Abstract
Dab2ip (DOC-2/DAB2 interacting protein) is a RasGAP protein which shows a growth-inhibitory effect in human prostate cancer cell lines. Recent studies have shown that Dab2ip also plays an important role in regulating dendrite development and neuronal migration during brain development. In this study, we provide a more complete description of the mouse Dab2ip (mDab2ip) gene locus and examined DNA methylation and expression of Dab2ip during cerebellar development. Analysis of cDNA sequences in public databases revealed a total of 20 possible exons for mDab2ip gene, spanning over 172 kb. Using Cap Analysis of Gene Expression (CAGE) data available through FANTOM5 project, we deduced five different transcription start sites for mDab2ip. Here, we characterized three different mDab2ip transcript variants beginning with exon 1. These transcripts varied by the presence or absence of exons 3 and 5, which encode a putative nuclear localization signal and the N-terminal region of a PH-domain, respectively. The 5′ region of the mDab2ip gene contains three putative CpG islands (CpG131, CpG54, and CpG85). Interestingly, CpG54 and CpG85 are localized on exons 3 and 5. Bisulfate DNA sequencing showed that methylation level of CpG54 remained constant whereas methylation of CpG85 increased during cerebellar development. Real-time PCR analysis showed that the proportion of PH-domain containing mDab2ip transcripts increased during cerebellar development, in correlation with the increase in CpG85 methylation. These data suggest that site-specific methylation of mDab2ip gene during cerebellar development may play a role in inclusion of exon 5, resulting in a Dab2ip transcript variant that encodes a full pleckstrin homology (PH) domain.
Keywords: Dab1, AIP1, cerebellum, GTPase Activating Protein
Introduction
Disabled homolog 2 interacting protein (Dab2ip), also known as apoptosis signal-regulating kinase 1 (ASK1)-interacting protein (AIP1) (Chen et al., 2002; Wang et al., 2002; Xie et al., 2009; Zhang et al., 2003), is a member of Ras GTPase-activating protein family (Wang et al., 2002; Zhang et al., 2003). Dab2ip is thought to be a tumor suppressor protein, regulating epithelial to mesenchymal transition (Min et al., 2010; Xie et al., 2010) and stem cell differentiation (Chang et al., 2013). In addition, down regulation of Dab2ip using shRNA enhances radio-resistance and proliferation in a metastatic prostate cancer cell line (Kong et al., 2010). Dab2ip also interacts with Dab1, a cytosolic adapter protein that controls neuronal migration and position during development. Dab2ip is widely expressed in specific regions of embryonic and adult mouse brain (Homayouni et al., 2003). Recent studies show mDab2ip regulates dendritic development and synapse formation in developing cerebellum (Qiao et al., 2013) and also plays crucial roles in neuronal migration in the developing neocortex (Lee et al., 2012; Qiao et al., 2015).
The mDab2ip gene is located on chromosome 2 (Chen et al., 2006). Originally, Chen et al. reported that mDab2ip spans 65 kb and contains 14 exons with one transcription start site (TSS) located on exon 1 (Chen et al., 2006). In addition, they showed that internal splice sites within exon 1 result in three variants, Ia, Ib and Ic. Since this initial report, several more Dab2ip cDNAs have been isolated from mouse and other species that contain additional 5′ exons. For example, the additional exons in human Dab2ip ortholog AIP1 (Acc# AY032952) encodes a complete pleckstrin homology (PH) domain (Von Bergh et al., 2004). Similarly, our group identified a longer mDab2ip cDNA (Acc# DQ473307), which encodes a complete N-terminal PH-domain (Homayouni et al., 2003). Notably, both of these clones encode a protein with a shorter C-terminal region, ending with the amino acid sequence SMH (serine-methionine-histidine), which may function as a PDZ-interacting site.
The promoter region of both mouse and human Dab2ip gene is GC-rich (Chen et al., 2006, 2003). Several groups have shown that hDab2ip promoter is hyper-methylated in cancer cell lines and tumor tissues including prostate, breast, gastrointestine, lung and liver (Chen et al., 2003; Dote et al., 2004; Yano et al., 2005, Dote et al., 2005; Qiu et al., 2007). In addition, both hDab2ip and mDab2ip promoters have been shown to be regulated by histone modification (Chen et al., 2006, 2003, 2002). Accumulating evidence suggests that promoter methylation plays a critical role during differentiation and maturation of the mammalian central nervous system (CNS) (Moore et al., 2012). To date, the methylation of Dab2ip promoter during development has not been investigated.
In the present study, we performed a thorough analysis of Dab2ip genomic organization using a variety of bioinformatic resources. We found at least five transcription start sites (TSS) and multiple putative translation initiation sites for mDab2ip. In addition, we identified four additional 5′ exons for mDab2ip than previously reported by Chen et al. (2006). We examined the expression of three Dab2ip 5′-splice variants, originating with the first TSS in exon one. Notably, mDab2ip contains three CpG islands located within exons 2, 3 and 5. Exon 5 CpG island was previously studied by Chen and colleagues and was shown to be the site of epigenetic control of mDab2ip expression (Chen et al., 2006). Here we show that methylation of exon 5 CpG (CpG85) increases during cerebellar development and is positively correlated with the inclusion of exon 5 in mDab2ip transcripts.
Materials and Methods
Bioinformatic analysis
We deduced the exon-intron junction of mDab2ip by aligning its genomic DNA sequence with hDab2ip cDNA clone (Acc# NP-115941). The mDab2ip gene contains 20 exons and 19 introns (Supplementary Table 1). Transcription start site analysis was performed using Zenbu visualization tool and mouse Cap Analysis of Gene Expression (CAGE) data available through FANTOM5 project website (http://fantom.gsc.riken.jp/zenbu/gLyphs/#config=FP3UWqGsJVPtWNfSfupRk;loc=mm9::chr2:35370843..35629647+) (Forrest et al., 2014; Severin et al., 2014). Dab2ip protein motifs were examined using Conserved Domain Database (CCD) at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and MyHits protein motif search tool (Pagni et al., 2007).
Reverse transcription PCR
Based on the sequence information acquired from UCSC genome browser (mouse genome assembly mm9, July 2007), PCR primers were designed against specific 5′ exons of mDab2ip (Table 1). Total RNA was extracted from mouse cerebellum at different ages (P8, P14, P21 and P30) using TRIzol reagent (Invitrogen). cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) with 1 μg of total RNA and oligo-dT primer. One-tenth volume of the first-strand reaction was used as a template for PCR amplification using the TaKaRa EX-Taq polymerase (Clontech Laboratories) and the primers listed in Table 1. Thermal cycling program started with an initial denaturation at 95 °C for 5 min, followed by 35 cycles (94 °C, 1 min; 61 °C, 1 min; 72 °C, 50 s) of amplification, followed by a 5 min extension at 72 °C.
Table 1.
List of oligonucleotide primers and TaqMan probe sequences used in this study
| Name | Sequence | Product size |
|---|---|---|
| Primers used for Dab2ip methylation | ||
| CpG 131/MSP/M/F | 5′-CGTTTTTTTTCGGTTTTAAACGTTTTTA-3′ | 243 |
| CpG 131/MSP//M/R | 5′-CGCCCTACAAACATCGTTCCCCGCGCGCT-3′ | 243 |
| CpG 131/MSP /UM/F | 5′-TGTTTTTTTTTGGTTTTAAATGTTTTTA-3′ | 243 |
| CpG 131/MSP/UM/R | 5′-CACCCTACAAACATCATTCCCCACACACT-3′ | 243 |
| CpG 131/BSP/F | 5′-GAATTTGGGGAATATGGAGTAGAATAG-3′ | 264 |
| CpG 131/BSP/R | 5′-TTCCCTACCTTCTTTACTATAACCCAA-3′ | 264 |
| CpG 54/MSP/M/F | 5′-TTAGGTTAGGATTTTGTTCGTATTGATTTTTC-3′ | 303 |
| CpG 54/MSP/M/R | 5′-CTCCCGATACTCTTCCTAACGTTACCGCCGAC-3′ | 303 |
| CpG 54/MSP/UM/F | 5′-TTAGGTTAGGATTTTGTTTGTATTGATTTTTT-3′ | 303 |
| CpG 54/MSP/UM/R | 5′-CTCCCAATACTCTTCCTAACATTACCACCAAC-3′ | 303 |
| CpG 54/BSP/F | 5′-GTTTAGATATGGTTGTTGGGTATATGTT-3′ | 243 |
| CpG 54/BSP/R | 5′-AAAACCAAAACTACCCTACAAAATAACT-3′ | 243 |
| CpG 85/MSP/M/F | 5′-GAGTTTTTCGTTGTTCGATATAAAAGGTATTTTC-3′ | 153 |
| CpG 85/MSP/M/R | 5′-CGAATCTTAAATTATACCCATTAACCGAACGCCT-3′ | 153 |
| CpG 85/MSP/UM/F | 5′-GAGTTTTTTGTTGTTTGATATAAAAGGTATTTTT-3′ | 153 |
| CpG 85/MSP/UM/R | 5′-CAAATCTTAAATTATACCCATTAACCAAACACCT-3′ | 153 |
| CpG 85/BSP/F | 5′-GAGGTGGGTATTGTTTTTTGAGTAG-3′ | 193 |
| CpG 85/BSP/R | 5′-TACTCCTCCCCTCCAAATATTC-3′ | 193 |
| Primers used for Dab2ip RT-PCR | ||
| E1-53.84-F | 5′-CGCTCATGGAGACGGCCTCGGTTCATAAATCA-3′ | |
| E7-117.149-R | 5′-AGCAGTAGTCCTGACCCAGGATGCTGCTGTGAA-3′ | |
| Primers used for qRT-PCR | ||
| GRD domain-F | 5′-GCCTTCTGCAAGATCATCAAC-3′ | |
| GRD domain-R | 5′-GCTGATGAGCCGTTCACTG-3′ | |
| PH domain-F | 5′-CGCGGACAATGAGAGGTC-3′ | |
| PH domain-R | 5′-GAGCAGGGACTCGTGTGAC-3′ | |
MSP, methyl specific primer; BSP, bisulfite specific primer; F, forward; R, reverse
Quantitative real-time PCR
Total RNA was isolated from mouse cerebellum at postnatal days 8, 14, 21 and 30 (P8, P14, P21 and P30) using TRIzol reagent (Invitrogen). Quantitative RT-PCR was performed on a LightCycler 480 Real-Time PCR System (Roche) using primers shown in Table 1 and Fig. 1A. Thermal cycling program started with an initial denaturation at 95°C for 5 minutes, followed by 50 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s, and final cooling at 40°C for 10 seconds. The product sizes were confirmed by agarose gel electrophoresis, and melting curves were analyzed to control the specificity of PCR reactions. Dab2ip expression levels were normalized to β-actin. The relative levels of Dab2ip expression were measured by a modified ΔΔCt (Pfaffl, 2001). A two-tailed Student’s t-test (unequal variance) was used to assess the significance of the change in three independent experiments.
FIG 1.
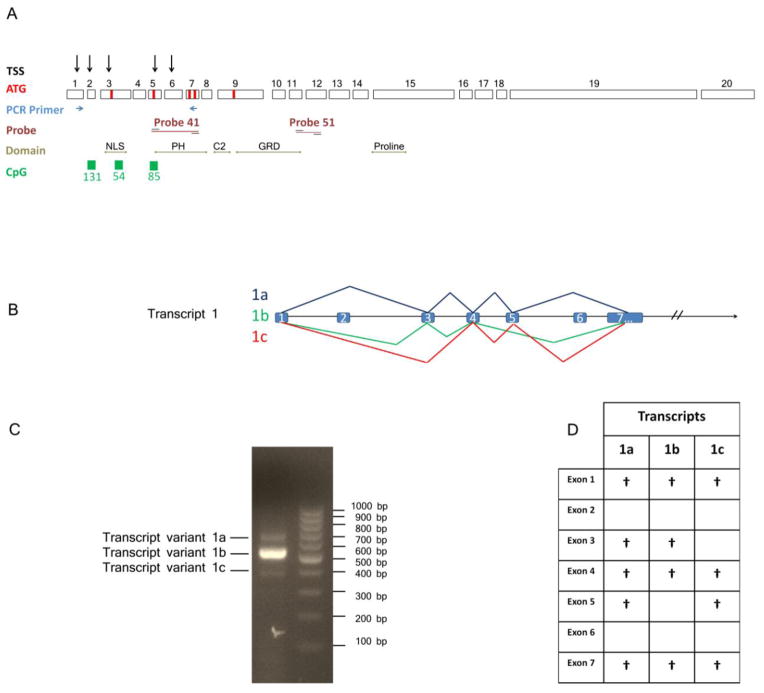
Evidence for alternatively spliced mDab2ip transcripts. (A) Schematic diagram of Dab2ip gene structure and the location of primers and probes used for RT-PCR and real-time PCR experiments in this study. The 5′-end of mDab2ip is associated with three CpG islands, five transcription start sites and seven translation initiation sites. (B) Schematic diagram showing three different mDab2ip splice variants beginning with the first TSS on exon 1. (C) RT-PCR amplification using primers on exon 1 (forward primer) and 7 (reverse primer) produced three distinct bands (688pb, 554bp, and 385bp). (D) A table summarizing the presence of specific exons in each of the three variants in C after DNA sequencing.
DNA purification and bisulphite modification
Genomic DNA was isolated from snap frozen mouse cerebella collected at different time points (P8, P14, P21 and P30) using DNA purification kit (Qiagen) according to the manufacturer’s instructions. Then, 500 ng of genomic DNA was denatured and treated with bisulfite, which converts all unmethylated cytosines to uracils without affecting methylated cytosine residues, using EZ DNA methylation-Gold kit (Zymo research) as described by manufacturer. The quality of bisulfite treatment was checked by measuring OD at 260/280 nm. The methylation status of each CpG island was analyzed separately by using bisulfite specific and methylation specific PCR. Bisulfite specific (BSP) and methyl specific (MSP) primers listed in Table 1, were designed using Methyl Primer Express Software v1.0 (Applied Biosystems), with some modifications (Table 1). Methyl specific PCR was performed by using two sets of primers for each CpG island. Each set of primers were able to distinguish between methylated (MSP/M) and unmethylated (MSP/UM) DNA sequence. PCR was carried out with approximately 30 ng of bisulfate treated genomic DNA in 30 μL total volume reaction containing 2.5 μl DMSO, 2.5 mM dNTP, and 0.20 U of TaKaRa EX-Taq DNA polymerase (Clontech Laboratories). Thermal cycling program started with an initial denaturation at 95 C for 5 min followed by 50 cycles of denaturation (95 C for 30 seconds), annealing (65 C for 45 sec, 60 C for 45 sec and 55 C for 45 seconds), extension (72 C for 5 min), followed by a final incubation at 72 C for 15 min. PCR products were examined by agarose gel electrophoresis. In another set of experiments, bisulfite specific PCR was performed as described above using 20 ng of bisulfite treated genomic DNA and primers (BSP), listed in Table 1. The PCR products were separated by agarose gel electrophoresis, purified using Qiagen PCR purification kit, and directly sequenced using ABI model 3130XL Genetic Analyser machine at University of Tennessee at Molecular Resource Center. Methylation sites within each CpG island were quantitated and a two-tailed Student’s t-test (unequal variance) was used to assess the significance of the change in three independent experiments.
Results
Genomic organization of Dab2ip
In this study, we report that the mDab2ip gene spans 172,535 bp on chromosome 2 and includes 20 exons. Using CAGE data available through the FANTOM5 project, we deduced five transcription start sites (TSS) located in exons 1, 2, 3, 5 and 6 (Supplementary Fig. 1). Three CpG islands (CpG131, CpG54 and CpG85) are located on exon 2, 3 and 5, respectively (Fig. 1A, Supplementary Fig. 1). The presence of multiple CpG islands around multiple TSSs suggests that mDab2ip gene may have alternative promoters, regulated by methylation of CpG islands, producing transcripts which encode different N-terminal protein domains.
Using UCSC genome browser, we were able to identify eight different putative translation initiation codons with likely Kozak sequences in exons 1, 3, 4, 5, 6, 7, 8 and 9 (Fig. 1A, Supplementary Table 1). However, only five translation initiation sites (located in exons 3, 5, 7 and 9) were in frame with the rest of the cDNA. Two alternative termination sites were also found on exon 19, followed by a large 3′ untranslated sequence. There are at least twelve mouse Dab2ip cDNA clones available through UCSC Genome Browser (Supplementary Table 1). While the middle region of all Dab2ip cDNA clones is identical, their 5′ and 3′ regions vary. The deduced protein sequence from the Dab2ip cDNAs revealed several functional domains, including Plekstrin Homology (PH) domain, PKC conserved 2 (C2) domain, GAP-related domain (GRD), NPXY motif and proline-rich region (Fig. 1A). Interesting, alternative splicing in the 5′ region of Dab2ip results in proteins that contain either a full-length PH domain, a partial PH-domain or no PH domain in the N-terminus. On the other hand, alternative splicing at the 3′ region results in a slightly longer isoform containing an additional 29 amino acids. Interestingly, the shorter Dab2ip protein isoform encodes an SMH class I PDZ-interacting motif (Sheng and Sala, 2001).
Several groups have reported that Dab2ip is highly expressed in the brain (Chen et al., 2006; Homayouni et al., 2003). In addition, more recent results from the FANTOM project show that the expression Dab2ip is highest in the cerebellum (Supplementary Fig. 1). To detect possible mDab2ip 5′ splice variants, we performed RT-PCR using RNA isolated from P30 cerebellum and specific forward and reverse primers targeting exons 1 and 7, respectively (Table 1). Three bands (688bp, 554bp, and 385bp) were detected by agarose gel electrophoresis, which were individually purified and sequenced (Fig. 1C). Sequencing revealed that the largest band (688bp, variant 1a) included exons 1, 3, 4, 5 and 7. In contrast, the mid-sized band (554bp, variant 1b) included exons 1, 3, 4, and 7 and the smallest band (385bp, variant 1c) included exons 1, 4, 5 and 7. Using Conserved Domain Database (CCD) at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) we found that exons 5, 7 and 8 encode a complete PH domain (112 aa). Thus, transcripts 1a and 1c, which contain exons 5 and 7, are able to code for an entire PH domain whereas transcript 1b lacks the N-terminal region of the PH domain (73 aa). Transcript 1a contains alternative translation initiation sites located in exon 3 and 5. In contrast, transcripts 1b and 1c have only one translation initiation site located in exons 3 and 5, respectively. Interestingly, exon 3 codes for an arginine-rich nuclear localization signal as detected by MyHits motif search tool. Finally, none of the three variants isolated from the brain encoded exons 2 and 6. Based on the cDNA sequences in GeneBank, it seems that mDab2ip transcripts containing exon 6 are possibly expressed in other tissues, such as the spleen (Supplementary Table 1).
mDab2ip methylation in the developing cerebellum
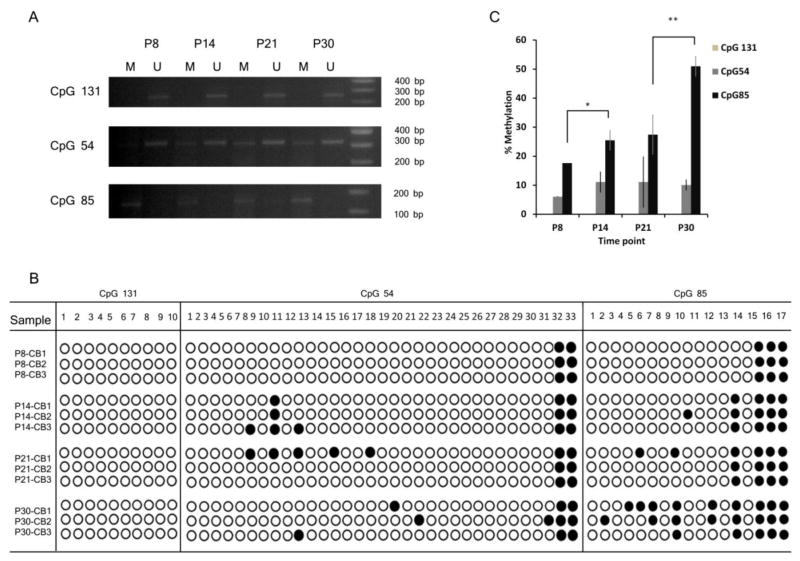
The presence of CpG islands in mDab2ip gene suggests that its expression may be regulated by DNA methylation (Supplementary Fig. 1). Indeed, hDab2ip promoter was shown to be methylated in prostate, breast, gasterointestinal, lung and liver cancers (Chen et al., 2003; Dote et al., 2004; Qiu et al., 2007; Yano et al., 2005). In this study, we examined methylation of mDab2ip gene during cerebellar development at postnatal days P8, P14, P21 and P30 (Fig. 2). CpG islands were examined by methylation-specific (MSP) PCR (Table 1) after bisulfite treatment of genomic DNA (Fig. 2A). We found that CpG131 was predominantly unmethylated whereas CpG54 appeared to be both methylated and un-methylated in all time points (Fig. 2A). In contrast, CpG85 appeared to be methylated at all time-points.
FIG 2.
Analysis of mDab2ip methylation during cerebellar development at postnatal days P8, P14, P21 and P30. (A) Methylation specific PCR of CpG131, CpG54, and CpG85 using PCR primers which specifically amplified methylated (M) or unmethylated (U) CpG islands. (B) Bisulfite sequencing of mDab2ip CpG131, CpG54, and CpG85 from cerebella (n=3) collected at indicated postnatal days. (C) Quantitation of CpG131, CpG54, and CpG85 methylation during cerebellar development by bisulfite sequencing. Methylation is shown as an average percentage of total CpG’s which were found to be methylated in three separate animals at each time point. Statistical analysis was performed using a two-tailed Student’s t-test (unequal variance). *, p<0.05, **, p < 0.01.
To further analyze the methylation status of mDab2ip gene, we performed PCR amplification using bisulfite specific primers (BSP) listed in Table 1, followed by direct sequencing and quantitation of methylation sites. Consistent with the results above, we did not detect any methylation of CpG131 (Fig 2B-C). CpG54 was methylated throughout cerebellar development. However its methylation did not change significantly across developmental time points (Fig. 2B-C). On the other hand, the methylation of CpG85 increased significantly between P8 and P14 (p < 0.05, t-test, n=3) as well as between P21 and P30 (p < 0.01, t-test, n=3; Fig 2B-C).
Expression of different mDab2ip transcripts in the developing cerebellum
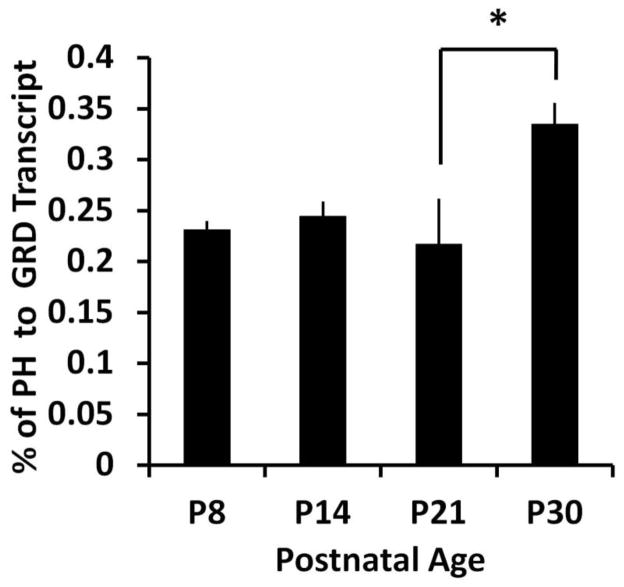
Recent evidence suggests that methylation of CpG sites located within exons may regulate their inclusion during splicing (Luco et al., 2011). Therefore, we hypothesized that methylation of CpG85 located in mDab2ip exon5 may positively correlate with expression of mDab2ip transcripts containing exon 5. We performed qRT-PCR experiments using a probe which specifically targeted exon 5 (TaqMan probe 41, Fig. 1A) compared with a probe that targeted exon 11, located in the core GRD region of Dab2ip (TaqMan probe 51, Fig 1A). Interestingly, we found that the ratio of exon 5 containing transcripts to the total mDab2ip transcripts significantly (p < 0.05, t-test, n=3) increased between P21 and P30 (Fig. 3). The increase in exon 5 containing transcripts positively correlated (r = 0.76, Pearson correlation) with CpG85 methylation. In contrast, there was no correlation between exon 5 containing transcript and CpG54 methylation (r = 0.04, Pearson correlation). These results suggest that methylation of CpG85 during cerebellar development promotes inclusion of exon 5 in mDab2ip transcripts.
FIG 3.
Dab2ip mRNA expression during cerebellar development at postnatal days P8, P14, P21 and P30. Quantitative RT-PCR was performed using a TaqMan probe which recognizes either exon 5 or exons 11–12 (encoding the core GRD domain. Ct values were normalized to β-actin levels at each time point. The graph shows the average percentage of exon 5 containing transcripts (PH domain) to total mDab2ip transcripts (GRD) in three separate mice. Statistical analysis was performed using a two-tailed Student’s t-test (unequal variance). *, p<0.05, **, p < 0.01.
Discussion
In this study, we performed a detailed analysis of the mouse Dab2ip gene, identified multiple 5′ Dab2ip transcript variants, and examined their expression levels with respect to Dab2ip DNA methylation during cerebellar development.
We report here that mDab2ip gene spans over 172 kb, which is much longer than the previously reported size by others (Chen et al., 2006) and that it contains three different CpG islands, located on exons 2, 3 and 5. Previous studies have examined methylation and histone acetylation of the region around exon 5 in Dab2ip (Chen et al., 2006, 2003). Mapping experiments revealed a basal promoter region upstream of exon 5 in mDab2ip which drives its expression and is regulated by both DNA methylation and histone acetylation (Chen et al., 2006). These results in combination with our observation that multiple exons and transcription start sites exist in mDab2ip gene (Fig. 1 and Supplementary Fig. 1) suggest that Dab2ip likely contains multiple promoters, resulting in expression of multiple transcript variants. In addition, Chen and colleagues showed that histone acetylation but not DNA methylation of the region around mDab2ip exon 5 correlated with gene induction in mouse prostate adenocarcinoma cell lines. Our results (Fig. 2 & 3) show that methylation of CpG85 in mDab2ip exon 5 is significantly increased between P21 and P30 in the developing cerebellum and positively correlates with the inclusion of exon 5 (encoding the N-terminal region of the PH domain). Our observations are consistent with recent studies in other systems that implicate a critical role for DNA methylation in regulating mRNA splicing and exon inclusion in the mature mRNA (Maunakea et al., 2013; Luco et al., 2011).
The notion of intergenic DNA-methylation-mediated mRNA splicing is relatively new. Recent genome-wide studies have shown an enrichment of CpG methylation in alternatively spliced exons (Anastasiadou et al., 2011; Maunakea et al., 2013). The interaction of methyl-CpG-binding protein (MeCP2) at DNA methylation sites recruits histone deacetylases, which promote the formation of nucleosomes. The interaction between DNA and nucleosomes limits its accessibility to RNA polymerase during transcription. Indeed, Shukla and colleagues showed that DNA methylation causes RNA polymerase II (POL II) to pause and promotes exon inclusion by enabling the splicing complexes to assemble co-transcriptionally (Shukla et al., 2011). Co-transcriptional splicing appears to be important for weak splice sites, particularly in long mammalian genes in which exons are flanked by very large introns (Luco 2011). It is interesting to point out that mDab2ip exons 2, 3, and 5, which contain CpG islands, were flanked by large introns ranging between 17–41 kb (Supplementary table 1). We identified transcripts in the brain that included exons 3 and 5, which also showed DNA methylation on CpG 54 and CpG85, respectively. In contrast, we did not detect methylation of CpG 131 nor a transcript containing exon 2 in the cerebellum.
The presence of the additional 5′ exons may have several important functional implications. First, we found two additional translation initiation sites (with Kozak consensus motif) on exons 3 and 5, which produce longer Dab2ip proteins. Exon 3 encodes a putative nuclear localization signal, which implies that Dab2ip may have a nuclear function which has not been characterized to date. In addition, Dab2ip exon 5 encodes the N-terminal region of a PH-domain, which suggests that both Dab2ip protein function and intracellular localization may be affected by the presence of exon 5 in this transcript variant. Dab2ip is a member of RasGAP family (Wang et al., 2002; Zhang et al., 2003). PH domains are commonly found in GAP proteins (Grewal et al., 2011; Lemmon et al., 2002; Rizo and Sudhof, 1998) and mediate interactions with lipids, which may allow GAP proteins to translocate to membranes in response to extracellular signals (Lemmon and Ferguson, 2000; Rebecchi and Scarlata, 1998; Shaw, 1996). In addition to lipid binding, the PH domains of some GEFs and GAPs also participate in intramolecular interactions, which can be regulated by PtdIns (3,4,5)P3 (Drugan et al., 2000; Han et al., 1998; Ma et al., 1998; Nimnual et al., 1998; Saito et al., 2001). Indeed, the PH domain of AIP1/Dab2ip was shown to undergo intramolecular interaction (Zhang et al., 2003). The presence of a full-length or partial PH domain in different Dab2ip transcripts may play an important role in regulation of its molecular and cellular functions.
The cerebellum is an ideal brain structure to study the mechanisms of neuronal development (Ha et al., 2015; Goldowitz and Hamre, 1998; Hatten et al., 1997). It has a distinct layered structure which is primarily composed of Purkinje Cells (PCs) and Granule Cells (GCs). PCs are born prenatally, but complete their migration and begin developing dendrites at P5-P8, during the time when GC are maximally proliferating (Goldowitz and Hamre, 1998). The newly born GCs migrate passed PCs and inhabit the inner layer of the cerebellum by P14. Finally, GCs extend axons (parallel fibers) that form synaptic connections with PC dendrites in the outer layer of the cerebellum. Importantly, maturation of the PC synaptic circuitry is completed between P21 and P30 (Miyazaki et al., 2003), during the time when we observed methylation of mDab2ip CpG85 and a concomitant increase in PH-domain encoding exon 5 in mDab2ip transcript. Interestingly, in our previous work, we showed that PC synaptic structures were affected in mice with a targeted disruption of the first five exons in mDab2ip gene (Qiao et al., 2013). Therefore, it is intriguing to speculate that methylation of mDab2ip CpG85 may play a critical role in producing a Dab2ip variant with a full PH-domain which may regulate synapse formation in the brain.
Supplementary Material
ZENBU genome browser view of the FANTOM5 promoter level mammalian expression atlas data. The tracks shown here include Entrez Gene mm9 gene boundaries, UCSC RefSeq mm9 gene boundaries, Phase I CTSS data and predictions, and the Phase I CTSS pooled sequence tags from various mouse tissues (bottom).
Highlights.
mDab2ip gene spans over 172kb and contains 20 exons.
mDab2ip gene contains 3 different CpG islands and multiple transcription start sites.
Three transcript variants were identified which include exon 1.
mDab2ip transcripts showed differential expression during cerebellar development.
Methylation of mDab2ip exon 5 during development correlates with its inclusion in the mRNA.
Acknowledgments
We thank Dr. Lynn A. Jones for assistance with the animal colonies. This work was supported by NIH MH068433 and the Assisi Foundation of Memphis.
List of abbreviations
- aa
amino acid
- cDNA
DNA complementary to RNA
- CNS
central nervous system
- DMSO
dimethylsulfoxide
- dNTP
deoxyribonucleoside triphosphate
- Dab2ip
Disabled homolog 2 interacting protein
- bp
base pair
- kb
kilobase
- mDab2ip
mouse Dab2ip
- hDab2ip
human Dab2ip
- AIP1
(ASK1)-interacting protein
- TSS
transcription start site
- PH domain
pleckstrin homology domain
- C2 domain
PKC conserved 2
- GRD
GAP-related domain
- SMH
serine-methionine-histidine
- CAGE
Cap Analysis of Gene Expression
- CCD
Conserved Domain Database
- BSP
bisulfite specific primers
- MSP
methyl specific primers
- P
postnatal day
- 5′ (five prime)
denotes a truncated gene at the 5′ end
- 3′ (five prime)
denotes a truncated gene at the 3′ end
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- PtdIns (3,4,5)P3
Phosphatidylinositol (3,4,5)-triphosphate
- PCs
Purkinje Cells
- GCs
granule Cells
- PCR
polymerase chain reaction
- RT-PCR
Reverse transcription-polymerase chain reaction
- qRT-PCR
Quantitative Reverse transcription-PCR
- NCBI
National Center for Biotechnology Information
- UCSC
University of California, Santa Cruz
Footnotes
Authors have no conflict of interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30:267–275. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- Chang SLY, Chou RH, Zeng HJ, Lin YH, Chiu TY, Yang DM, Hung SC, Lai CH, Hsieh JT, Shyu WC, Yu YL. Downregulation of DAB2IP Promotes Mesenchymal-To-Neuroepithelial Transition and Neuronal Differentiation of Human Mesenchymal Stem Cells. PLoS One. 2013;8:e75884. doi: 10.1371/journal.pone.0075884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Karam JA, Schultz R, Zhang Z, Duncan C, Hsieh JT. Cloning of mouse Dab2ip gene, a novel member of the RasGTPase-activating protein family and characterization of its regulatory region in prostate. DNA Cell Biol. 2006;25:232–245. doi: 10.1089/dna.2006.25.232. [DOI] [PubMed] [Google Scholar]
- Chen H, Pong RC, Wang Z, Hsieh JT. Differential regulation of the human gene DAB2IP in normal and malignant prostatic epithelia: cloning and characterization. Genomics. 2002;79:573–581. doi: 10.1006/geno.2002.6739. [DOI] [PubMed] [Google Scholar]
- Chen H, Toyooka S, Gazdar AF, Hsieh JT. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J Biol Chem. 2003;278:3121–30. doi: 10.1074/jbc.M208230200. [DOI] [PubMed] [Google Scholar]
- Dote H, Toyooka S, Tsukuda K. Aberrant Promoter Methylation in Human DAB2 Interactive Protein ( hDAB2IP ) Gene in Breast Cancer Aberrant Promoter Methylation in Human DAB2 Interactive Protein ( hDAB2IP ) Gene in Breast Cancer. 2004:2082–2089. doi: 10.1158/1078-0432.ccr-03-0236. [DOI] [PubMed] [Google Scholar]
- Drugan JK, Rogers-Graham K, Gilmer T, Campbell S, Clark GJ. The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J Biol Chem. 2000;275:35021–35027. doi: 10.1074/jbc.M004386200. [DOI] [PubMed] [Google Scholar]
- Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, Lassmann T, Itoh M, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume Da, Carninci P, Hayashizaki Y. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998 doi: 10.1016/S0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Grewal T, Koese M, Tebar F, Enrich C. Differential Regulation of RasGAPs in Cancer. Genes Cancer. 2011;2:288–297. doi: 10.1177/1947601911407330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, Krishna UM, Falck JR, White MA, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol. 1997 doi: 10.1016/S0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- Homayouni R, Magdaleno S, Keshvara L, Rice DS, Curran T. Interaction of Disabled-1 and the GTPase activating protein Dab2IP in mouse brain. Mol Brain Res. 2003;115:121–129. doi: 10.1016/s0169-328x(03)00176-1. [DOI] [PubMed] [Google Scholar]
- Kong Z, Xie D, Boike T, Raghavan P, Burma S, Chen DJ, Habib Aa, Chakraborty A, Hsieh JT, Saha D. Downregulation of human DAB2IP gene expression in prostate cancer cells results in resistance to ionizing radiation. Cancer Res. 2010;70:2829–39. doi: 10.1158/0008-5472.CAN-09-2919. [DOI] [PubMed] [Google Scholar]
- Lee GH, Kim SH, Homayouni R, D’Arcangelo G. Dab2ip Regulates Neuronal Migration and Neurite Outgrowth in the Developing Neocortex. PLoS One. 2012;7:e46592. doi: 10.1371/journal.pone.0046592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350(Pt 1):1–18. doi: 10.1042/0264-6021:3500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002 doi: 10.1016/S0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011 doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AD, Metjian A, Bagrodia S, Taylor S, Abrams CS. Cytoskeletal reorganization by G protein-coupled receptors is dependent on phosphoinositide 3-kinase gamma, a Rac guanosine exchange factor, and Rac. Mol Cell Biol. 1998;18:4744–4751. doi: 10.1128/mcb.18.8.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–69. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, Bronson RT, Ryeom S, Hahn WC, Loda M, Cichowski K. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Fukaya M, Shimizu H, Watanabe M. Subtype switching of vesicular glutamate transporters at parallel fibre-Purkinje cell synapses in developing mouse cerebellum. Eur J Neurosci. 2003;17:2563–2572. doi: 10.1046/j.1460-9568.2003.02698.x. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology. 2012 doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual AS, Yatsula BA, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: Improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Kim SH, Heck D, Goldowitz D, LeDoux MS, Homayouni R. Dab2IP GTPase Activating Protein Regulates Dendrite Development and Synapse Number in Cerebellum. PLoS One. 2013;8:e53635. doi: 10.1371/journal.pone.0053635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu GH, Xie H, Wheelhouse N, Harrison D, Chen GG, Salto-Tellez M, Lai P, Ross Ja, Hooi SC. Differential expression of hDAB2IPA and hDAB2IPB in normal tissues and promoter methylation of hDAB2IPA in hepatocellular carcinoma. J Hepatol. 2007;46:655–663. doi: 10.1016/j.jhep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Rebecchi MJ, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- Saito K, Scharenberg AM, Kinet JP. Interaction between the Btk PH domain and phosphatidylinositol-3,4,5-trisphosphate directly regulates Btk. J Biol Chem. 2001;276:16201–16206. doi: 10.1074/jbc.M100873200. [DOI] [PubMed] [Google Scholar]
- Severin J, Lizio M, Harshbarger J, Kawaji H, Daub CO, Hayashizaki Y, Consortium TF, Bertin N, Forres ARR. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol. 2014;32:217–219. doi: 10.1038/nbt.2840. [DOI] [PubMed] [Google Scholar]
- Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bergh ARM, Wijers PM, Groot AJ, Van Zelderen-Bhola S, Falkenburg JHF, Kluin PM, Schuuring E. Identification of a Novel RAS GTPpase-Activating Protein (RASGAP) Gene at 9q34 as an MLL Fusion Partner in a Patient with De Novo Acute Leukemia. Genes Chromosom Cancer. 2004;39:324–334. doi: 10.1002/gcc.20004. [DOI] [PubMed] [Google Scholar]
- Wang Z, Tseng C-P, Pong R-C, Chen H, McConnell JD, Navone N, Hsieh J-T. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. The Journal of biological chemistry. 2002 doi: 10.1074/jbc.M110568200. [DOI] [PubMed] [Google Scholar]
- Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G, Long M, Kabbani W, Yu L, Zhang H, Chen H, Sun X, Boothman DA, Min W, Hsieh JT. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci U S A. 2010;107:2485–2490. doi: 10.1073/pnas.0908133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Gore C, Zhou J, Pong RC, Zhang H, Yu L, Vessella RL, Min W, Hsieh JT. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc Natl Acad Sci U S A. 2009;106:19878–83. doi: 10.1073/pnas.0908458106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Toyooka S, Tsukuda K, Dote H, Ouchida M, Hanabata T, Aoe M, Date H, Gazdar AF, Shimizu N. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005a;113:59–66. doi: 10.1038/sj.bjc.6602458. [DOI] [PubMed] [Google Scholar]
- Yano M, Toyooka S, Tsukuda K, Dote H, Ouchida M, Hanabata T, Aoe M, Date H, Gazdar AF, Shimizu N. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005b;113:59–66. doi: 10.1002/ijc.20531. [DOI] [PubMed] [Google Scholar]
- Yano M, Toyooka S, Tsukuda K, Dote H, Ouchida M, Hanabata T, Aoe M, Date H, Gazdar AF, Shimizu N. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005c;113:59–66. doi: 10.1002/ijc.20531. [DOI] [PubMed] [Google Scholar]
- Zhang R, He X, Liu W, Lu M, Hsieh JT, Min W. AIP1 mediates TNF-alpha-induced ASK1 activation by facilitating dissociation of ASK1 from its inhibitor 14-3-3. J Clin Invest. 2003;111:1933–1943. doi: 10.1172/JCI200317790.Introduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ZENBU genome browser view of the FANTOM5 promoter level mammalian expression atlas data. The tracks shown here include Entrez Gene mm9 gene boundaries, UCSC RefSeq mm9 gene boundaries, Phase I CTSS data and predictions, and the Phase I CTSS pooled sequence tags from various mouse tissues (bottom).