling sites. Moreover, the partnership with the Smithsonian Institute’s Global Genome Initiative for long-term bioarchiving of all OSD samples enables the community to re-analyze the samples in the future.
OSD beyond 2014
The OSD Consortium aims to expand in terms of sites and methods, as well as towards multicellular organisms. Future key tasks are to align closely with the Genomic Observatories (GOs) Network [8] towards biocoding the ocean, as well as to secure long-term resources and commitments to create an OSD time-series. The mid-term vision of the OSD Consortium is to generate microbial Essential Biodiversity Variables (EBV) data [9]. The envisioned regular OSD events would qualify for the candidate EBVs “Species populations” and “Community composition” to indicate, for example, vulnerability of ecosystems and climatic impacts on community composition. In the long term such indicators may be incorporated into the Ocean Health Index (OHI) [10], which currently excludes microorganisms from biodiversity assessment due to the lack of reliable data. OSD has the potential to close that gap and amend EBV and OHI by expanding oceanic monitoring towards microbes. This could lead to a global system of harmonized observations to inform scientists and policy-makers.
Conclusions
This commentary outlines the process for creating, managing and formalizing the OSD Consortium and describes its vision for a sustainable study of marine microbes. As we move forward, we will continue to explore and expand the scope of OSD beyond 2014. The idea of an OSD time-series is still in its early days but incorporating the OSD data set as EBVs and in the OHI is a strong source of motivation since this could pave the way to prioritize scientific research and raise public awareness for the unseen majority of the world’s oceans.
Acknowledgements
We wish to acknowledge our extensive range of Micro B3/OSD participants, partners, advisors and supporters who have made the OSD possible. The generosity and tremendous support from our partners and participants has not only enriched the data set but also allowed the OSD Consortium to save resources for future OSD activities. We would also like to thank Sandra Nowack, Hilke Döpke, Greta Reintjes, Timmy Schweer and the technicians of the Max Planck Institute for Marine Microbiology for their tremendous support with on-site logistics. This work was supported by the Micro B3 project, which is funded from the European Union’s Seventh Framework Programme (FP7; Joint Call OCEAN.2011‐2: Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential) under the grant agreement no 287589. This manuscript is NOAA-GLERL contribution number 1763.
Abbreviations
- ABS
Access and benefit sharing
- CIESM
Mediterranean Science Commission (Commission Internationale pour l'Exploration Scientifique de la Méditerranée)
- EBV
Essential biodiversity variables
- EEZ
Exclusive economic zone
- EMP
Earth microbiome project
- GOs
Genomic observatories
- GOS
Global ocean sampling expedition
- Micro B3
Marine Microbial Biodiversity, Bioinformatics, Biotechnology
- Micro B3-IS
Micro B3 information system
- M2B3
Marine microbial biodiversity, bioinformatics and biotechnology data reporting and service standards
- EBV
Essential biodiversity variables
- OHI
Ocean health index
- OSD
Ocean sampling day
- rRNA
ribosomal RNA
Footnotes
Anna Kopf and Mesude Bicak contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AK drafted the original text with detailed input from MB, RK, JS, IK, KL and FOG. All authors have read and approved the final manuscript and/or participated in OSD.
Contributor Information
Anna Kopf, Email: aklindwo@mpi-bremen.de.
Mesude Bicak, Email: mesude.bicak@oerc.ox.ac.uk.
Renzo Kottmann, Email: rkottman@mpi-bremen.de.
Julia Schnetzer, Email: jschnetz@mpi-bremen.de.
Ivaylo Kostadinov, Email: i.kostadinov@jacobs-university.de.
Katja Lehmann, Email: kleh@ceh.ac.uk.
Antonio Fernandez-Guerra, Email: afernand@mpi-bremen.de.
Christian Jeanthon, Email: jeanthon@sb-roscoff.fr.
Eyal Rahav, Email: eyal.rahav@ocean.org.il.
Matthias Ullrich, Email: m.ullrich@jacobs-university.de.
Antje Wichels, Email: antje.wichels@awi.de.
Gunnar Gerdts, Email: Gunnar.Gerdts@awi.de.
Paraskevi Polymenakou, Email: polymen@hcmr.gr.
Giorgos Kotoulas, Email: kotoulas@hcmr.gr.
Rania Siam, Email: rsiam@aucegypt.edu.
Rehab Z Abdallah, Email: r.abdallah@aucegypt.edu.
Eva C Sonnenschein, Email: evaso@bio.dtu.dk.
Thierry Cariou, Email: cariou@sb-roscoff.fr.
Fergal O’Gara, Email: f.ogara@ucc.ie.
Stephen Jackson, Email: sjackson@ucc.ie.
Sandi Orlic, Email: sorlic@irb.hr.
Michael Steinke, Email: msteinke@essex.ac.uk.
Julia Busch, Email: julia.busch@uni-oldenburg.de.
Bernardo Duarte, Email: baduarte@fc.ul.pt.
Isabel Caçador, Email: micacador@fc.ul.pt.
João Canning-Clode, Email: Canning-ClodeJ@si.edu.
Oleksandra Bobrova, Email: Aleks-bobrovaOd@yandex.ru.
Viggo Marteinsson, Email: viggo@matis.is.
Eyjolfur Reynisson, Email: eyjolfur@matis.is.
Clara Magalhães Loureiro, Email: c.angela.m.loureiro@gmail.com.
Gian Marco Luna, Email: gianmarco.luna@ve.ismar.cnr.it.
Grazia Marina Quero, Email: grazia.quero@ve.ismar.cnr.it.
Carolin R Löscher, Email: cloescher@geomar.de.
Anke Kremp, Email: anke.kremp@ymparisto.fi.
Marie E DeLorenzo, Email: marie.delorenzo@noaa.gov.
Lise Øvreås, Email: lise.ovreas@bio.uib.no.
Jennifer Tolman, Email: j.tolman@dal.ca.
Julie LaRoche, Email: Julie.Laroche@Dal.Ca.
Antonella Penna, Email: antonella.penna@uniurb.it.
Marc Frischer, Email: marc.frischer@skio.uga.edu.
Timothy Davis, Email: timothy.davis@noaa.gov.
Barker Katherine, Email: barkerk@si.edu.
Christopher P Meyer, Email: meyerc@si.edu.
Sandra Ramos, Email: ssramos@ciimar.up.pt.
Catarina Magalhães, Email: cmagalhaes@ciimar.up.pt.
Florence Jude-Lemeilleur, Email: florence.jude@u-bordeaux.fr.
Ma Leopoldina Aguirre-Macedo, Email: leo@mda.cinvestav.mx.
Shiao Wang, Email: shiao.wang@usm.edu.
Nicole Poulton, Email: npoulton@bigelow.org.
Scott Jones, Email: jonesms@si.edu.
Rachel Collin, Email: CollinR@si.edu.
Jed A Fuhrman, Email: fuhrman@usc.edu.
Pascal Conan, Email: pascal.conan@obs-banyuls.fr.
Cecilia Alonso, Email: calonso@cure.edu.uy.
Noga Stambler, Email: drnogas@gmail.com.
Kelly Goodwin, Email: kelly.goodwin@noaa.gov.
Michael M Yakimov, Email: michail.yakimov@iamc.cnr.it.
Federico Baltar, Email: federico.baltar@otago.ac.nz.
Levente Bodrossy, Email: lev.bodrossy@csiro.au.
Jodie Van De Kamp, Email: jodie.vandekamp@csiro.au.
Dion MF Frampton, Email: dion.frampton@csiro.au.
Martin Ostrowski, Email: martin.ostrowski@mq.edu.au.
Paul Van Ruth, Email: Paul.VanRuth@sa.gov.au.
Paul Malthouse, Email: paul.malthouse@sa.gov.au.
Simon Claus, Email: simon.claus@vliz.be.
Klaas Deneudt, Email: klaas.deneudt@vliz.be.
Jonas Mortelmans, Email: jonas.mortelmans@vliz.be.
Sophie Pitois, Email: sophie.pitois@cefas.co.uk.
David Wallom, Email: david.wallom@oerc.ox.ac.uk.
Ian Salter, Email: ian.salter@awi.de.
Rodrigo Costa, Email: rscosta@ualg.pt.
Declan C Schroeder, Email: dsch@mba.ac.uk.
Mahrous M Kandil, Email: mahrouskan@yahoo.com.
Valentina Amaral, Email: vamaral@cure.edu.uy.
Florencia Biancalana, Email: biancaf@criba.edu.ar.
Rafael Santana, Email: wuaffa@hotmail.com.
Maria Luiza Pedrotti, Email: pedrotti@obs-vlfr.fr.
Takashi Yoshida, Email: yoshidaten@gmail.com.
Hiroyuki Ogata, Email: ogata@kuicr.kyoto-u.ac.jp.
Tim Ingleton, Email: tim.ingleton@environment.nsw.gov.au.
Kate Munnik, Email: kate@lwandle.co.za.
Naiara Rodriguez-Ezpeleta, Email: nrodriguez@azti.es.
Veronique Berteaux-Lecellier, Email: veronique.berteaux@criobe.pf.
Patricia Wecker, Email: patricia.wecker@criobe.pf.
Ibon Cancio, Email: ibon.cancio@ehu.es.
Daniel Vaulot, Email: vaulot@sb-roscoff.fr.
Christina Bienhold, Email: cbienhol@mpi-bremen.de.
Hassan Ghazal, Email: hassan.ghazal@fulbrightmail.org.
Bouchra Chaouni, Email: bouchrachaouni@gmail.com.
Soumya Essayeh, Email: fpn.sessayeh@gmail.com.
Sara Ettamimi, Email: sara-ettamimi@hotmail.fr.
El Houcine Zaid, Email: ijazaid@yahoo.fr.
Noureddine Boukhatem, Email: noureddineboukhatem@yahoo.fr.
Abderrahim Bouali, Email: boualiabderrahime@yahoo.fr.
Rajaa Chahboune, Email: chahraja@hotmail.com.
Said Barrijal, Email: barrijal@yahoo.fr.
Mohammed Timinouni, Email: mohammed.timinouni@pasteur.ma.
Fatima El Otmani, Email: elotmanifatima@yahoo.fr.
Mohamed Bennani, Email: mohamed.bennani@pasteur.ma.
Marianna Mea, Email: mmea@mpi-bremen.de.
Nadezhda Todorova, Email: nadeshda@abv.bg.
Ventzislav Karamfilov, Email: karamfilov.v@gmail.com.
Petra ten Hoopen, Email: petra@ebi.ac.uk.
Guy Cochrane, Email: cochrane@ebi.ac.uk.
Stephane L’Haridon, Email: stephane.lharidon@univ-brest.fr.
Kemal Can Bizsel, Email: can.bizsel@deu.edu.tr.
Alessandro Vezzi, Email: alessandro.vezzi@unipd.it.
Federico M Lauro, Email: flauro@ntu.edu.sg.
Patrick Martin, Email: pmartin@ntu.edu.sg.
Rachelle M Jensen, Email: rachelle@indigovexpeditions.com.
Jamie Hinks, Email: jhinks@ntu.edu.sg.
Susan Gebbels, Email: susan.gebbels@ncl.ac.uk.
Riccardo Rosselli, Email: riccardo.rosselli@cribi.unipd.it.
Fabio De Pascale, Email: fabio.depascale@unipd.it.
Riccardo Schiavon, Email: riccardo.schiavon@unipd.it.
Antonina dos Santos, Email: antonina@ipma.pt.
Emilie Villar, Email: villar@igs.cnrs-mrs.fr.
Stéphane Pesant, Email: spesant@marum.de.
Bruno Cataletto, Email: bcataletto@inogs.it.
Francesca Malfatti, Email: fmalfatti@inogs.it.
Ranjith Edirisinghe, Email: ranjith_e@hotmail.com.
Jorge A Herrera Silveira, Email: jherrera@mda.cinvestav.mx.
Michele Barbier, Email: mbarbier@ciesm.org.
Valentina Turk, Email: valentina.turk@mbss.org.
Tinkara Tinta, Email: tinta@mbss.org.
Wayne J Fuller, Email: wjf4965@gmail.com.
Ilkay Salihoglu, Email: ilkay.salihoglu@neu.edu.tr.
Nedime Serakinci, Email: nedimeserakinci@gmail.com.
Mahmut Cerkez Ergoren, Email: mahmutcerkez@gmail.com.
Eileen Bresnan, Email: Bresnane@marlab.ac.uk.
Juan Iriberri, Email: juan.iriberri@ehu.es.
Paul Anders Fronth Nyhus, Email: Kind.of.Blue@mac.com.
Edvardsen Bente, Email: bente.edvardsen@ibv.uio.no.
Hans Erik Karlsen, Email: h.e.karlsen@bio.uio.no.
Peter N Golyshin, Email: p.golyshin@bangor.ac.uk.
Josep M Gasol, Email: pepgasol@icm.cat.
Snejana Moncheva, Email: snejanam@abv.bg.
Nina Dzhembekova, Email: sonata_bg@yahoo.com.
Zackary Johnson, Email: zij@duke.edu.
Christopher David Sinigalliano, Email: christopher.sinigalliano@noaa.gov.
Maribeth Louise Gidley, Email: maribeth.gidley@noaa.gov.
Adriana Zingone, Email: zingone@szn.it.
Roberto Danovaro, Email: r.danovaro@univpm.it.
George Tsiamis, Email: gtsiamis@upatras.gr.
Melody S Clark, Email: mscl@bas.ac.uk.
Ana Cristina Costa, Email: accosta@uac.pt.
Monia El Bour, Email: monia.elbour@instm.rnrt.tn.
Ana M Martins, Email: anamartins@uac.pt.
R Eric Collins, Email: recollins@alaska.edu.
Anne-Lise Ducluzeau, Email: aducluzeau@alaska.edu.
Jonathan Martinez, Email: jonathan.martinez@hawaii.edu.
Mark J Costello, Email: m.costello@auckland.ac.nz.
Linda A Amaral-Zettler, Email: amaral@mbl.edu.
Jack A Gilbert, Email: gilbertjack@gmail.com.
Neil Davies, Email: ndavies@moorea.berkeley.edu.
Dawn Field, Email: fiedawn@gmail.com.
Frank Oliver Glöckner, Email: fog@mpi-bremen.de.
References
- 1.Seymour JR. A sea of microbes: the diversity and activity of marine microorganisms. Microbiology Australia. 2014. [Google Scholar]
- 2.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS One. 2007;5(3) doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karsenti E, Acinas SG, Bork P, Bowler C, De Vargas C, Raes J, et al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9(10) doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ten Hoopen P, Cochrane G, Micro B3 Consortium. Ocean Sampling Day Handbook - Version of June 2014. Available at: http://www.microb3.eu/sites/default/files/osd/OSD_Handbook_v2.0.pdf. 2014.
- 5.Gilbert JA, Sommerfield P, Temperton B, Huse S, Joint I, Field D. Day-length is central to maintaining consistent seasonal diversity in marine bacterioplankton. Available from Nature Precedings: http://precedings.nature.com/documents/4406/version/1. 2010.
- 6.The Wellcome Trust (Fort Lauderdale January 14 – 15, 2003). Sharing Data from Large-Scale Biological Research Projects: A System of Tripartite Responsibility. Available at: https://www.genome.gov/Pages/Research/WellcomeReport0303.pdf. 2003 [Accessed December 18, 2014].
- 7.Gilbert JA, Jansson JK, Knight R. The earth microbiome project: successes and aspirations. BMC Biol. 2014;12(1):69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies N, Field D, Amaral-Zettler L, Clark MS, Deck J, Drummond A, et al. The founding charter of the genomic observatories network. Gigascience. 2014;3(1):2. doi: 10.1186/2047-217X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira HM, Ferrier S, Walters M, Geller GN, Jongman RH, Scholes RJ, et al. Ecology. Essential biodiversity variables. Science. 2013;339(6117):277–8. doi: 10.1126/science.1229931. [DOI] [PubMed] [Google Scholar]
- 10.Halpern BS, Longo C, Hardy D, McLeod KL, Samhouri JF, Katona SK, et al. An index to assess the health and benefits of the global ocean. Nature. 2012;488(7413):615–20. doi: 10.1038/nature11397. [DOI] [PubMed] [Google Scholar]


