Highlight
A new role of phytosulfokine in the regulation of ethylene production was identified.
Key words: Cu, deficiency, ethylene, PSK, root elongation, TPST.
Abstract
To increase our understanding of the adaptation for copper (Cu) deficiency, Arabidopsis mutants with apparent alterations under Cu deficiency were identified. In this report, a novel mutant, tpst-2, was found to be more sensitive than wild-type (Col-0) plants to Cu deficiency during root elongation. The positional cloning of tpst-2 revealed that this gene encodes a tyrosylprotein sulfotransferase (TPST). Moreover, the ethylene production of tpst-2 mutant was higher than that of Col-0 under Cu deficiency, and adding the ethylene response inhibitor AgNO3 partially rescued defects in root elongation. Interestingly, peptide hormone phytosulfokine (PSK) treatment also repressed the ethylene production of tpst-2 mutant plants. Our results revealed that TPST suppressed ethylene production through the action of PSK.
Introduction
Cell-to-cell communication plays an important role in the response to developmental and environmental cues in multicellular organisms. In plants, phytohormones (such as auxin, cytokinins, and ethylene) play critical roles in long-distance cell-to-cell interactions. In addition, secreted peptides [e.g., phytosulfokine (PSK), plant peptide containing sulfated tyrosine 1 (PSY1), and root growth factor] have recently emerged as a novel type of plant hormone (Matsubayashi and Sakagami, 1996; Matsubayashi, 2011, 2014; Yamada and Sawa, 2013). These peptides function as intercellular signalling molecules that coordinate and specify cellular functions in plants. Some of the secreted peptide hormones undergo complex posttranslational modifications mediated by specific enzymes that recognize particular sequences of multiple target peptides (Matsuzaki et al., 2010). Protein tyrosine sulfation is a form of posttranslational modification and is mediated by tyrosylprotein sulfotransferases (TPSTs). The transfer of sulfate from 3′-phosphoadenosine 5′-phosphosulfate to the hydroxyl group of peptidyl tyrosine residues is catalysed by TPSTs to form a tyrosine O4-sulfate ester (Bettelheim, 1954; Huttner, 1982). In animals, TPSTs are located in the trans-Golgi network and distributed in various tissues and cells. Several of these enzymes play important roles in inflammation, haemostasis, and immunity (Moore, 2003). However, plants have evolved structurally different TPSTs from their animal counterparts, which are critical for the biological activities of the peptide hormones PSK and PSY1 in plants (Matsubayashi, 2011). A recombinant version of an Arabidopsis thaliana TPST, At1g08030, expressed in yeast catalysed tyrosine sulfation of both PSK and PSY1 precursor polypeptides in vitro (Komori et al., 2009). AtTPST, a type I transmembrane protein localized in cis-Golgi, is expressed throughout the plant body, with the highest expression observed in the root apical meristem (Komori et al., 2009). Moreover, TPST is associated with auxin and brassinosteroid, which exhibit diverse roles for sulfated peptides in plant growth and development (Zhou et al., 2010; Hartmann et al., 2013). Although the role of the tyrosine sulfation motif in peptides remains unclear, characterizing peptide signalling is important for future plant research.
Copper (Cu) is an essential micronutrient for the growth and development of plants. Many metabolic processes, such as photosynthesis, mitochondrial electron transport, respiration, reactive oxygen metabolism, cell wall remodelling, and ethylene perception, also require Cu as an essential cofactor (Pilon et al., 2006; Burkhead et al., 2009). The Cu contents of plants vary among species, and environmental Cu conditions affect these Cu contents (Cohu and Pilon, 2007). The Cu content in plants typically ranges from 2 µg g−1 to 50 µg g−1 dry weight (DW) (ppm), and a Cu content of 6 µg g−1 is considered adequate for the normal growth and development of plants (Epstein and Bloom, 2005). However, when Cu content is lower than 5 µg g−1 DW or higher than 20 µg g−1 DW, symptoms of deficiency or toxicity appear (Marschner, 1995; Burkhead et al., 2009). Cu deficiency (-Cu) varies by agricultural soil types or crops; organic soil, including peat soils, and mineral soils with high levels of organic matter (6–10%) are most likely to be deficient in available Cu. -Cu develops on organic soils mainly because of Cu binding to organic matter (Marschner, 1995; Shorrocks and Alloway, 1988). Symptoms of -Cu in plants include reduced growth rates, leaf bleaching, pollen infertility, and a reduction in seed set and yield (Marschner, 1995).
In response to -Cu, plants control Cu homeostasis by regulating Cu uptake and allocation (Puig and Thiele, 2002; De Freitas et al., 2003). One of the mechanisms activated by Arabidopsis plants in response to -Cu is elevating Cu uptake. In Arabidopsis, the expression of some Cu transporter genes is upregulated in response to -Cu (Wintz et al., 2003; Mukherjee et al., 2006). The genes for Cu transporters zinc-regulated transporter iron-regulated transporter protein 2 (ZIP2) and ZIP4 are upregulated under -Cu (Wintz et al., 2003). Ferric reductase oxidase 3 (FRO3) expression is also induced in roots and shoots by Cu limitation (Mukherjee et al., 2006). In addition, under Cu-deficient conditions, copper chaperone (CCH) may be involved in mobilizing limited Cu among tissues during senescence (Mira et al., 2002). The production of CCH is also upregulated under -Cu. Another mechanism of plant response to -Cu is modifying Cu allocation for improved Cu economy, which has been well characterized in Chlamydomonas reinhardtii (Merchant et al., 2006). However, A. thaliana plants have a Cu allocation pattern different from that of C. reinhardtii. In A. thaliana, the expression patterns of copper/zinc superoxide dismutase1 (CSD1) and CSD2, as well as copper chaperone for SOD (CCS), show similar responses to the availability of Cu; i.e., these genes are downregulated under -Cu (Wintz et al., 2003; Abdel-Ghany et al., 2005). Given that CSD1 and CSD2 are downregulated under -Cu, ferric superoxide dismutase 1 (FSD1) becomes active, and the function of Cu/ZnSOD is replaced by FeSOD. Cu is then stored for the most essential functions under -Cu conditions (Abdel-Ghany et al., 2005; Cohu and Pilon, 2007). Identifying Cu-microRNAs whose expression is upregulated by low Cu increased our understanding of the mechanism of Cu homeostasis. miR398 regulates the mRNA stability of major Cu-containing proteins, such as CSD1, CSD2, and COX5b-1, under Cu-limited conditions. miR398 can directly regulate CSD mRNA degradation under -Cu (Yamasaki et al., 2007). In addition to miR398, three microRNAs, namely, miR397, miR408, and miR857, target genes that encode Cu-containing proteins (Abdel-Ghany and Pilon, 2008). -Cu activates miR398b and miR398c transcriptions via these GTAC motifs in A. thaliana, and the GTAC motif is sufficient for the response to -Cu (Yamasaki et al., 2009). The sequence analysis revealed that A. thaliana transcription factor squamosa promoter binding protein-like 7 (SPL7) is a candidate homologue of C. reinhardtii CRR1 protein, which is involved in Cu homeostasis in Chlamydomonas (Cardon et al., 1999; Kropat et al., 2005). Further analysis indicated that SPL7 is a conserved central regulator of Cu-deficiency response in plants. SPL7 can bind directly to GATC motifs in miR398 promoter (Yamasaki et al., 2009). In addition, some genes that encode Cu transporters (COPT1, COPT2, ZIP2, and YSL2), Cu chaperones (CCH and CCS), FRO3, FRO4, FRO5, and FSD1 are also misregulated in spl7 plants under -Cu (Yamasaki et al., 2009; Bernal et al., 2012). Moreover, many new genes in response to -Cu are regulated by SPL7 (Yamasaki et al., 2009). Despite the importance of preventing -Cu in plants, our understanding of the mechanisms controlling Cu homeostasis and trafficking inside the cell remains limited.
An attempt is made here to identify mutants with apparent alterations under -Cu to gain further insight into the molecular requirements of Cu homeostasis. In this paper, a novel mutant, tpst-2, is described as sensitive to -Cu in root length. This tpst-2 mutant has a considerably shorter root phenotype under -Cu conditions, whereas a high concentration of Cu (50 μM Cu) can partially recover its root elongation. Cu content of tpst-2 was lower than that of wild type Col-0. Positional cloning of tpst-2 revealed that this gene encodes a tyrosylprotein sulfotransferase (TPST). Moreover, tpst-2 was sensitive to ethylene, and adding AgNO3 to tpst-2 plants could partially rescue the root elongation of the mutant under -Cu. The ethylene production of tpst-2 mutant was higher than that of Col-0 under -Cu. Moreover, peptide hormone PSK treatment also repressed the ethylene production of tpst-2 mutant plants. The results obtained in this study reveal that TPST suppresses ethylene production through PSK action.
Materials and methods
Plant materials and growth conditions
Ethyl-methanesulfate (EMS)-mutagenized Columbia (Col-0 gl1-1) seeds were purchased from Lehle Seeds (Cat. No. M2E-01A-07; Roud Rock TX, USA). The seeds of the homozygous lines of T-DNA insertion SALK_009847 (tpst-1) and 35S:AtTPST/tpst-1 were kindly provided by Prof. Matsubayashi (Komori et al., 2009). F1 seeds used in the allelism test were obtained from a cross between tpst-1 and 33–4. 35S:AtTPST/33–4 seeds were obtained from F3 of a cross between 35S:AtTPST/tpst-1 and 33–4. The point mutations in 33–4 lines carrying the 35S:AtTPST gene were detected using CAPS primers with the DdeI restriction site (Supplementary Table S1). The seeds were surface-sterilized and sowed on an MGRL growth medium (Fujiwara et al., 1992) solidified with 1.5% gellan gum (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and supplemented with 1% sucrose. After the seeds were incubated for 2 d at 4°C, the plates were placed vertically and the plants were grown at 22°C under a 16h light/8h dark photoperiod. The plants were supplied with high Cu nutrient (50 μM Cu) containing 1.512mM NaH2PO4·2H2O, 0.257mM Na2HPO4·12H2O, 1.5mM MgSO4·7H2O, 4mM Ca(NO3)2·4H2O, 6mM KNO3, 8.6 μM C10H12FeN2NaO8·3H2O, 10.3 μM MnSO4, 50 μM CuSO4·5H2O, 30 μM H3BO3, 24nM Na6Mo7O24·4H2O, and 130nM CoCl2·6H2O (Fujiwara et al., 1992). The plants were also grown under a -Cu condition, in which no Cu was supplied in the medium.
Mutant selection and genetic mapping
EMS-mutagenized M2 seeds [50 seeds/plate × 20 plates per batch; a total of 26 batches (ca. 26,000 seeds)] was screened and grown on -Cu (MGRL without CuSO4) medium. The plants with short roots were transferred to MGRL with 50 µM CuSO4 (50 Cu), and the position of the root tips was marked. After 5 d, the plants with root elongations recovered by 50 Cu were transferred to the -Cu medium. After 5 d, the plants with terminated or slowed root elongations were selected. M3 seeds were obtained and then sown in -Cu and 50 Cu media; the corresponding phenotype was confirmed. After the second screening was performed, the 33–4 mutant was obtained.
In mapping, the 33–4 mutant was crossed with Ler, and F2 seeds were obtained. Genomic DNA was isolated from F2 plants exhibiting a mutant phenotype under -Cu condition; the gene was mapped using simple sequence length polymorphism (SSLP) markers (Sakamoto et al., 2011). After rough mapping was completed, the following primer pairs were used in the final step of the mapping: F24B9-F (5′-GGGGTCATTTGTGATCGAAG-3′) and F24B9-R (5′- TAA ATCACGTATGCCGCTCA-3′); F22O13-F (5′-TACGCAATGAG CCCTCAAAT-3′) and F22O13-R (5′- CTTGCATTGGGTTCA TTCCT-3′). Whole-genome SOLiD sequencing was also performed as described by Tabata et al. (2013).
Quantitative reverse transcription PCR analysis
Total RNA was prepared from the plants using an RNeasy plant mini kit (Qiagen, Valencia, CA, USA) and treated with RNase-free DNase (Qiagen). Total RNA was reverse-transcribed to prepare cDNA by using a PrimeScript RT reagent kit, diluted 2-fold, and used for real-time PCR analysis with a Thermal Cycler Dice system (Takara, Ohtsu, Japan) and SYBR Premix Ex Taq II (Takara). The sequences of the primers used in this study are provided in Supplementary Table S1. The primers used for elongation factor 1a (EF1a, internal standard) were described previously by Takano et al. (2006).
Analysis of Cu concentration
Whole roots and shoots of Col-0 and tpst-2 plants grown on -Cu or 50 Cu conditions for 10 d were harvested; samples were washed with distilled water thrice, dried at 65°C, and digested with concentrated nitric acid. The Cu concentration was then determined using inductively coupled plasma mass spectrometry (SPQ9700; SII, Chiba, Japan) as described previously (Takano et al., 2001).
Determination of sensitivity to Cu, AgNO3, 1-aminocyclopropane-1-carboxylic acid, and PSK
The seeds were sown on a medium containing various concentrations of Cu and combined Cu and 1 µM AgNO3, 1 µM 1-aminocyclopropane-1-carboxylic acid (ACC), or 10nM PSK to determine the sensitivity of tpst-2 mutant plants to AgNO3 and ACC. The root length of 10-day-old plants was determined using ImageJ (http://rsb.info.nih.gov/ij/).
Root length measurements
Sterilized seeds were incubated at 4°C for 2 d; afterwards, the plates were placed vertically and the plants were grown at 22°C in a 16h light/8h dark photoperiod. Root length was measured using ImageJ. Data are presented as means ± standard deviation. At least 30 seedlings were subjected to statistical analysis in each case. Tukey’s test was conducted for statistical analysis (P < 0.05).
Ethylene measurements
Seedlings (approximately 30 per vial) were grown in 22-mL gas chromatograph vials containing 3mL of MS medium or MGRL medium with various concentrations of Cu. The vials were flushed with hydrocarbon-free air and then capped at indicated times. The amount of accumulated ethylene was measured using a GC-14B gas chromatograph (Shimazu Corporation, Kyoto, Japan) equipped with an alumina column and a flame ionization detector. Ethylene peaks were quantified using Sic 480 II DataStation software (System Instruments Co., Ltd., Tokyo, Japan) and compared with a 10.06 μL/L ethylene standard. Ethylene production was normalized in terms of the number of seedlings in each vial and the time between capping and sampling. The basal levels of ethylene from Col-0, tpst-1, and tpst-2 were determined from three independent experiments of at least three replicates.
Results
Isolation and phenotypic characterization of a Cu-deficiency-sensitive mutant, 33–4
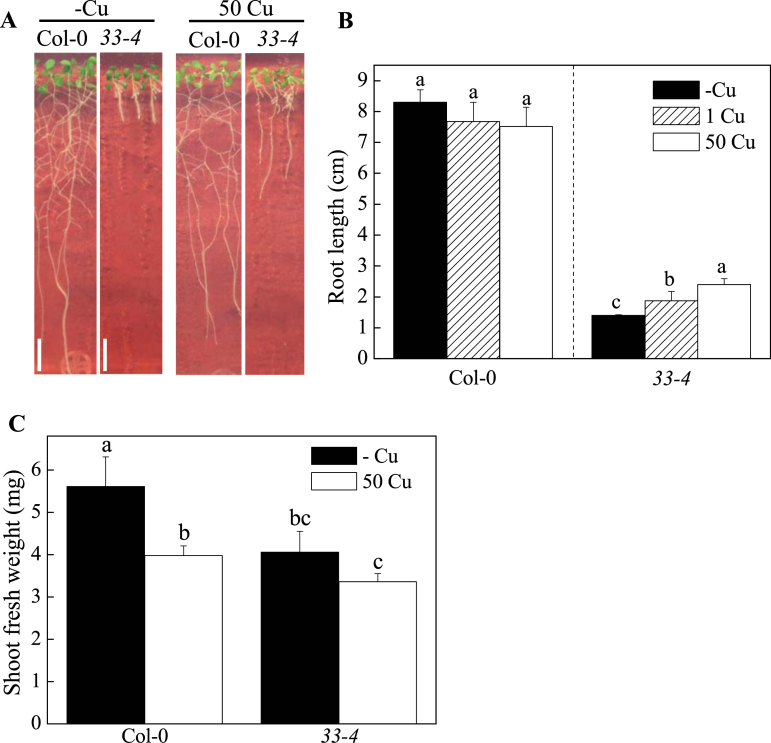
In the mutant screening, mutated seeds were repeatedly grown between -Cu medium and 50 Cu medium, to obtain Cu-deficiency-specific sensitive mutant. EMS-mutagenized M2 seeds (n = 26,000 seeds in total, Col-0 gl1-1) were sown onto -Cu medium to identify novel mutants under -Cu condition. Plants with short roots were selected and transferred to 50 Cu medium. The concentration of Cu in the standard medium was 1 µM. After 5 d, the plants with recovered root elongation were transferred to -Cu medium. After another 5 d, the plants with halted or reduced root elongation were selected and M3 seeds were harvested from individual plants. M3 seeds were sown onto -Cu and 50 Cu media and the lines with differential growth were selected. After this screening was completed, one mutant, namely 33–4, was obtained (Fig. 1A). The measured root lengths confirmed that root elongation was impeded (Fig. 1B). Under -Cu condition, the root lengths of Col-0 and 33–4 seedlings were 8.28±0.11 and 1.42±0.11cm, respectively. Under 50 Cu condition, the root lengths of Col-0 and 33–4 were 7.0±0.33 and 2.6±0.07cm, respectively. These data revealed that 33–4 was a Cu-deficiency-sensitive mutant. The shoot phenotype of 33–4 was indistinguishable from that observed in Col-0 under -Cu and 50 Cu conditions (Fig. 1A); however, Col-0 plants yielded heavier fresh shoot weights than the 33–4 mutant under 50 Cu and -Cu conditions (Fig. 1C). In addition, Col-0 plants showed heavier fresh shoot weights under -Cu condition than the plants under 50 Cu condition; by contrast, 33–4 plants showed similar shoot fresh weights under both -Cu and 50 Cu conditions (Fig. 1C).
Fig. 1.
Phenotypes of Arabidopsis 33–4 mutants in response to Cu conditions. (A) Col-0, 33–4 seedlings grown in -Cu or 50 Cu for 10 d. Bar = 1cm. (B) Root lengths of Col-0 and 33–4 seedlings. The primary root lengths of 10-day-old seedlings grown in -Cu, 1 Cu (normal medium), or 50 Cu are expressed as means ± SE (n = 10). Letters represent significant differences at the 0.05 level based on Tukey’s test. (C) Shoot fresh weight of Col-0 and 33–4 seedlings. The shoot fresh weights of 10-day-old seedlings grown in -Cu or 50 Cu are expressed as means ± SE (n = 10). Letters represent significant differences at the 0.05 level based on Tukey’s test.
Identification of genes responsible for the increased low-Cu sensitivity of the 33–4 mutant
The 33–4 mutant in the Col-0 background was crossed with Ler wild-type plants for genetic analysis and mapping. The resultant F2 progeny showed a segregation of the short- and long-root phenotypes. Among 134 plants, 25 exhibited mutant short-root phenotype and 109 displayed wild-type long-root phenotype. This result was not significantly different from the segregation ratio of 1:3 (P = 0.09, chi-square test), indicating that 33–4 carries a single recessive mutation responsible for the short-root phenotype. In mapping, genomic DNA was isolated from 200 individual F2 plants that exhibited the mutant phenotype under -Cu condition; the mutation was then mapped using SSLP markers. The causal gene was roughly mapped to chromosome 1 and then finely mapped between SSLP markers F24B9 (2.42Mb) and F22O13 (2.78Mb; Supplementary Fig. S1A). This region contained 93 putative genes annotated in TAIR10 (Fig. S1A). To identify point mutations, the genomic DNA of the 33–4 mutant was sequenced using the SOLiD sequencer. Two nucleotide substitutions (SNPs) were identified from the sequencing data. One SNP was found in At1g07910, resulting in an amino acid substitution. Another SNP was found in an intron of At1g08030 (Fig. S1A). At1g08030 is annotated as tyrosyl protein sulfotransferase (TPST); a G-to-A transition at nucleotide no. 2491575 of the genomic sequence (the consensus splicing site of fourth intron) was found. This mutation likely disrupted splicing. The phenotype of tpst-1 mutants has been described previously (Komori et al., 2009); the abnormal root elongation phenotype was similar to the 33–4 mutant.
A mutant line (tpst-1, SALK_009847) homozygous to a T-DNA insertion in the AtTPST gene was examined (Fig. S1B) to determine whether TPST is the causal gene of 33–4. tpst-1 and F1 plants derived from crosses between 33–4 and tpst-1 showed the same phenotype as 33–4 under the -Cu conditions; similar to the case of 33–4, root elongation was partially recovered by treatment with 50 Cu (Fig. S1C). These findings suggest that the phenotypes of 33–4 and tpst-1 are caused by a mutation in the same locus. Furthermore, the introduction of 35S:AtTPST to the 33–4 mutant restored the wild-type phenotype under both -Cu and 50 Cu growth conditions (Fig. S1C). These results indicate that the causal gene of 33–4 is indeed TPST. In this study, 33–4 is referred to as tpst-2 hereafter.
A mutation in TPST in tpst-2 disrupts splicing
The point mutation in tpst-2 is in the splicing consensus sequence in the fourth intron. The mRNA of TPST in tpst-2 was examined to study the effect of the point mutation on its expression. RT-PCR analysis was performed using primers that amplified the PCR products covering the mutation region (Supplementary Table S1). Based on the sequence information of TPST, the PCR product using the designed primers should have been 409bp in length. A single band of the expected size was produced by RT-PCR using the wild-type mRNA. The mRNA of tpst-2 resulted in two PCR products (Supplementary Fig. S2). The PCR products were then sequenced. The size of the band from the wild type was 409bp; by contrast, the bands from the mutant were 517 and 381bp in length. Based on the sequence, the structure of each transcript was determined (Fig. S2B,C,D). The wild-type transcript yielded the expected structure; by contrast, the transcripts from the mutant showed either a deletion or an insertion of extra sequences. These transcripts were unlikely to produce a wild-type protein.
Mutation of TPST specifically affects the Cu response
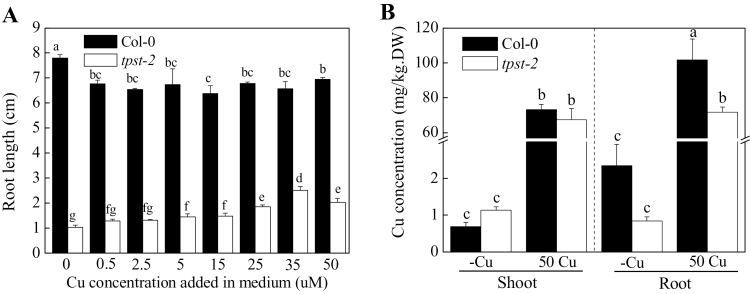
At increased concentrations, Cu induces the generation of harmful reactive oxygen species, which cause oxidative damage to cells at lipid, protein, and nucleic acid levels (Halliwell and Gutteridge, 1984; Drazkiewicz et al., 2004). The seedlings of Col-0 and tpst-2 were grown in various Cu conditions and root length was measured to further examine Cu sensitivity of tpst-2 root elongation. The seedlings of Col-0 showed a longer root under -Cu and 50 Cu conditions than other Cu conditions (Fig. 2A). The root elongation of tpst-2 was responsive to external Cu in a concentration-dependent manner (up to 35 µM Cu, referred to as 35 Cu); furthermore, 35 Cu elicited the strongest effect on the root elongation of tpst-2 (Fig. 2A). By contrast, the addition of other divalent transition metals, such as Fe, Mn, Ni, and Zn, failed to recover normal tpst-2 root elongation (Fig. S3). The root lengths of Col-0 and tpst-2 were restrained under Fe deficiency, 50 µM Fe, and 100 µM Ni conditions (Fig. S3). In Cu treatment, CuSO4 was added to the media to increase the Cu and sulfate concentrations. CuCl2 was analysed to confirm that Cu was responsible for root elongation recovery of tpst-2. CuCl2 treatment elicited an effect on the root elongation of tpst-2 similar to CuSO4 (Fig. 1A, Fig. S4); this result suggests that tpst-2 is a mutant affecting Cu homeostasis.
Fig. 2.
The tpst-2 mutant plants are sensitive to Cu deficiency. (A) Root lengths of Col-0 and tpst-2 mutant plants under various Cu concentrations. Plants were grown for 10 d on vertically placed MGRL medium. CuSO4 (0.5, 2.5, 5, 15, 25, 35, and 50 µM) was added to the medium (excluding -Cu medium). Letters represent significant differences at the 0.05 level based on Tukey’s test. Bar = SE, n=10. (B) Cu concentrations in roots and shoots of Col-0 and tpst-2 mutant plants. Plants were grown for 11 d on vertically placed MGRL medium. CuSO4 (50 µM) was added to 50 Cu medium but not to -Cu medium. Letters represent significant differences at the 0.05 level based on Tukey’s test. Bar = SE, n = 10.
TPST mutation affects Cu homeostasis in tpst-2 plants
Plants can sense internal and external Cu status and adapt to changing Cu conditions by modifying gene expression. Cu concentrations of tpst-2 seedlings under -Cu and 50 Cu conditions were measured to determine whether Cu sensitivity of tpst-2 mutant seedlings was affected. The results showed that tpst-2 roots exhibited a lower Cu concentration than Col-0 under both the -Cu and 50 Cu conditions; however, this finding was not observed in the shoots. Thus, this result cannot be considered as a generalized observation (Fig. 2B). Under the -Cu condition, root Cu concentrations of tpst-2 and Col-0 seedlings were 0.84±0.11 and 2.35±0.58mg/kg DW, respectively. Under the 50 Cu condition, root Cu concentrations of tpst-2 and Col-0 were 71.9±2.9 and 102±12mg/kg DW, respectively (Fig. 2B). However, no significant difference in shoot Cu concentration was observed between Col-0 and tpst-2 mutant plants grown under either -Cu or 50 Cu conditions (Fig. 2B). The altered root Cu concentration suggested that the TPST mutation is involved in Cu homeostasis.
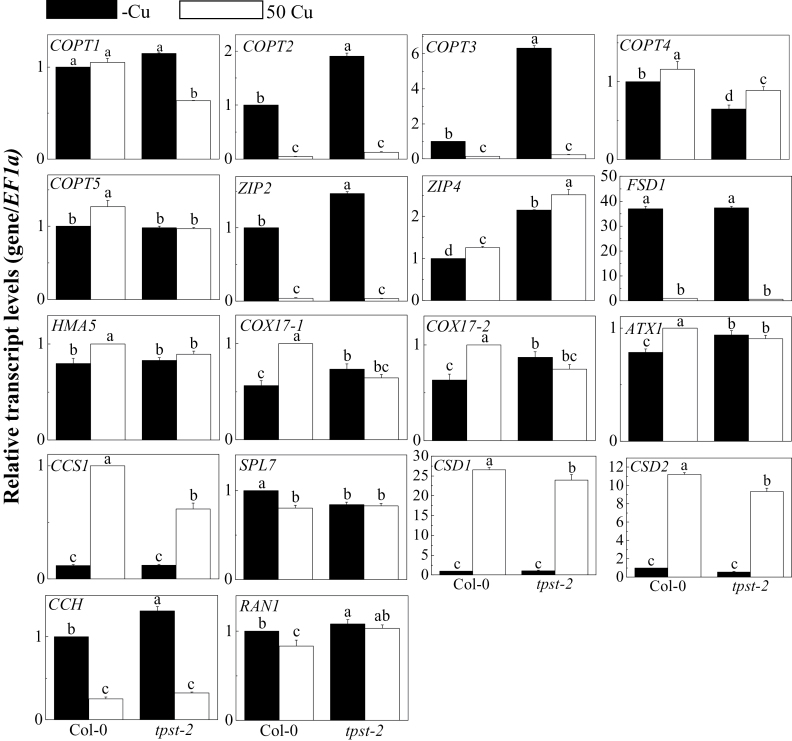
To confirm this hypothesis, the mRNA levels of several genes involved in Cu homeostasis were determined in tpst-2 mutant plants. This analysis included genes encoding Cu transporters (COPT1, COPT2, COPT3, COPT4, COPT5, ZIP2, ZIP4, and RAN1), Cu chaperones (CCH, CCS, ATX, COX17-1, COX17-2), -Cu indicators (FSD1, CSD1, CSD2), P-type ATPases (HMA5), and a master Cu regulator, SPL7 (Fig. 3). Interestingly, the mRNAs of COPT3 and COPT2 accumulated to a considerably higher level (6.3-fold for COPT3, and 1.9-fold for COPT2) in tpst-2 roots than in Col-0 roots under -Cu condition, although both mRNAs were upregulated in Col-0 and tpst-2 roots by -Cu. The expression of COPT1 was upregulated in tpst-2 but remained unaffected in Col-0 roots by -Cu. Although the expression of ZIP4 was not altered by -Cu in either Col-0 or tpst-2 roots, ZIP4 was constitutively expressed to a higher level in tpst-2 than in Col-0. COX17-1 and COX17-2 expressions were downregulated in Col-0, but were not regulated in tpst-2 roots by -Cu. Furthermore, the expressions of SPL7, RAN1, ATX1, HMA5, COPT4, and COPT5 were not evidently upregulated by -Cu, even in Col-0 (Fig. 3). The expressions of CCH, FSD1, and ZIP2 were upregulated by -Cu in both Col-0 and tpst-2 roots (Fig. 3); by contrast, the expressions of CSD1, CSD2, and CCS1 expression were downregulated by -Cu in both Col-0 and tpst-2 roots (Fig. 3).
Fig. 3.
The expression levels of Cu-related genes in the root of Col-0 and tpst-2 under -Cu/50 Cu conditions. Plants were grown for 10 d on vertically placed MGRL medium with or without 50 µM CuSO4. Total RNA was extracted from the whole roots of seedlings. At least 10 plants were used per replicate. The mRNA levels were normalized to the EF1α mRNA levels in the same samples. The data are expressed as means ± SE (n = 3, technical repeats) relative to the Col-0 value (defined as 1). Letters represent significant differences at the 0.05 level based on Tukey’s test.
TPST mutation increases ethylene production under -Cu condition
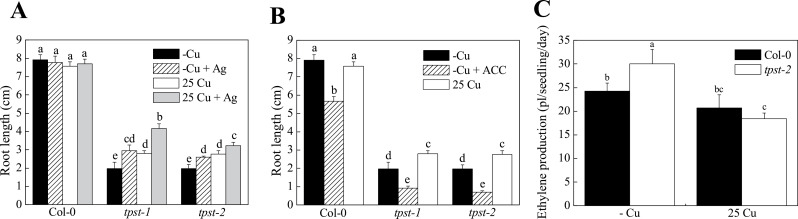
The role of Cu in the binding of ethylene to its receptors supports the role of Cu homeostasis in plant signalling and developmental regulation (Rodríguez et al., 1999). In Cu transport across intracellular membranes, RAN1 is a P-type ATPase that transports Cu to a late secretory compartment for delivery to ethylene receptors (Hirayama et al., 1999). Considering that TPST is involved in Cu response, detailed study was performed to determine whether the disruption of TPST affects ethylene sensing and/or signalling. For this purpose, root elongation of Col-0 and tpst-2 plants was determined under -Cu conditions with additional AgNO3 or ACC treatment, which are respectively known as an inhibitor of ethylene binding and a substrate of ACC oxidase, which makes ethylene (Lurssen et al., 1979; Strader et al., 2009). This result showed that the root elongation of tpst-2 was responsive to AgNO3 and ACC treatment (Fig. 4A,B). Under -Cu conditions, tpst-2 root elongation was partially rescued by additional AgNO3 treatment. Furthermore, the combination of AgNO3 and Cu elicited a greater effect on the root elongation of tpst-2 (Fig. 4A). The response of tpst-1 was similar to that of tpst-2. ACC also affected the root elongation of tpst-2, but this effect differed from that of AgNO3. With additional ACC treatment, tpst-2 seedlings showed an 85% reduction in root length, whereas Col-0 showed a 42% reduction in root length (Fig. 4B). These results can be explained by the increased ethylene production in the tpst-2 mutant and inhibition of the ethylene response by Cu and Ag.
Fig. 4.
Ethylene measurement and response of tpst-1 and tpst-2 root to AgNO3 and ACC under different Cu conditions. (A) Combination treatment of plants with AgNO3 and Cu. Col-0, tpst-1, and tpst-2 plants were grown for 14 d on vertically placed MGRL medium with or without additional 1 μM AgNO3. CuSO4 (25 µM) was added to 25 Cu medium but not to -Cu medium. Letters represent significant differences at the 0.05 level based on Tukey’s test. Bar = SE, n = 10. (B) Combination treatment of plants with ACC and Cu. Col-0, tpst-1, and tpst-2 plants were grown for 14 d on vertically placed MGRL medium with or without additional 1 μM ACC. CuSO4 (25 µM) was added to 25 Cu medium but not to -Cu medium. Letters represent significant differences at the 0.05 level based on Tukey’s test. Bar = SE, n = 10. (C) Ethylene production of plants. Col-0 and tpst-2 plants were grown for 5 d on vertically placed MGRL medium. CuSO4 (25 µM) was added to 25 Cu medium but not to -Cu medium. Letters represent significant differences at the 0.05 level based on Tukey’s test. Bar = SE, n = 30.
To confirm this hypothesis, ethylene production was determined under various Cu conditions. Under normal Cu (1 µM) conditions, tpst-2 and tpst-1 exhibited higher ethylene production than Col-0 (Supplementary Fig. S5A). In addition, tpst-2 yielded higher ethylene production than Col-0 under -Cu conditions; conversely, tpst-2 displayed ethylene production similar to wild-type plants under 25 Cu conditions (Fig. 4C). These results confirmed that ethylene production by tpst mutants was higher than that by wild-type plants under -Cu conditions.
TPST mutation affects genes associated with ethylene response, biosynthesis, and signal transduction
The expression of an ethylene-responsive marker gene was determined to further characterize the relationship between TPST and ethylene. Expression of the basic chitinase gene is induced by ethylene in adult Arabidopsis plants (Samac et al., 1990). In the present study, the basic chitinase gene exhibited a higher expression level in the roots of tpst-2 and tpst-1 under -Cu conditions than under 50 Cu conditions (Supplementary Fig. S5B). These results confirm that TPST mutation increases ethylene production.
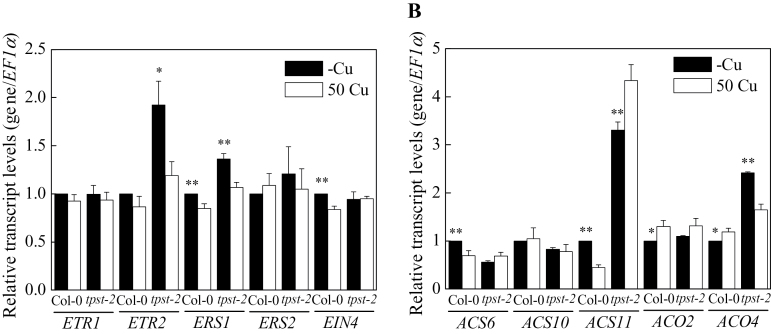
To examine how the TPST mutation disrupts ethylene production, the expression of several ethylene signal transduction and biosynthesis genes was examined by quantitative RT-PCR (Fig. 5) to investigate the mechanism by which TPST mutation disrupts ethylene production. Interestingly, the mRNA of ACO4, an ethylene biosynthesis gene, accumulated to a much higher level in the roots of tpst-2 than in the roots of Col-0 under -Cu; by contrast, ACO4 expression differed slightly between tpst-2 and Col-0 under 50 Cu conditions. The ethylene receptor gene ETR2 showed expression patterns similar to ACO4. This result suggests that TPST mediates the accumulation of high ethylene levels and induces the expression of signal transduction genes. However, only some of the genes associated with ethylene production and signal transduction were affected by TPST. The expression of ACS11, a biosynthesis gene, showed an opposite pattern between Col-0 and tpst-2 under Cu treatment; in particular, ACS11 was more highly expressed in tpst-2 than in Col-0. A marked upregulation of other biosynthesis and signal transduction genes, including ACS6, ACS10, ACO2, ETR1, ERS1, ERS2, and EIN4, was not detected, even in wild-type roots (Fig. 5). These results suggest that TPST is involved in the regulation of some ethylene biosynthesis and signal transduction genes under -Cu.
Fig. 5.
Expression of (A) ethylene receptor and (B) biosynthesis genes in roots of Col-0 and tpst-2 by quantitative RT-PCR. Col-0 and tpst-2 mutant plants were grown for 10 d on vertically placed MGRL medium. CuSO4 (50 µM) was added to 50 Cu medium, but not to -Cu medium. Total RNA was extracted from the whole roots of seedlings. At least 10 plants were used per replicate. The mRNA levels were normalized to those of EF1α in the same samples. The data are expressed as means ± SE (n = 3, technical repeats) relative to the Col-0 value (defined as 1). Asterisks represent significant differences from the 50 Cu conditions (**P < 0.01; *P < 0.05, Student’s t-test).
Expression of TPST is not regulated by ethylene or Cu
Ethylene is involved in the sensitivity of root elongation to -Cu in tpst-2 mutants, so determining whether the expression of TPST is regulated by Cu or ethylene is important. To answer this question, TPST expression was measured under various Cu conditions combined with AgNO3 and ACC treatment. The quantitative RT-PCR results revealed that TPST transcript accumulation was not regulated by Cu, AgNO3, or ACC treatment (Supplementary Fig. S6).
Treatment with the peptide hormone PSK suppresses ethylene production by tpst-2 seedlings
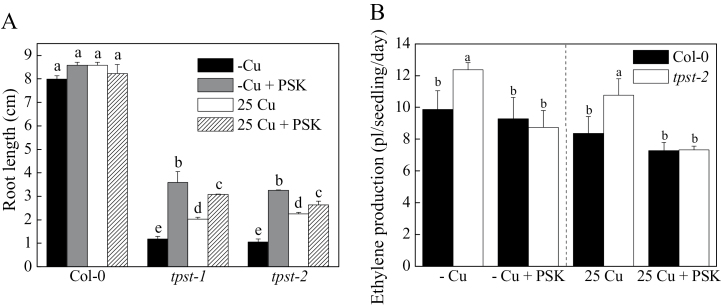
TPST encodes tyrosylprotein sulfotransferases responsible for the sulfation of the peptide hormone PSK. The fact that ethylene production of tpst-2 was elevated (Fig. 4C) suggests that PSK suppresses ethylene production in plants. To confirm this hypothesis, Col-0, tpst-2, and tpst-1 plants were treated with 10nM PSK, and root length and ethylene production were measured. PSK treatment of tpst-2 and tpst-1 partially rescued root elongation (Fig. 6A). The extent of the rescue was considerably higher under -Cu than 25 Cu conditions. In addition, PSK treatment had a greater recovery effect on the root elongation of the tpst mutants than did Cu (Fig. 6A). Ethylene production by tpst-2 plants decreased in the presence of PSK under -Cu or 25 Cu conditions, whereas ethylene production by Col-0 was unaffected by PSK treatment (Fig. 6B). These results suggest that PSK suppresses ethylene production in tpst mutants.
Fig. 6.
Root length and ethylene content measurement of Col-0 and tpst-2 plants. (A) Ethylene content measurement of Col-0 and tpst-2 plants. Col-0 and tpst-2 plants were grown for 5 d on -Cu and 25 Cu MGRL medium with or without additional 10nM PSK. Letters represent significant differences at the 0.05 level using Tukey’s test. Bar = SE, n = 30. (B) Root elongation of tpst-2 plants under Cu and PSK treatments. Col-0, tpst-1, and tpst-2 plants were grown for 10 d on vertically placed MGRL medium with or without 10nM PSK. CuSO4 (25 µM) was added to 25 Cu medium, but not to -Cu medium. Letters represent significant differences at the 0.05 level using Tukey’s test. Bar = SE, n = 10.
Discussion
In the present study, A. thaliana mutants sensitive to -Cu conditions were identified and used to determine that TPST is involved in Cu-dependent root elongation (Fig. 1 and Supplementary Fig. S1). Furthermore, results demonstrate that TPST is involved in ethylene production and PSK suppresses ethylene production under -Cu conditions (Figs. 4 and 6). This represents a novel link between two phytohormones, PSK and ethylene, in a Cu-dependent manner.
Cell-to-cell communication is important for plants. Phytohormones (including auxin, cytokinins, ethylene, and brassinosteroids) and secreted peptides, which are a novel type of plant hormone, play critical roles in cell-to-cell communication in plants (Matsubayashi, 2011; Yamada and Sawa, 2013). In higher plants, many genes encoding small, secreted peptides have been identified (Matsubayashi, 2011). Modifications of these peptides, such as posttranslational sulfation of PSK, are important for their function (Komori et al., 2009; Matsuzaki et al., 2010; Zhou et al., 2010).
In the present study, evidence was obtained that suggests a novel relationship between PSK and ethylene. The plant hormone ethylene influences various plant developmental processes, including germination, tissue senescence, fruit ripening, sex determination, and the response to a wide variety of stresses (Chang and Bleecker, 2004; Wu et al., 2010). Ethylene is also part of the signalling pathway that modulates cell division in the quiescent centre in the stem cell niche during the postembryonic development of the root system (Ortega-Martínez et al., 2007). In this study, several lines of evidence indicated that TPST suppresses ethylene production through the action of PSK (Figs. 4A,B and 6, and Supplementary Fig. S5). Based on these results and the hypotheses of this study combined with previous reports, a mechanism for the root elongation of tpst-2 mutant plants is proposed. In wild-type Arabidopsis plants, active PSK is thought to repress ethylene production, which results in normal root elongation. The tpst-2 mutant lacks the ability for the sulfation of the PSK precursor peptides, and ethylene production increases because of the lack of PSK. The high ethylene levels then decrease root elongation. Alternatively, since there is not a difference in ethylene production between tpst-2 and Col-0 in all conditions, many sickly mutants and even WT plants subjected to stressful conditions (-Cu, in this case) may also overproduce ethylene, so it might also be reasonable that the tpst mutants have elevated levels of ethylene, and, thus, a subset of the tpst phenotypes is a consequence of enhanced ethylene signalling. Accordingly, it is also logical that silver ions can partially rescue some aspects of the tpst mutant phenotype that are caused by extra ethylene. Likewise, it is expectable that exogenous application of the peptide hormone PSK (the molecular target of the TPST-mediated enzymatic activity) can bypass the requirement for functional TPST and reduces the severity of the tpst mutant defects, thus lessening ethylene overproduction in this mutant.
Notably, root elongation of the mutant was partially rescued by high Cu (Fig. 1). Cu is required not only for ethylene binding but also for the signalling function of ethylene receptors. Cu likely downregulated ethylene signalling pathways in these experiments, rescuing root elongation by preventing the development of ethylene response phenotypes. Moreover, root elongation of the tpst-2 mutant phenotype was partially recovered by AgNO3 (Fig. 4A), which could also affect auxin efflux independently of ethylene response (Strader et al., 2009). Although ethylene production is affected by various stresses, and the link between PSK and ethylene may not be a direct link, the evidence suggests a new role of PSK in the regulation of ethylene production.
Cu is an essential micronutrient for normal growth and development, and deficiency of this metal has a severe effect on plant development. Thus, to respond to -Cu, plants have evolved cellular and molecular mechanisms to maintain Cu homeostasis. Despite the importance of preventing -Cu in plants, much remains to be elucidated about the mechanisms controlling Cu homeostasis and trafficking inside the cell.
Root length is a highly sensitive parameter frequently used to test plant Cu sensitivity (Sancenón et al., 2004; Andrés-Colás et al., 2006). A defect in Cu homeostasis dramatically affects root elongation in Arabidopsis. Arabidopsis seedlings defective in COPT1 expression display a significant decrease in Cu acquisition and accumulation, and increased root length under Cu-limited conditions, whereas the roots could return to wild-type length when treated with 30 µM Cu sulfate (Sancenón et al., 2004). The same situation has been observed with hma5, atx1, and cch-atx1 double mutant (Andrés-Colás et al., 2006; Shin et al., 2012). The root length in 8-day-old seedlings is dramatically decreased in both hma5 mutant alleles treated with 20 µM Cu, whereas no defect was observed in MS plates. Root length was shorter for atx1 and cch-atx1 than for the wild type and the cch mutant with excess Cu; additionally, with 25, 35, and 50 µM Cu, the root length was about 80%, 76%, and 57%, respectively, in the wild type. Moreover, Arabidopsis seedlings overexpressing COPT1 or COPT3 displayed increased endogenous Cu concentration, and they did not appreciably differ from the wild-type ones when they were grown on MS. However, the seedlings grown on 10 µM CuSO4 displayed dramatic inhibition in root growth.
The tpst-2 mutant identified in this study showed a new and opposite phenotype to these observations mentioned above. The tpst-2 mutant defective in TPST not only displayed a significant decrease in root Cu concentration (Fig. 2B), but also a significant decrease in root length under -Cu conditions. At the same time, the root elongation could be partially rescued by 50 µM Cu sulfate (Fig. 1). Notably, endogenous low Cu concentration in tpst-2 mutant plants was perceived by the Cu sensor system, as suggested by the changes in the expression of the well-known Cu transporters COPT2 and COPT3 (Fig. 3). However, the Cu transcriptional factor SPL7 mRNA level was not influenced by Cu in tpst-2 mutant (Fig. 3). This difference in root elongation among tpst-2 and other Cu-related mutants suggests that the effect of Cu on root elongation in tpst-2, which has point mutation of TPST, is different from the mechanisms of known Cu transporters (COPT1 and COPT3) and chaperones (ATX1 and CCH).
The present study was performed using a Cu response mutant. The finding that tpst mutants exhibit Cu responses and differential Cu accumulation in roots suggests a novel regulatory mechanism of Cu homeostasis, i.e., the involvement of PSK. The tpst-2 mutant showed a significant decrease in root Cu concentration (Fig. 2B) and root length (Fig. 1) under -Cu conditions. Moreover, root elongation was partially rescued by 50 Cu (Fig. 1). Expression of the well-known Cu transporters COPT2 and COPT3 was higher in the mutant (Fig. 3). However, the mRNA level of the Cu transcriptional factor SPL7 was unaffected by tpst-2 mutation (Fig. 3). Such differences in the effect of tpst-2 mutations among the Cu-inducible genes suggest that the roles of PSK in the Cu response are limited to Cu homeostasis or Cu sensing/signal transduction pathways. The TPST mRNA level was not regulated by Cu (Supplementary Fig. S6), suggesting that the relationship between PSK and Cu homeostasis/signalling is uni-directional. PSK affects Cu homeostasis/signalling, but Cu does not affect PSK.
At this time, it is not possible to demonstrate the mechanisms underlying PSK-mediated regulation of Cu homeostasis, but the data revealed a possible site of action. In the tpst-2 mutant, the Cu concentration in roots was lower than that in the wild type (Fig. 2B), whereas expression of the Cu transporter genes COPT2 and COPT3 was significantly upregulated (Fig. 3). Arabidopsis COPT3 proteins have been implicated in Cu transport (Sancenón et al., 2004), and overexpression of COPT3 increased the total content of Cu in the plant. COPT2 is a plasma membrane protein that functions in Cu acquisition and distribution, whose expression is regulated most strongly in -Cu (Sancenón et al., 2003; Yamasaki et al., 2009; del Pozo et al., 2010). It is reasonable to assume that the roots of tpst-2 mutants with high COPT2 and COPT3 expression have higher Cu concentration. However, the results shown in Fig. 2B contradict this hypothesis. Possibly, the tpst-2 mutation affects the link between the high expression of COPT2/3 and the high accumulation of Cu in the roots. PSK may affect the activity of COPT2/3 and maintain the high COPT2/3 expression and low Cu contents.
In summary, in the present study, a novel link between PSK and ethylene production has been identified, as well as a novel factor that regulates Cu homeostasis in Arabidopsis.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used for RT-PCR and quantitative RT-PCR analyses.
Fig. S1. Mapping and cloning of the causal gene of Cu-deficiency-sensitive mutant 33–4.
(A) Identification of the causal gene of 33–4 mutant by rough mapping and SOLiD. The left panel indicates rough mapping of the mutation. The markers and their positions are according to TAIR 10 database. The number of recombinants at each marker is indicated on the left side of the schematic chromosome. The right panel represents the result of sequencing analysis using SOLiD. Based on the analysis of the mapped region for SNP identification, the criterion is at least three reads with more than 60% point mutations. Using this criterion, two mutations were identified in the mapped region at positions 2448324 and 2491575 on chromosome 1 based on the TAIR 10 database.
(B) Schematic of the exon-intron structure of AT1G08030 based on the TAIR 10 database and the location of the point mutation (2491575) in 33–4 and T-DNA insertion in tpst-1. Boxes and lines represent exons and introns, respectively. White boxes represent 5′ and 3′ UTRs. The T-DNA is not drawn to scale. The point mutation disrupts splicing consensus from AG to AA.
(C) Genotyping of the mutant and transgenic plants. Col-0, 33–4, tpst-1, F1 (tpst-1 × 33–4), and 35S:AtTPST/33–4 plants were grown for 10 d on vertically placed solid medium containing 50 µM CuSO4 (50 Cu) or no additional Cu (-Cu) medium. Bars = 1cm.
Fig. S2. The point mutation in the consensus splicing sequence of TPST fourth intron results in two different proteins. (A) RT-PCR analysis of TPST in Col-0 and tpst-2. M: 100-bp DNA ladder. (B) Original TPST gene structure. M: point mutation of TPST in tpst-2 mutant plants; RT-F, RT-R and arrowheads indicate the positions and orientations of the primers used for RT-PCR. (C) Mutation in TPST intron splicing site results in two variant forms. The 1bp represents the first nucleic acid of the start codon (ATG) in the TPST sequence based on the TAIR 10 database.
Fig. S3. Root lengths of Col-0 and tpst-2 seedlings under different metal conditions. The primary root length of 10-day-old seedlings grown in medium without Zn, Fe, Mn, Ni, or medium containing 50 or 100 µM of each element. The control values of root length were obtained in normal MGRL medium containing 1 µM Cu, 1 µM Zn, 8.6 µM Fe, and 10.3 µM Mn. The root lengths were expressed as means ± SE (n = 10).
Fig. S4. The response of tpst-2 root elongation to CuCl2. Col-0 and tpst-2 mutant plants were grown for 11 d on vertically placed MGRL medium with or without CuCl2 (50 µM). n = 10. Letters represent significant differences at the 0.05 level based on Tukey’s test.
Fig. S5. Ethylene production and expression of the ethylene marker gene basic chitinase. (A) Col-0, tpst-1, and tpst-2 plants were grown on MS medium (1 μM Cu) for 5 d. Letters represent significant differences at the 0.05 level by Tukey’s test. Bar = SE, n = 30. (B) Quantitative RT-PCR analysis of basic chitinase expression in the roots of Col-0, tpst-1, and tpst-2 seedlings. Col-0, tpst-1, and tpst-2 plants were grown on vertically placed MGRL medium. CuSO4 (50 µM) was added to 50 Cu medium, but not to -Cu medium. Total RNA was extracted from the whole roots of 10-day-old seedlings. At least 10 plants were used per replicate. Levels of basic chitinase mRNA were normalized to those of EF1α in the same samples. The data were expressed as means ± SE (n = 3, technical repeats) relative to the Col-0 (-Cu) value (defined as 1). Asterisks represent significant differences from the 50 Cu condition (**P < 0.01, Student’s t-test).
Fig. S6. The expression pattern of TPST under various Cu conditions with or without AgNO3 and ACC treatment. Col-0 plants were grown for 10 d on vertically placed MGRL medium with -Cu or various Cu concentrations (1, 25, 35, and 50 µM). Next, 1 μM AgNO3 and 1 μM ACC were added to the 1 Cu medium. Total RNA was extracted from the whole roots of seedlings. At least 10 plants were used per replicate. Levels of TPST mRNA were normalized to those of EF1α in the same samples. The data were expressed as means ± SE (n = 3, technical repeats) relative to the value of Col-0 plants grown on 1 Cu medium (defined as 1). No significant differences were found based on Tukey’s test.
Acknowledgements
TW was supported by the JSPS fellowship for foreigners (P11394). This work was supported by a Grant-in-Aid for Scientific Research (S) to TF (grant no. 25221202) and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to TF (grant no. 22119002).
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. 2005. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Pilon M. 2008. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis . Journal of Biological Chemistry 283, 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. 2006. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. The Plant Journal 45, 225–236. [DOI] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, Merchant SS, Krämer U. 2012. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis . Plant Cell 24, 738–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelheim FR. 1954. Tyrosine O-sulfate in a peptide from fibrinogen. Journal of The American Chemical Society 76, 2838–2839. [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. 2009. Copper homeostasis. New Phytologist 182, 799–816. [DOI] [PubMed] [Google Scholar]
- Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P. 1999. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. [DOI] [PubMed] [Google Scholar]
- Chang C, Bleecker AB. 2004. Ethylene biology. More than a gas. Plant Physiology 136, 2895–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohu CM, Pilon M. 2007. Regulation of superoxide dismutase expression by copper availability. Physiologia Plantarum 129, 747–755. [Google Scholar]
- De Freitas J, Wintz H, Kim JH, Poynton H, Fox T, Vulpe C. 2003. Yeast, a model organism for iron and copper metabolism studies. Biometals 16, 185–197. [DOI] [PubMed] [Google Scholar]
- del Pozo T, Cambiazo V, González M. 2010. Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana . Biochemical and Biophysical Research Communications 393, 248–252. [DOI] [PubMed] [Google Scholar]
- Drazkiewicz M, Skórzyńska-Polit E, Krupa Z. 2004. Copper-induced oxidative stress and antioxidant defence in Arabidopsis thaliana . Biometals 17, 379–387. [DOI] [PubMed] [Google Scholar]
- Epstein E, Bloom AJ. 2005. Mineral Nutrition of Plants: Principles and Perspectives. Ed 2 Sunderland, MA: Sinauer Associates, 17–39. [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S. 1992. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiology 99, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. 1984. Lipid peroxidation, oxygen radicals, cell damage, and antioxidant therapy. Lancet 1, 1396–1397. [DOI] [PubMed] [Google Scholar]
- Hartmann J, Stührwohldt N, Dahlke RI, Sauter M. 2013. Phytosulfokine control of growth occurs in the epidermis, is likely to be non-cell autonomous and is dependent on brassinosteroids. The Plant Journal 73, 579–590. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR. 1999. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis . Cell 97, 383–393. [DOI] [PubMed] [Google Scholar]
- Huttner WB. 1982. Sulphation of tyrosine residues: a widespread modification of proteins. Nature 299, 273–276. [DOI] [PubMed] [Google Scholar]
- Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. 2009. Identification of tyrosylprotein sulfotransferase in Arabidopsis . Proceedings of The National Academy of Sciences of The United States of America 106, 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depege N, Huijser P, Merchant S. 2005. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proceedings of The National Academy of Sciences of The United States of America 102, 18730–18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurssen K, Naumann K, Shroder R. 1979. 1-aminocyclopropane-1-carboxylic acid: an intermediate of ethylene biosynthesis in higher plants. Zeitschrift für Pflanzenphysiologie 92, 285–294. [Google Scholar]
- Marschner H. 1995. Mineral Nutrition of Higher Plants. Boston, MA: Academic Press. [Google Scholar]
- Matsubayashi Y. 2011. Post-translational modifications in secreted peptide hormones in plants. Plant and Cell Physiology 52, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis . Science 329, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. 1996. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proceedings of The National Academy of Sciences of The United States of America 93, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Allen MD, Kropat J, Moseley JL, Long JC, Tottey S, Terauchi AM. 2006. Between a rock and a hard place: trace element nutrition in Chlamydomonas . Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1763, 578–594. [DOI] [PubMed] [Google Scholar]
- Mira H, Martínez N, Peñarrubia L. 2002. Expression of a vegetative-storage-protein gene from Arabidopsis is regulated by copper, senescence and ozone. Planta 14, 939–946. [DOI] [PubMed] [Google Scholar]
- Moore KL. 2003. The biology and enzymology of protein tyrosine O-sulfation. Journal of Biological Chemistry 278, 24243–24246. [DOI] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL. 2006. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223, 1178–1190. [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O, Pernas M, Carol RJ, Dolan L. 2007. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510. [DOI] [PubMed] [Google Scholar]
- Pilon M, Abdel-Ghany SE, Cohu CM, Gogolin KA, Ye H. 2006. Copper cofactor delivery in plant cells. Current Opinion in Plant Biology 9, 256–263. [DOI] [PubMed] [Google Scholar]
- Puig S, Thiele DJ. 2002. Molecular mechanisms of copper uptake and distribution. Current Opinion in Chemical Biology 6, 171–180. [DOI] [PubMed] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. 1999. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis . Science 283, 996–998. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T. 2011. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis . Plant Cell 23, 3533–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallaly PE, Shah DM. Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana . Plant Physiology 93, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mateu-Andrés I, Dorcey E, Thiele DJ, Peñarrubia L. 2004. The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. Journal of Biological Chemistry 279, 15348–15355. [DOI] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L. 2003. Identification of a copper transporter family in Arabidopsis thaliana . Plant Molecular Biology 51, 577–587. [DOI] [PubMed] [Google Scholar]
- Shin LJ, Lo JC, Yeh KC. 2012. Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiology 159, 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrocks VM, Alloway BJ. 1988. Copper in Plant, Animal and Human Nutrition. Potters Bar, Hertfordshire: Copper Development Association. [Google Scholar]
- Strader LC, Beisner ER, Bartel B. 2009. Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell 21, 3585–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Kamiya T, Shigenobu S, Yamaguchi K, Yamada M, Hasebe M, Fujiwara T, Sawa S. 2013. Identification of an EMS-induced causal mutation in a gene required for boron-mediated root development by low-coverage genome re-sequencing in Arabidopsis . Plant Signaling and Behavior 8, e22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Wada M, Ludewig U, Schaaf G, von Wirén N, Fujiwara T. 2006. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18, 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Yamagami M, Noguchi K, Hayashi H, Fujiwara T. 2001. Preferential translocation of boron to young rosette leaves in Arabidopsis thaliana regulated by the BOR1 gene. Soil Science and Plant Nutrition 47, 345–357. [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. 2003. Expression profiles of A. thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. Journal of Biological Chemistry 278, 47644–47653. [DOI] [PubMed] [Google Scholar]
- Wu T, Qin Z, Zhou X, Feng Z, Du Y. 2010. Transcriptome profile analysis of floral sex determination in cucumber. Journal of Plant Physiology 167, 905–913. [DOI] [PubMed] [Google Scholar]
- Yamada M, Sawa S. 2013. The roles of peptide hormones during plant root development. Current Opinion in Plant Biology 16, 56–61. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. 2007. Regulation of copper homeostasis by micro-RNA in Arabidopsis . Journal of Biological Chemistry 282, 16369–16378. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. 2009. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis . Plant Cell 21, 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, Zhai Q, Jiang H, Chen R, Chen Q, Sun J, Chu J, Zhu L, Liu CM, Li C. 2010. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22, 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.