Highlight
Mannans present in the coleorhiza and root of Brachypodium seeds disappear early during germination sensu-stricto, while mannanase activity increases and three BdMAN transcripts are localized in these tissues.
Key words: BdMAN gene family, Brachypodium distachyon, coleorhiza, endo-β-mannanases, germination, MAN gene expression, mannan immunolocalization, mRNA in situ hybridization.
Abstract
Immunolocalization of mannans in the seeds of Brachypodium distachyon reveals the presence of these polysaccharides in the root embryo and in the coleorhiza in the early stages of germination (12h), decreasing thereafter to the point of being hardly detected at 27h. Concurrently, the activity of endo-β-mannanases (MANs; EC 3.2.1.78) that catalyse the hydrolysis of β-1,4 bonds in mannan polymers, increases as germination progresses. The MAN gene family is represented by six members in the Brachypodium genome, and their expression has been explored in different organs and especially in germinating seeds. Transcripts of BdMAN2, BdMAN4 and BdMAN6 accumulate in embryos, with a maximum at 24–30h, and are detected in the coleorhiza and in the root by in situ hybridization analyses, before root protrusion (germination sensu stricto). BdMAN4 is not only present in the embryo root and coleorhiza, but is abundant in the de-embryonated (endosperm) imbibed seeds, while BdMAN2 and BdMAN6 are faintly expressed in endosperm during post-germination (36–42h). BdMAN4 and BdMAN6 transcripts are detected in the aleurone layer. These data indicate that BdMAN2, BdMAN4 and BdMAN6 are important for germination sensu stricto and that BdMAN4 and BdMAN6 may also influence reserve mobilization. Whether the coleorhiza in monocots and the micropylar endosperm in eudicots have similar functions, is discussed.
Introduction
Poaceae grains (caryopses) include the seed proper, formed by a triploid endosperm and a diploid embryo, surrounded by the maternal tissues of the seed coat (testa) and the pericarp. The coleorhiza is a non-vascularized multicellular embryonic tissue, covering the seminal roots of Poaceae seeds. The coleorhiza has been thought to have a role in protecting the emerging root (Sargent and Osborne, 1980) and, more recently, it has been also associated with the regulation of dormancy, since abscisic acid (ABA) sensitivity is reduced in this tissue during germination of non-dormant barley seeds and the gene encoding the HvABA8′OH-1 enzyme, that is critical for ABA degradation, is expressed in the coleorhiza. During germination, both in barley and in Brachypodium seeds, the coleorhiza is the first structure that protrudes after the pericarp and testa rupture (coleorhiza emergence), followed by the coleorhiza rupture that allows root emergence (root emergence), and indicates the end of germination sensu stricto (Millar et al., 2006; Barrero et al., 2009, 2012; Gao and Ayele, 2014).
The germination process can be separated into germination sensu stricto and subsequent reserve mobilization (post-germination) and has been more deeply investigated in eudicotyledonous than in monocotyledonous seeds. In Arabidopsis thaliana, Sisymbrium officinale, Lepidium sativum and Nicotiana tabacum, the germination sensu stricto occurs in two different steps; first, the testa ruptures and, afterwards, the micropylar endosperm breakage takes place, allowing the radicle to emerge (Leubner-Metzger and Meins, 2000; Nonogaki et al., 2000; Müller et al., 2006; Iglesias-Fernández and Matilla, 2010; Iglesias-Fernández et al., 2011a , b ; Weitbrecht et al., 2011; Nonogaki, 2014). Addition of ABA to the imbibition medium specifically blocks endosperm weakening and prevents its rupture (Müller et al., 2006; Piskurewicz et al., 2009; Carrillo-Barral et al., 2014). It is assumed that testa rupture is influenced by the driving force of the imbibed elongating radicle and that the endosperm rupture is mainly produced by the weakening of the endosperm cell walls (CWs) by enzymes, specifically those localized to the micropylar endosperm, such as endo-β-1,4-mannanases (MANs), endo-ß-1,3-glucanases, expansins, xyloglucan-transglycosylases/hydrolases (XTHs) and pectin-methylesterases (Leubner-Metzger, 2005; Nonogaki et al., 2007; Iglesias-Fernández et al., 2011a , b ; Endo et al., 2012; Martínez-Andújar et al., 2012; Rodríguez-Gacio et al., 2012; Scheler et al., 2014).
Since the endosperm CWs of several eudicot seeds are rich in mannans (Lee et al., 2012), the MAN activity and the expression of MAN genes upon seed germination have been further characterized and their transcriptional regulation studied. In A. thaliana, four MAN genes (AtMAN2, AtMAN5, AtMAN6 and AtMAN7) are expressed in germinating seeds and their transcripts are restricted to the micropylar endosperm and to the radicle, disappearing as soon as the radicle emerges. Moreover, knock-out mutants in the AtMAN5, AtMAN6 and AtMAN7 genes, as well as, in the AtbZIP44 gene encoding an important activating transcription factor of AtMAN7, have a significantly retarded germination as compared to that of wild-type seeds, indicating a role for these MAN genes and their regulators during germination sensu stricto (Iglesias-Fernández et al., 2011a , b , 2013; Rodríguez-Gacio et al., 2012; Yan et al., 2014).
Brachypodium distachyon is being considered a model species for the genetics and molecular genomics of cereals, due in part to its small sequenced genome (~355 Mbp), short life cycle, self-fertility, diploidy and its close phylogenetic relationship with important crop plants of the tribe Triticeae within the Poaceae family, such as wheat and barley (International Brachypodium Initiative, 2010; Mochida and Shinozaki, 2013; Girin et al., 2014). In Poaceae seeds, the β-1,3-1,4-glucans are abundant in the endosperm cell walls (Burton and Fincher, 2009; Guillon et al., 2011) and genes encoding hydrolytic enzymes involved in their degradation, such as endo-β-1,3-glucanases and endo-β-1,3-1,4-glucanases, have been associated with rice post-germination events (2–4 days of imbibition; Akiyama et al., 2004) and with the elongation of barley coleoptiles (Takeda et al., 2010). Although mannan content is lower than glucan content in Brachypodium seeds (Rancour et al., 2012), the function of mannans and MANs may be relevant in its germinating seeds.
In this work, mannan polysaccharides were immunolocalized to the root and the coleorhiza of germinating seeds early in imbibition, decreased thereafter at later stages, and the enzymatic activity of endo-β-mannanases increased as germination progressed. The MAN gene family of B. distachyon was annotated and the expression of its six members explored in vegetative and reproductive organs. Interestingly, genes BdMAN2, BdMAN4 and BdMAN6 were clearly induced upon seed germination and mRNA in situ hybridization analyses demonstrated that these transcripts were found in the coleorhiza and the root during germination sensu stricto. BdMAN4 and BdMAN6 were also expressed in the aleurone layer, and may also be involved in post-germinative reserve mobilization.
Materials and Methods
Biological material, growth conditions and germination assays
The diploid inbred Brachypodium distachyon strain Bd21 (kindly provided by Prof. Garvin from the University of Minnesota, USA; International Brachypodium Initiative, 2010) was used in this work. Seeds were surface-sterilized with 1% NaOCl for 10min and washed in sterile water, before germinating on Petri dishes, containing two filter papers (Whatman 3) moistened with 8ml of sterile water, at 22ºC in the dark for 2 d. They were then transferred to pots in the greenhouse under long-day conditions (16h/8h, light/darkness; light intensity 155 µmol photons m−2 s−1) for sampling roots (6-week-old plants), young and old leaves (6- and 12-week-old plants) and spikes. For the germination experiments that lasted up to 42h, seeds were incubated in the dark at 22ºC, in Petri dishes with moistened filter papers, using triplicate lots of 25 non-stratified after-ripened seeds (stored at 22ºC and 30% relative humidity in the dark for 3 months). Seed samples were separated into embryo and endosperm (de-embryonated seeds) at 0, 12, 24, 30, 36 and 42h of imbibition, and used for RNA quantification and for protein extraction to determine MAN enzymatic activity.
Endo-β-1,4-mannanase (MAN) activity assays
Seed samples, obtained as described above, were homogenized in 100mM sodium acetate buffer (pH 4.5) containing 1M NaCl and 0.5% ascorbic acid, at 4ºC for 2h in an orbital shaker (VWR International Eurolab, Barcelona, Spain). The homogenates were centrifuged at 15,000 × g for 45min, and 80 μl of this supernatant was mixed with 150 μl of 0.25% mannan (1,4-β-D-mannan from carob; Megazyme International Ireland Ltd., Wicklow, Ireland). Incubation was at 30ºC for different periods of time and the enzymatic activity was determined by the increase in reducing sugar production per mg of protein, as determined by the 4-OH-Benzoic Acid Hydrazide (PAH-BAH; Sigma-Aldrich) method (Lever, 1977). The MAN from Aspergillus niger (Megazyme) was used to establish the control curve (one unit of MAN activity defined as the amount of enzyme that releases 1 nmol of reducing sugar per minute under the experimental conditions). Protein concentration was determined with the Bradford reagent (Bio-Rad Laboratories, Munich, Germany) using bovine serum albumin (BSA) as a standard.
Bioinformatic tools: BdMANs identification and phylogenetic analysis
The deduced protein sequences of the six MAN genes were obtained from the B. distachyon genome using the TBLASTN tool at the Phytozome v8.0 Database (Goodstein et al., 2012; www.phytozome.net), using the eight OsMAN proteins from the Oryza sativa genome as query sequences (Yuan et al., 2007). The Interpro Program (PFAM database; Bateman et al., 2002; http://pfam.sanger.ac.uk) was used to confirm the presence of the MAN conserved domain (glycosyl-hydrolase family 5). The complete amino acid sequences deduced from B. distachyon, O. sativa and Arabidopsis thaliana MAN genes were aligned by means of the CLUSTAL W program (Thompson et al., 1994) and utilized to construct a phylogenetic dendrogram, using the neighbor-joining algorithm, a bootstrap analysis with 1,000 replicates, complete deletion and the Jones Taylor Thornton matrix, as settings. The MEME program software version 4.0 (Tamura et al., 2007) was used to identify conserved motifs within the deduced MAN proteins and to validate the phylogenetic tree (Table 1). Default parameters were used with the following exceptions: the maximum number of motifs to find was set to 22 and the minimum width was set to eight amino-acid residues (Bailey et al., 2009; http://meme.sdsc.edu/meme4_6_0/intro.htlm). A single capital letter represents a single residue relative frequency, if this is greater than 50% than twice that of the second most frequent residue in the same position. If no single residue matches these criteria, a pair of residues, represented by capital letters in brackets, is given if the sum of their relative frequencies exceeds 75%. If none of these characteristics are satisfied, a lowercase letter is given when the relative frequency of a residue is greater than 40%, if not, x is set.
Table 1.
Conserved amino acid motifs obtained by means of MEME (Bailey et al., 2009) from the analysis of the endo-β-mannanase proteins of Brachypodium distachyon, Oryza sativa, and Arabidopsis thaliana
The conserved amino acids, critical for enzyme activity, are in bold and the signature sequence for endo-β-mannanase enzymes is underlined.
| Motif | E value | Consensus sequences |
|---|---|---|
| 1 | 5.6e-1050 | L[TS]S[DN]D[DS]FF[TS][DN]PT[IV][KR][DS][YF][YF] KN[HY]VK[AT]VLTR[VK]NT[LV]TG[VI]AY[KR]D[DE] PTI[FL] AWEL[MI]NEPRCxSDP[ST]GDTLQAW[IV] EEMAAYVKS[ILV]DP[NK]H |
| 2 | 6.5e-718 | DGGY[RN][AP]LQI[SA]PG[VR][YF][DN]ED[VM]F[QK] [GA]LDFV[IVL][AS]EA[RK][RK]HG[IV][RK]L[IL]L[SC] LVNN [WL][DE][DA][YF]GGK[AK]QYVRWA |
| 3 | 1.40E-307 | x[LY]GTDF[IV][AR]N[HNS]Q[VA]PGIDFA[ST][VI]H[SV] YPDxW[LF]P |
| 4 | 7.90E-213 | NGRPFY[VAI]NG[FW]N[AST]YWLMxxA[ASV]DP[AS]T |
| 5 | 2.80E-202 | [KRN]W[ML][DQ][AS]H[IV]ED[AG][AEQ][NA][IE]L[GKR] [KM]P[LVI]L[VFIL][TAG]EFG[KL]S |
| 6 | 3.40E-199 | WKxPG[YF][NST]T[AS]QRDA[FL][LFY]R[AT]VYD[KA] IYASAR[RK]GG[APS][GA][AV]G[AG]L[FV]WQ[LV]L |
| 7 | 2.10E-172 | [TS][AE][AMV][FL][RQ]QA[AS]A[MH]GL[TN]V[CA] RTWAF[SN] |
| 8 | 2.20E-161 | L[LV][ET][VI]GLEGFYGP[SG]SPER[KL]xVN |
| 9 | 2.50E-146 | GM[ED]x[YFM]DDG[YF][ESA][IV][VI][LF][AS]ES[PS] STAS[IL][IL]x[EN][HQ]S[RC] |
| 10 | 2.90E-64 | [PAG][EGS][DGW]G[FM]V[RE]R[NR]GT[QR][FL]V[LV] |
| 11 | 8.20E-37 | KDGKF[GD][NS][EG]FRE[DT]FM[KE]T[VI]Y[RN][IN] FLSSW[KE][EG]GVIGGGCLLWQLFP |
| 12 | 4.40E-11 | [YM][KHS][ILC]L[GC][FL][LAF][LSV][LC]LA[IVF][VI][YI] [LAF][SQ][FLSW] |
| 13 | 4.40E-05 | FNS[LR]C[AS]W[RS]CRWGC[KN]K |
| 14 | 1.00E-02 | [HL][KY][GK]EGDPGWQC[ST]IPP |
| 15 | 2.60E+01 | [CR]F[IV]SLSRSISSFI[QV][DQ]NF |
| 16 | 3.70E+01 | [HR]L[KL][DE][KQ]K[LS][IK]E[LM]CSHR[HP] |
| 17 | 1.50E+02 | QLA[AES]L[ND]G[QK][DF][AD][DE][AGV][LV][RC] RR[RA][RS][RS] |
| 18 | 1.00E+03 | [AG][AG][GP]GG[GW][LV]KLPVPWLQ |
| 19 | 4.30E+04 | KAFARA[ER][QR][AE][QR][PS]ARGKG |
| 20 | 6.60E+04 | [AI]VL[CH][ES][AS][SV][FY][IW][EI][LW][RT][CQ]NR |
| 21 | 2.70E+05 | SS[HP]RK[IT][GR][LS][GT]SGG[DS][SW]D |
| 22 | 3.00E+05 | L[GH][AH][GH][AR][AV]ALLVL[AL]CV[HV] |
The major biochemical parameters of the deduced MAN proteins from B. distachyon and O. sativa are listed in Supplementary Table S1. Both isoeletric point (pI) and molecular weight (MW) were predicted using the Compute pI/MW tool (Gasteiger et al., 2005; http://www.expasy.ch/tools/pi_tool.html) and the putative signal peptide cleavage site and sub-cellular localization were deduced by the SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP) and TargetP 1.1 tools (http://www.cbs.dtu.dk/services/TargetP/), respectively (Emanuelsson et al., 2007).
Real time quantitative PCR (RT-qPCR) analyses
Total RNA was purified from roots (6-week-old plants), young and old leaves (6- and 12–week-old plants) and spikes by the phenol/chloroform method (Lagrimini et al., 1987). For the isolation of RNA from seeds at different stages of development (0–10 d after pollination: dap) and at different time points of germination (12, 24, 30, 36 and 42h), the protocol described by Oñate-Sánchez and Vicente-Carbajosa (2008) was followed. RNA samples were treated with DNAse I, RNAse-free (Roche Applied Science, Manheim, Germany) to avoid genomic DNA contamination. First-strand cDNA was synthesized with random hexamers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manusfacturer’s recommendations. Samples were stored at −20ºC until used.
PCR-amplification was performed in an Eco Real-Time PCR System (Illumina, San Diego, CA, USA). For each 10 µl reaction, 2 µl of DNA sample was mixed with 5 µl of FastStart SYBR Green Master (Roche Applied Science) and 0.25 µl of each primer (final concentration 500nM) plus sterile water up to final volume. Samples were subjected to thermal-cycling conditions of 95ºC for 10min and 40 cycles of 10 s at 95ºC and 30 s at 60ºC for annealing and extension, respectively. The melting curve was designed to increase from 55ºC to 95ºC, and the melting temperatures for each amplicon and primer efficiencies (Supplementary Table S2) were estimated using a calibration dilution curve and slope calculation (E=10(−1/slope)). The specific primers used are shown in Supplementary Table S2 and they were designed on the 3′-non-coding region using the Primer3Plus program (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The BdGAPDH gene (encoding glyceraldehyde 3-phospate dehydrogenase; Hong et al., 2008) was used to normalize the data, since the expression of this gene was previously demonstrated to be constant throughout the period studied (Hernando-Amado et al., 2012; González-Calle et al., 2014; Supplementary Fig. S1). Expression levels were calculated as the number of cycles needed for the amplification to reach a cycle threshold fixed in the exponential phase of the PCR (Ct; Pfaffl, 2001). All analyses used three different biological replicates for each time-point and each one was made in triplicate. Means ± standard error (SE) of three independent experiments are indicated in the corresponding figures.
Preparation of embedded material for microscopy
Samples were treated according to a modified version of the protocol described in Ferrandiz et al. (2000). After-ripened dry seeds and germinating seeds of B. distachyon (12h, 27h and 36h) were collected and, after removal of the lemma and palea, were infiltrated with the FAE solution (formaldehyde: acetic acid: ethanol: water, 3.5:5:50:41.5 by volume) for 40min in 25mm Hg vacuum; the seeds were then incubated at 4ºC for 3 d with gentle shaking. The samples were dehydrated through a graded series of aqueous ethanol mixtures and progressively embedded in paraffin after the replacement of ethanol with HistoClear (National Diagnostics, Hessle Hull, England). Thin sections of 8 µm were collected on glass slides and de-waxed.
Heteromannan immunolocalization
The protocol used was a modification of those described in Marcus et al. (2010) and Guillon et al. (2011). In a pre-immunolabelling step, sections of embedded material as described above, were incubated in phosphate buffer sodium solution (PBS) and treated with 1mg/ml proteinase-K (Roche Applied Science). In order for the specific antibodies to have access to the heteromannans (mannans, glucomannans, and galactomannans) of the cell walls, β-1,3-1,4- glucans were removed by incubating the sections with a solution of 4 µg/ml lichenase [β-1,3-1,4- glucanase; Megazyme] for 2h at 37ºC, and then rinsed with de-ionized water. For heteromannan immunodetection, sections were first incubated at room temperature for 30min in a blocking solution (3% BSA, 1× PBS, and 5mM sodium azide; pH7), and then treated with primary anti-heteromannan antibody LM21 (PlantProbes, Leeds, UK) at a dilution of 1:5 in the same blocking solution but only containing 1% BSA for 2h. Sections were thoroughly washed in PBS containing 5mM sodium azide and then incubated for 2h in the same buffer containing the secondary rabbit antibody Anti-Rat IgG-FITC (Sigma-Aldrich) at a dilution of 1:100. The sections were extensively washed in PBS buffer and in water, mounted and examined in a confocal microscope (absorption 494nm; emission 521nm; Leica TCS-SP8, Leica, Wetzlar, Germany).
mRNA in situ hybridization analyses
Pre-hybridization was carried out by incubating the sections in 0.2M HCl, neutralizing them and then treating them with 1mg/ml proteinase-K (Roche Applied Science). Samples were then dehydrated in an aqueous ethanol dilution series and hybridized with sense and anti-sense digoxigenin (DIG)-labelled RNA probes, corresponding to DNA fragments (200–300bp) derived from the 3′-non coding regions of the BdMAN2, BdMAN4 and BdMAN6 genes (Supplementary Table S3), synthesized with the DIG RNA labelling mix according to the manufacturer’s specifications (Roche Applied Science). Probes were hybridized at 52ºC overnight followed by two washes in 2× SSC (150mM NaCl, 15mM Na3-citrate) and 50% formamide for 90min at the same temperature. Incubation with the alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Applied Science) and colour detection was carried out according to the manufacturer’s instructions (Ferrandiz et al., 2000). Sections were dried and examined on a Zeiss Axiophot Microscope (Carl Zeiss, Oberkochen, Germany), and images were captured and processed with the Leica Application Suite 2.8.1 build software (Leica).
Protein and polysaccharide histological determinations
B. distachyon dry and germinating seeds were stained with 5% (w/v) toluidine blue (Merck, Darmstadt, Germany) for checking tissue integrity (Fig. 1A). Samples were stained with PAS reagent (0.5% w/v periodic acid-Schiff reagent) (Merck) to detect polysaccharides and with 1% (w/v) Naphthol Blue Black (Sigma-Aldrich) for proteins (Iglesias-Fernández and Matilla, 2010). Visualization was done on a Zeiss Axiophot Microscope (Carl Zeiss) and the images were captured and processed with the Leica Application Suite 2.8.1 build software (Leica).
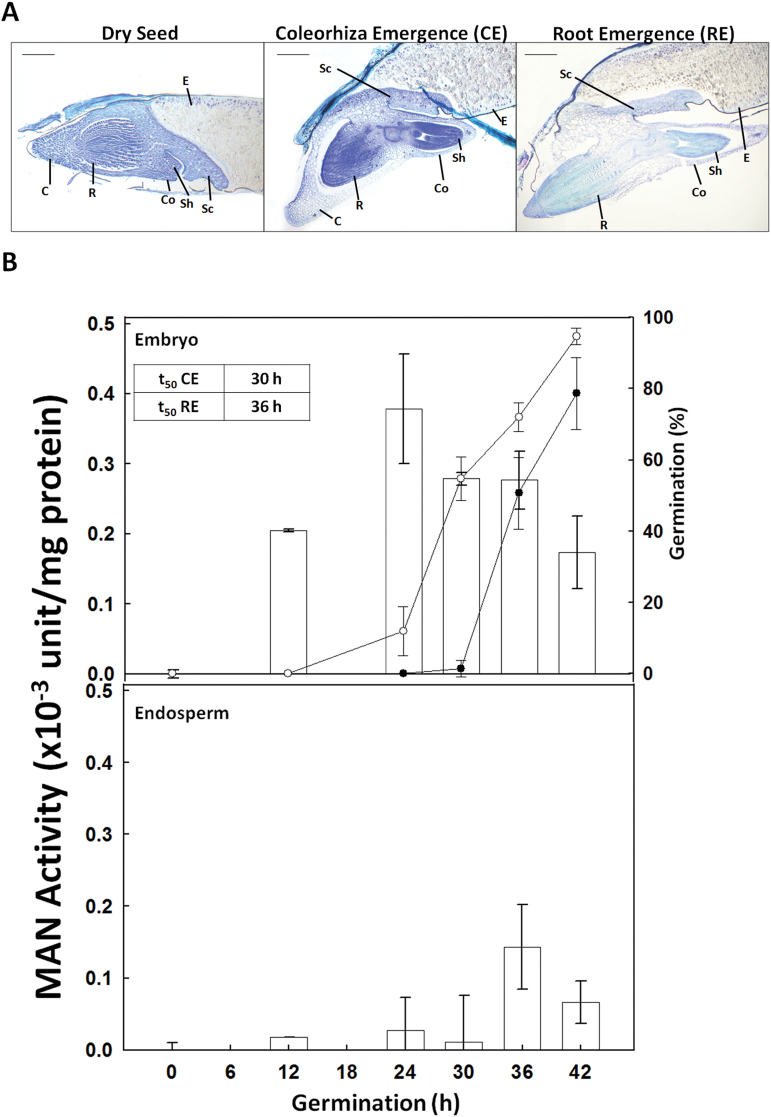
Fig. 1.
(A) Longitudinal sections of the different phases of Brachypodium distachyon germination sensu stricto, stained with toluidine blue. C, coleorhiza; Co, coleoptile; E, endosperm; Sc, scutellum; Sh, shoot; R, root. Scale bar, 200 μm. (B) Endo-β-mannanase activity (white bars) in embryo and endosperm (de-embryonated seed) upon B. distachyon seed germination (0–24h). One unit of MAN activity is defined as the amount of enzyme that releases 1 nmol of reducing sugar per minute and per mg of protein. Percentage germination evaluated as coleorhiza emergence (CE; open circles) and root emergence (RE; close circles) are represented. In the inset, the time needed for 50% of CE (t50CE) and RE (t50RE) is indicated. Data are means ± standard error (SE) of three technical replicates of three biological samples.
Results
Enzymatic β-mannanase activity during Brachypodium seed germination
The time course of sensu stricto germination of B. distachyon seeds occurs in two different steps: first, the coleorhiza emerges (CE), and in a second step, the root emergence (RE) takes place (Fig. 1A). The enzymatic activity of MAN upon germination has been analysed separately in the embryo and in the de-embryonated seed (endosperm). As shown in Fig. 1B, dried seeds have no detectable MAN activity, but this progressively increases with germination, peaking at 24h in embryos, containing the coleorhiza (~0.4×10–3 units/mg protein), and decreasing to half this value at 42h (~0.2×10–3 units/mg protein). In endosperms, MAN activity is much lower than in embryos, and reaches its maximum level (~0.15×10–3 units/mg protein) at 36h of germination. Data from Fig. 1B indicate that MAN activity is maximum in embryos, just before reaching 50% of germination sensu stricto (t50CE=30h; t50RE=36h), suggesting that MAN is important for facilitating both coleorhiza and root emergence.
Heteromannans are preferentially localized to the root tip and the coleorhiza in germinating seed embryos
Mannan polymers have been detected in longitudinal sections of B. distachyon germinating seeds (at 12 and 27h of imbibition) by in situ immunofluorescence labelling, using the LM21 antibody that specifically recognizes mannan polysaccharides (gluco- and galacto-mannans). To facilitate accession of the antibody to mannans in plant CWs, the seed sections have been previously treated with lichenase (β-1,3-1,4- glucanase; Marcus et al., 2010).
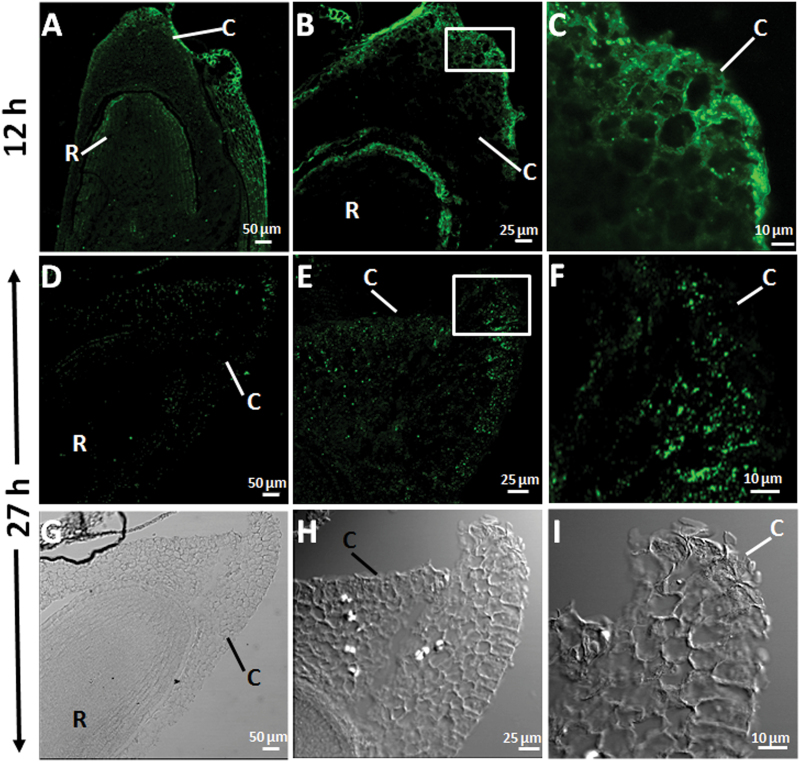
As shown in Fig. 2, at 12h of seed imbibition, seed mannan polymers are mainly localized to the periphery cells of the coleorhiza (C) and to the epidermis of the root tip (R) (Fig. 2A–C). Interestingly, these mannans are barely detected at later stages of germination (27h of imbibition; Fig. 2D–F). Differential interference contrast (DIC) images are shown in Fig. 2G–I. This observation together with data of MAN enzymatic activity (Fig. 1B) with a maximum at 24h in embryos, may suggest that the disappearance of the mannan polymers is due to the hydrolysis catalysed by endo-β-mannanases.
Fig. 2.
Mannan polymer immunolocalization at the root tip and the coleorhiza in longitudinal sections of Brachypodium germinating embryos at (A–C) 12h and at (D–F) 27h. (G–I) DIC images of D, E and F. (C, F, I) Close-up of the coleorhizae. C, coleorhiza; R, root. Scale bars: (A, D, G), 50 μm; (B, E, H), 25 μm; (C, F, I), 10 μm.
The Brachypodium endo-β-mannanase gene family
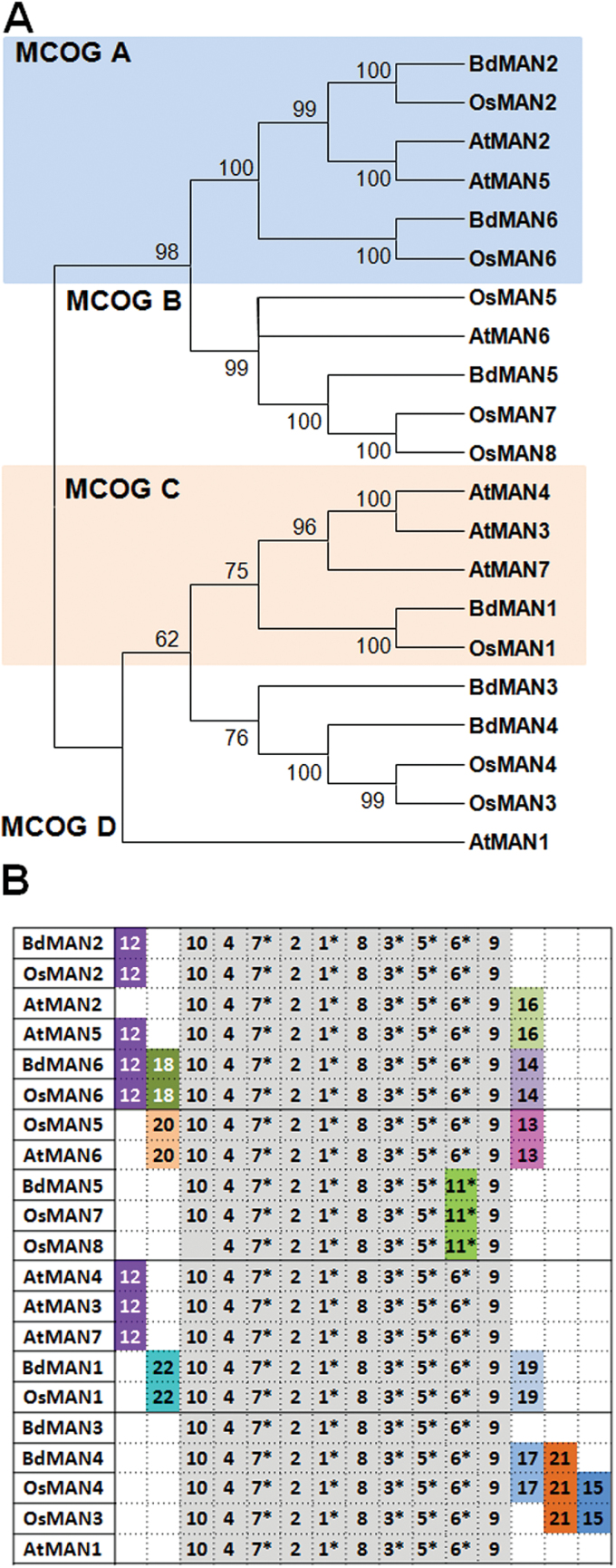
In order to get a deeper insight into the MAN function upon B. distachyon germination, it was decided to annotate and characterize further the BdMAN family. The already described MAN family from O. sativa (Yuan et al., 2007) has been used to perform a TBLASTN against the whole Brachypodium genome (http://www.phytozome.net). Six predicted non-redundant MAN deduced proteins, with MW 43–52 KDa, and Ip 4.4–8.8, three of them with predicted signal peptides, have been identified and named according to their orthologues in rice (Supplementary Table S1). The MAN protein sequences from A. thaliana (AtMAN1-7) and O. sativa (OsMAN1-8) together with those from B. distachyon (BdMAN1-6) have been used to construct a phylogenetic unrooted tree by using the neighbor-joining algorithm. Four major clusters of orthologous groups (MCOGs) have been defined (A, B, C, D), supported by bootstrapping values higher than 62% (Fig. 3A) and by the occurrence of common motifs (Fig. 3B; MEME).
Fig. 3.
(A) Phylogenetic dendrogram with deduced protein sequence of the mannanase gene families form Brachypodium distachyon, Oryza sativa and Arabidopsis thaliana; bootstrapping values are indicated in the branches. (B) Schematic distribution of conserved motifs among the deduced protein sequences in the phylogenetic tree (A), identified by means of the MEME analysis. Asterisks indicate those motifs important for enzymatic activity. Motifs in grey share >85% of similar amino acid residues. This figure is available in colour at JXB online.
The search for conserved amino-acid motifs using the MEME software (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) reveals that all MAN sequences have in common motifs described as critical for the enzymatic activity, such as 1, 3, 5, 6/11 and 7 (Fig. 3B, Table 1). The deduced signature sequence [AWEL(MI)NEPRC] of Arabidopsis and rice MANs (Yuan et al., 2007), included in motif 1, is also present in Brachypodium MAN. Besides, members in MCOG A, BdMAN2, OsMAN2, BdMAN6 and OsMAN6 share motif 12, and BdMAN6 and OsMAN6 also share motifs14 and 18. In MCOG C, BdMAN1 shares with OsMAN1 motifs 19 and 22, and in MCOG D BdMAN4 shares motifs 17 and 21 with OsMAN4, but lacks motif 15 shared by the rice paralogues OsMAN3 and OsMAN4. Similarly, BdMAN5, OsMAN7 and OsMAN8 (MCOG B) have in common motif 11, but they do not share with OsMAN5 motifs 20 and 13. The MAN protein motifs from A. thaliana have been included for comparison (Iglesias-Fernández et al., 2011a ).
Expression kinetics of selected BdMAN genes during seed maturation and germination
The expression pattern of the six BdMAN genes has been explored by RT-qPCR analysis in different organs: young (6 d) and old (12 d) leaves, roots (6 d) and spikes (mix of different stages; Supplementary Fig. S2). While BdMAN1, 2, 3, 5 genes are not detected in leaves, BdMAN4 gene expression in old leaves is ~10 times lower than in young leaves, and BdMAN6 has the same low expression in young and old leaves. In roots, BdMAN2, BdMAN4 and BdMAN6 are expressed at low levels, and BdMAN1, BdMAN2, BdMAN4 and BdMAN6 transcripts are detected in spikes (Supplementary Fig. S2).
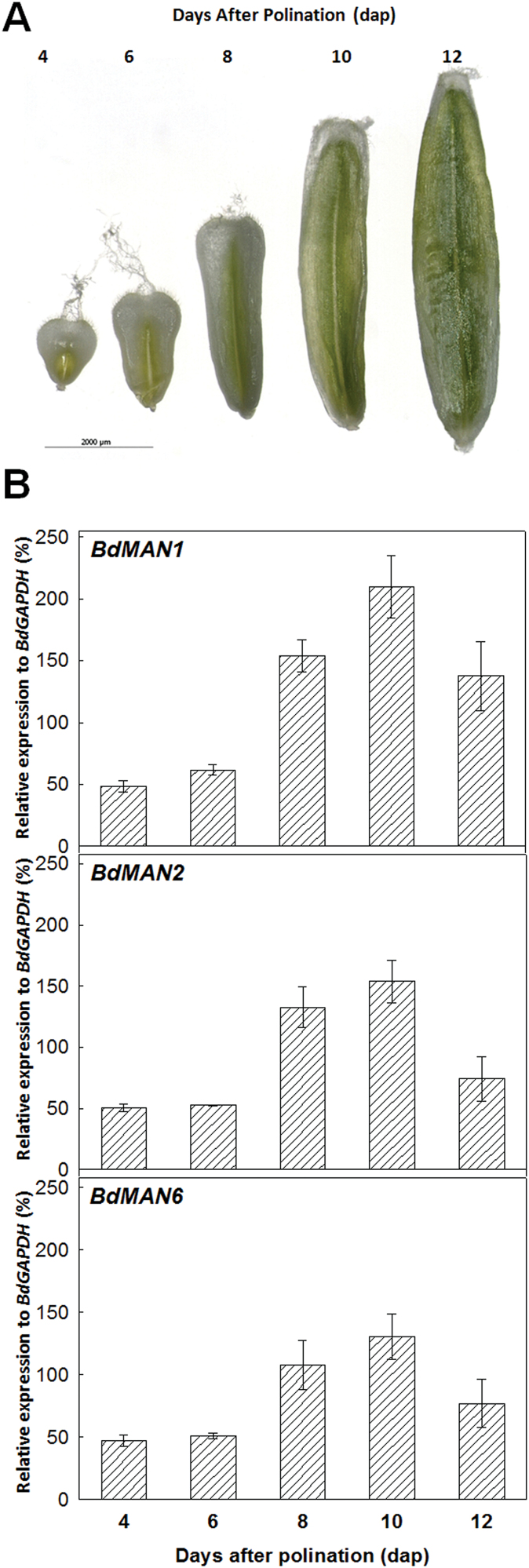
Since our preliminary data indicate that BdMAN1, BdMAN2 and BdMAN6 transcripts are abundant in developing seeds (see Supplementary Fig. S3A), the expression kinetics of these three genes has been established throughout seed maturation, at 4, 6, 8, 10 and 12 d after pollination (dap) (Fig. 4A). Although the gene BdMAN1 is the most highly expressed during the late phases of seed development (8, 10, 12 dap), the expression patterns of BdMAN1, BdMAN2 and BdMAN6 show a progressive increase from 4 to 10 dap, reaching all of them their maximum expression at 10 dap when maturation is almost completed (Fig. 4B).
Fig. 4.
(A) Different stages of Brachypodium distachyon seed development (4, 6, 8, 10, 12 d after pollination; dap). (B) Expression of the BdMAN1, BdMAN2 and BdMAN6 genes by RT-qPCR during seed development. Data are means ± standard error (SE) of three technical replicates of three biological samples.
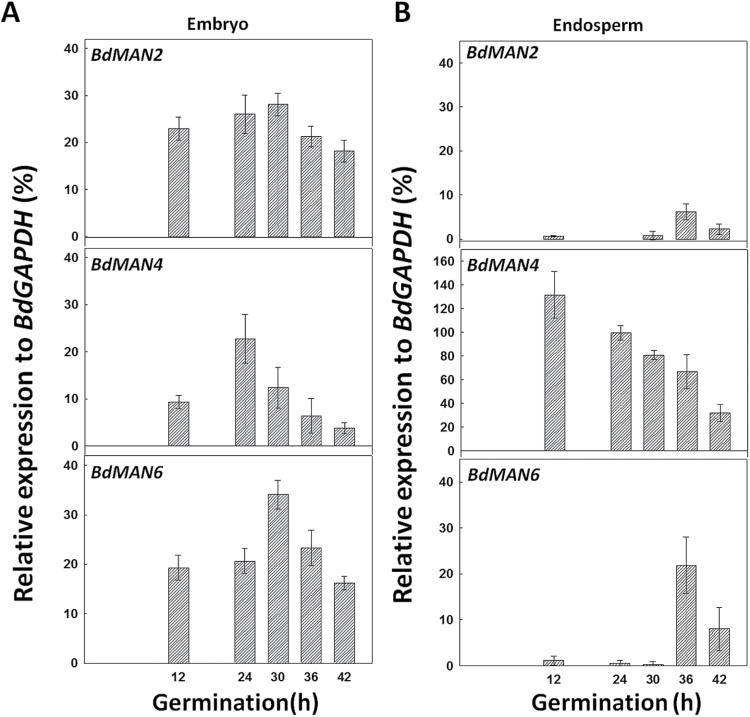
Data in Supplementary Fig. S3B indicate that genes BdMAN2, BdMAN4 and BdMAN6 are the most abundantly expressed ones during seed germination, and their expression kinetics has been more thoroughly analysed (Fig. 5). Germinating seeds taken at 12, 24, 30, 36 and 42h of imbibition, have been sectioned into embryos and de-embryonated seeds (endosperm). In germinating embryos, BdMAN2, BdMAN4 and BdMAN6 transcripts appear early upon imbibition (12h), before CE, and their maximum expression is attained between 24–30h (t50CE=30h) and it decreases as germination progresses (42h; Fig. 5A). However, the expression of BdMAN4 in endosperms is high at early imbibition times (12h; ~140% relative to BdGAPDH), decreasing thereafter. The BdMAN2 and BdMAN6 transcripts have low expression in the endosperms of germinating seeds with a maximum at 36h of imbibition (<10 % for BdMAN2 and ~25 % for BdMAN6 relative to BdGAPDH, respectively), indicating a possible role in reserve mobilization for these MAN genes, and perhaps also for BdMAN4 post-germination (Fig. 5B).
Fig. 5.
Transcript accumulation of BdMAN2, BdMAN4 and BdMAN6 in (A) embryos, and in (B) endosperms (de-embryonated seeds) upon B. distachyon seed germination. Data are means ± standard error (SE) of three technical replicates of three biological samples.
BdMAN2, BdMAN4 and BdMAN6 transcripts are localized to different seed tissues during B. distachyon seed germination
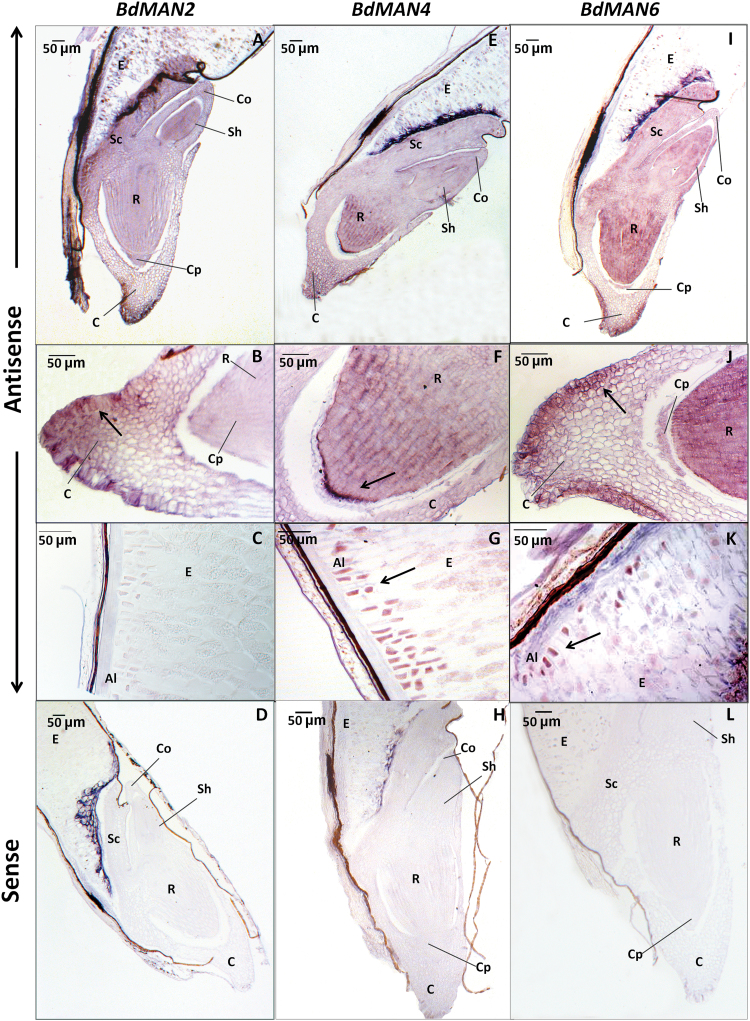
To determine the spatial expression of MAN genes within the B. distachyon germinating seeds, mRNA in situ hybridization experiments have been done (Fig. 6). Longitudinal sections of seeds at 27h of imbibition have been hybridized to specific antisense and sense (as negative controls) probes for BdMAN2, BdMAN4 and BdMAN6. The BdMAN2 transcripts are mainly expressed in the periphery cells of the coleorhiza and are not detected in the aleurone layer (Fig. 6A–D). BdMAN4 mRNA is localized preferentially to the tip and the apical meristem of the root, to the coleorhiza and to the aleurone layer (Fig. 6E–H), and the BdMAN6 transcripts are detected not only at the coleorhiza, but also throughout the embryo and faintly also at the aleurone layer (Fig. 6I–L). BdMAN6 transcripts are detected in the aleurone layer at 27h of imbibition by mRNA in situ hybridization, but they are scarcely detected in 24–30h germinating endosperms by RTqPCR (Fig. 5B), indicating that its expression could be diluted by the remaining endosperm during the RNA isolation process. As expected, no signal has been detected when sections have been hybridized with the corresponding sense probes for BdMAN2, BdMAN4 and BdMAN6 (negative controls; Figs 6D, 6H and 6L).
Fig. 6.
In situ mRNA hybridization analysis of BdMAN2, BdMAN4 and BdMAN6 in 27h germinating Brachypodium seeds. (A–D) BdMAN2, (E–H) BdMAN4, (I–L) BdMAN6. (A, E, I) Longitudinal sections of germinating embryos. (B, F, J) Close-up of the coleorhiza and the root tip. (C, G, K) Close-up of the endosperm and the aleurone. (D, H, L) Control sense probes. Al, aleurone layer; C, coleorhiza; Co, coleoptile; Cp, calyptra; E, endosperm; Sc, scutellum; Sh, shoot; R, root. The black arrow indicates the localization of transcripts. Scale bar, 50 μm.
Other histological observations of germinating B. distachyon seeds
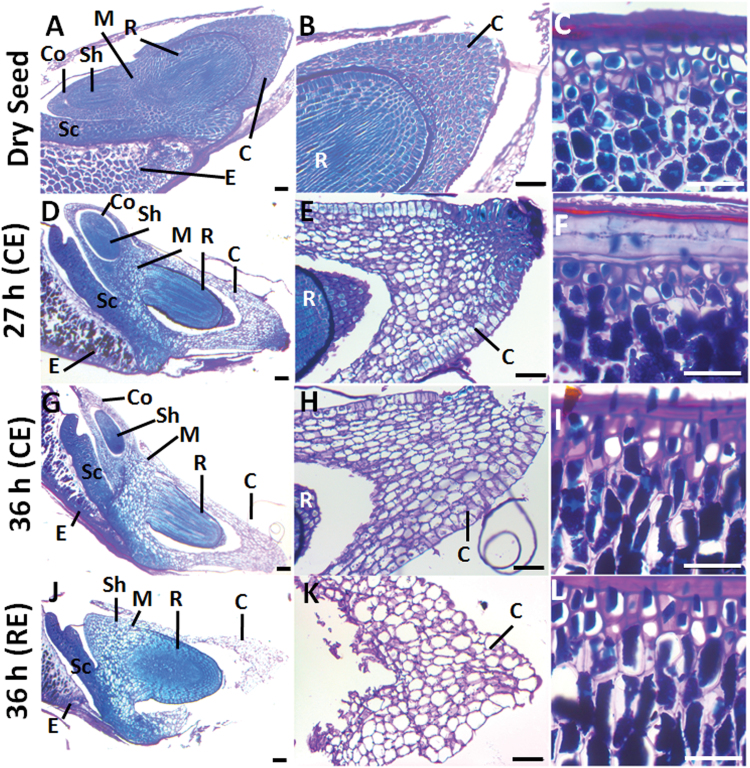
Longitudinal sections of B. distachyon seeds (dry and water imbibed at 27 and 36h; including seeds before and after root emergence) have been stained with PAS reagent to detect insoluble polysaccharides (mainly cellulose and starch) and with Naphthol Blue Black for proteins. As shown in Fig. 7A–C, dry seeds have abundant protein bodies (PBs; blue stained) and thick cell walls (CWs; pink stained) in the root, coleorhiza and endosperm cells. When the coleorhiza emerges (27h of imbibition) the PBs in the coleorhiza (C) and in the mesocotyl (M) cells start to hydrolyze, while the endosperm cells are full of reserves at this stage (Fig. 7D–F). After 36h, when the seed coat ruptures but before root emergence (RE), the coleorhiza cells start to elongate and its protein bodies (PBs), as well as, those of the mesocotyl (M) are almost completely consumed, while those of the coleoptile (Co) initiate their degradation (Fig. 7G–I) and those of the endosperm remain as in the dry seeds (Fig. 7C, 7I). Finally, when root emergence (RE) takes place, the lateral part of the coleorhiza breaks (≥36h of imbibition) and its PBs are fully degraded, while those in the endosperm cells are almost intact (Fig. 7J–L).
Fig. 7.
Polysaccharide and protein mobilization upon B. distachyon seed germination. Bright field microscopy of longitudinal seed sections stained with PAS-Naphthol Blue Black. (A, D, G, J) Longitudinal sections from dry and water-imbibed seeds at 27h and 36h. (B, E, H, K) Close-up of the coleorhiza in A, D, G, J and (C, F, I, L) close-up of the endosperm in A, D, G, J, respectively. Proteins stain in blue and polysaccharide-rich cell walls in pink. C, coleorhiza; Co, coleoptile; E, endosperm; M, mesocotile; Sc, scutellum; Sh, shoot; R, root. Scale bar: 50 μm.
Discussion
In this work, mannans and endo-β-mannanases (MAN) in Brachypodium distachyon have been investigated in order to establish whether they are important in the germination of these monocotyledonous seeds. Mannans have been immunolocalized in the embryo root and the coleorhiza in the early stages of germination and these polymers decrease upon imbibition while the enzymatic activity of MAN increases. The MAN gene family in B. distachyon has been annotated and the gene expression of the six members of this family has been explored in different vegetative and reproductive organs, and, more specifically, in germinating seeds. Three of these genes, BdMAN2, BdMAN4 and BdMAN6, are highly induced in germinating embryos and their transcripts are localized to the coleorhiza and the root, and BdMAN4 and BdMAN6 appear also in the aleurone layer. These facts indicate that the BdMAN enzymes should be spatially distributed in the seed in the vicinity of their putative substrates, thus contributing to the mannan hydrolysis and to the loosening of the coleorhiza cell walls, thereby facilitating root protrusion (germination sensu stricto).
During seed development, BdMAN1, BdMAN2 and BdMAN6 genes are expressed, and their mRNAs are abundant at the middle and late maturation stages. Upon cereal seed maturation, several tissues undergo a progressive enlargement of their cells, a process that involves nutrient remobilization and CW softening to allow cell expansion; to this aim, the participation of a complex set of hydrolytic enzymes have been described (Domínguez and Cejudo, 2014). These data indicate that the BdMAN1, BdMAN2 and BdMAN6 proteins could contribute to such a process during Brachypodium seed development.
The enzymatic analysis in the embryos of germinating seeds shows a maximum of MAN activity at 24h of imbibition, just before the coleorhiza emergence (CE50=30h), and this enzymatic activity progressively decreases to 50% at 42h. However, MAN activity in de-embryonated seeds (endosperms) is low, with a maximum at 36h of imbibition, when germination sensu stricto is almost completed [~100% coleorhiza emergence (CE); ~80% root emergence (RE) at 42 h]. These data point out to a more important role of the MAN activity in the embryo than in the endosperm during germination sensu stricto. In rice, MAN activity and expression of the OsMAN1, OsMAN2 and OsMAN6 genes have been detected in the aleurone layer only after 48h of imbibition when 100% of germination has been achieved. This MAN activity is associated with reserve mobilization, a clear post-germinative event (Ren et al., 2008). In barley, the HvMAN1 enzyme has been purified from 10-day-old seedlings and its catalytic parameters established, although its physiological role has not been investigated (Hrmova et al., 2006).
The function of the coleorhiza tissue in the grasses has been classically associated to a protective function of the growing root during germination, but other physiological functions are being uncovered, such as our observations of the hydrolysis of proteins (disappearance of PBs) or the decrease in mannan content detected within the coleorhiza cells during Brachypodium germination sensu stricto. Nowadays, and similarly to what has been proposed for the endosperm of eudicot seeds (Piskurewicz et al., 2009), the coleorhiza is being considered to be a key tissue preventing root emergence in dormant barley seeds (Millar et al., 2006; Barrero et al., 2009). These authors have hypothesized that root emergence may not depend only on the softening of the coleorhiza, driven by CW remodelling enzymes, but also by the expansive force of the imbibing root cells. Important transcriptional changes in the barley coleorhiza associated to the dormancy degree have been found and these differences affect mainly the expression of CW modifying genes (mannanases among them), nitrate and nitrite reductase genes etc. (Barrero et al., 2009). The cytosolic nitrate reductase is an important source of the hormone nitric oxide (NO) that is involved in promoting seed germination (Arc et al., 2013). Therefore, the coexistence at the coleorhiza of NO and mannanases, and perharps proteases of the CatepsinB3 type (Iglesias-Fernández et al., 2014), should have an influence in the seed germination of the grasses.
The Arabidopsis radicle tip has been described as the primary location of growth-promoting genes and its surrounding-cells the centre for CW expansion (Bassel et al., 2014). Moreover, a dual enzymatic activity for MAN (hydrolase and transglycosylase activities) has been described; the transglycosylase activity being more related to cell expansion, as occurs in the radicle before protrusion, and the hydrolytic activity could be relevant for weakening of the CWs of the embryo-surrounding tissues (Schröder et al., 2009; Iglesias-Fernández et al., 2011a , b). It is remarkable that the BdMAN4 transcripts are localized to the aleurone layer during imbibition (27h), when practically no MAN activity is detected in the de-embryonated (endosperm) seed, suggesting a possible accumulation of these transcripts and their corresponding proteins as inactive forms in the aleurone cells. Interestingly, the BdMAN4 deduced protein sequence has a predicted signal peptide for the secretory pathway and it is possible that the BdMAN4 isozyme could be transported later on during post-germinative reserve mobilization from the aleurone to the endosperm cells through the apoplastic space. In Arabidopsis the AtMAN7 and in poplar the PtrMAN6 proteins also contain signal peptides and have been localized to the apoplast, indicating that the mature MANs could be mobilized to the outer space (Iglesias-Fernández et al., 2013; Zhao et al., 2013).
Several authors have proposed that polysaccharides (β-1,3-1,4-glucans, mannans and others) present at the endosperm CWs of the Poaceae grains, not only have a structural function, but also a storage role (Guillón et al., 2012). Heteromannans, although globally less abundant in these seeds than the β-glucans, are concentrated not only in the coleorhiza and in the root but also these polymers are found in the aleurone layer and storage endosperm of B. distachyon (Guillón et al., 2011). In this context, our data demonstrate that MAN activity is important for the weakening of the coleorhiza cell walls and for the expansion of the root cells, thus facilitating germination sensu stricto, but it may be also important for the mannan hydrolysis in endosperm during post-germinative reserve mobilization.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Transcription levels of the housekeeping (BdGAPDH gene), presented as Ct mean values, in different organs, in developing seeds and during seed germination of B. distachyon.
Supplementary Fig. S2. Transcripts analysis by RTqPCR of the BdMAN1-6 genes in different organs.
Supplementary Fig. S3. Expression analysis by RT-qPCR of BdMAN1-6 genes in developing seeds and germinating seeds.
Supplementary Table S1. Major biochemical characteristics of Brachypodium distachyon and Oryza sativa endo-β-mannanase proteins.
Supplementary Table S2. Oligonucleotide sequences, amplicon length and PCR efficiency of the primers used for RT-qPCR analyses.
Supplementary Table S3. Primers used for the synthesis of the in situ mRNA hybridization probes.
Acknowledgments
Financial support from MINECO, Spain (Project BFU2009-11809; Principal Investigator PC) is gratefully acknowledged. We thank Prof. AJ Matilla (Universidad de Santiago de Compostela, USC, Spain) for critical reading of the manuscript. VG-C is the recipient of a pre-doctoral contract from Universidad Politécnica de Madrid (Spain).
References
- Akiyama T, Pillai MA, Sentoku N. 2004. Cloning, characterization and expression of OsGLN2, a rice endo-1,3-beta-glucanase gene regulated developmentally in flowers and hormonally in germinating seeds. Planta 220, 129–139. [DOI] [PubMed] [Google Scholar]
- Arc E, Galland M, Godin B, Cueff G, Rajjou L. 2013. Nitric oxide implication in the control of seed dormancy and germination. Frontiers in Plant Science 4, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motifs discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Jacobsen JV, Talbot MJ, White RG, Swain SM, Garvin DF, Gubler F. 2012. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytologist 93, 376–386. [DOI] [PubMed] [Google Scholar]
- Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F. 2009. Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiology 150, 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Stamm P, Mosca G, Barbier de Reuille P, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, Smith RS. 2014. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proceedings of the National Academy of Sciences USA 111, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. 2002. The Pfam protein families database. Nucleic Acids Research 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Fincher GB. 2009. (1,3;1,4)-beta-D-glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages. Molecular Plant 2, 873–882. [DOI] [PubMed] [Google Scholar]
- Carrillo-Barral N, Matilla AJ, Rodríguez-Gacio Mdel C, Iglesias-Fernández R. 2014. Nitrate affects sensu-stricto germination of after-ripened Sisymbrium officinale seeds by modifying expression of SoNCED5, SoCYP707A2 and SoGA3ox2 genes. Plant Science 217–218, 99–108. [DOI] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ. 2014. Programmed cell death (PCD): an essential process of cereal seed development and germination. Frontiers in Plant Science 5, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunnak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 643–652. [DOI] [PubMed] [Google Scholar]
- Endo A, Tatematsu K, Hanada K, Duermeyer L, Okamoto M, Yonekura-Sakakibara K, Saito K, Toyoda T, Kawakami N, Kamiya Y, Seki M, Nambara E. 2012. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiology 53, 16–27. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Liljegren S, Yanofsky M. 2000. FRUITFULL negatively regulates the SHATTERPROOF genes during Arabidopsis fruit development. Science 289, 436–438. [DOI] [PubMed] [Google Scholar]
- Gao F, Ayele BT. 2014. Functional genomics of seed dormancy in wheat: advances and prospects. Frontiers in Plant Science 5, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server. In: Walker JK, ed. The Proteomics Protocols Handbook. New York: Humana Press, 571–607. [Google Scholar]
- Girin T, David LC, Chardin C, Sibout R, Krapp A, Ferrario-Méry S, Daniel-Vedele F. 2014. Brachypodium: a promising hub between model species and cereals. Journal of Experimental Botany 65, 5683–5696. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. 2012. Phytozome: a comparartive platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Calle V, Iglesias-Fernández R, Carbonero P, Barrero-Sicilia C. 2014. The BdGAMYB protein from Brachypodium distachyon interacts with BdDOF24 and regulates transcription of the BdCathB gene upon seed germination. Planta 240, 539–552. [DOI] [PubMed] [Google Scholar]
- Guillon F, Bouchet B, Jamme F, Robert P, Quemener B, Barron C, Larre C, Dumas P, Saulnier L. 2011. Brachypodium distachyon grain: characterization of endosperm cell walls. Journal of Experimental Botany 62, 1001–1015. [DOI] [PubMed] [Google Scholar]
- Guillon F, Larré C, Petipas F, Berger A, Moussawi J, Rogniaux H, Santoni A, Saulnier L, Jamme F, Miquel M, Lepiniec L, Dubreucq B. 2012. A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. Journal of Experimental Botany 63, 739–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C. 2012. The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biology 12, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Ynag MS, Xiang F, Park CM. 2008. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Burton RA, Biely P, Lahnstein J, Fincher GB. 2006. Hydrolysis of (1,4)-β-D-mannans in barley (Hordeum vulgare L.) is mediated by the concerted action of (1,4)- β-D-mannan endohydrolase and β-D-mannosidase. Biochemical Journal 399, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Matilla AJ. 2010. Genes involved in ethylene and gibberellins metabolism are required for endosperm-limited germination of Sisymbrium officinale L. seeds. Planta 231, 653–664. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sanchez L, Carbonero P. 2013. Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7 . The Plant Journal 74, 767–780. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodríguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla AJ. 2011. a Three endo-β-mannanase genes expressed in the micropilar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233, 25–36. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodríguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A. 2011. b Molecular analysis of endo-β-mannanase genes upon seed imbibition suggest a cross-talk between radicle and micropylar endosperm during germination of Arabidopsis thaliana . Plant Signaling & Behavior 6, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Wozny D, Iriondo-de Hond M, Oñate-Sánchez L, Carbonero P, Barrero-Sicilia C. 2014. The AtCathB3 gene, encoding a cathepsin B-like protease, is expressed during germination of Arabidopsis thaliana and transcriptionally repressed by the basic leucine zipper protein GBF1. Journal of Experimental Botany 65, 2009–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463, 736–768. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. 1987. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proceedings of the National Academy of Sciences USA 84, 7542–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Dekkers BJ, Steinbrecher T, Walsh CT, Bacic A, Bentsink L, Leubner-Metzger G, Knox JP. 2012. Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiology 160, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G. 2005. Beta-1,3-glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. The Plant Journal 41, 133–145. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Meins F., Jr 2000. Sense transformation reveals a novel role for class I beta-1, 3-glucanase in tobacco seed germination. The Plant Journal 23, 215–221. [DOI] [PubMed] [Google Scholar]
- Lever M. 1977. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAH-BAH): Effect of bismuth on the reaction. Analytical Biochemistry 81, 21–27. [DOI] [PubMed] [Google Scholar]
- Marcus SE, Blake AW, Benians TA, et al. 2010. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. The Plant Journal 64, 191–203. [DOI] [PubMed] [Google Scholar]
- Martínez-Andújar C, Pluskota WE, Bassel GW, et al. 2012. Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. The Plant Journal 71, 575–586. [DOI] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F. 2006. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8’-hydroxylase. The Plant Journal 45, 942–954. [DOI] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. 2013. Unlocking Triticeae genomics to sustainably feed the future. Plant Cell Physiology 54, 1931–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. 2006. Endosperm-limited Brassicacae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana . Plant Cell Physiology 47, 864–977. [DOI] [PubMed] [Google Scholar]
- Nonogaki H. 2014. Seed dormancy and germination-emerging mechanisms and new hypotheses. Frontiers in Plant Science 5, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Chen F, Bradford K. 2007. Mechanisms and genes involved in germination sensu stricto . In: Bradford K, Nonogaki H, eds. Seed Development, Dormancy and Germination. Oxford: Blackwell, 264–304. [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ. 2000. A germination-specific endo-beta-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiology 123, 1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sanchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including sedes and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Tureckova V, Lacombe E, Lopez-Molina L. 2009. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO Journal 28, 2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Marita JM, Hatfield RD. 2012. Cell wall composition throughout development for the model grass Brachypodium distachyon . Frontiers in Plant Science 3, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YF, Bewley JD, Wang XF. 2008. Protein and gene expression patterns of endo-ß-mannanase following germination of rice. Seed Science Research 8, 139–149. [Google Scholar]
- Rodríguez-Gacio MC, Iglesias-Fernández R, Carbonero P, Matilla AJ. 2012. Softening-up mannan-rich cell walls. Journal of Experimental Botany 63, 3975–3988. [DOI] [PubMed] [Google Scholar]
- Sargent JA, Osborne DJ. 1980. A comparative study of the fine structure of coleorhiza and root cells during the early hours of germination of rye embryos. Protoplasma 104, 91–103. [Google Scholar]
- Scheler C, Weitbrecht K, Pearce SP, et al. 2014. Promotion of testa rupture during Lepidium sativum germination involves seed compartment-specific expression and activity of pectin methylesterases. Plant Physiology pp.114.247429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder R, Atkinson RG, Redgwell RJ. 2009. Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Annals of Botany 104, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Sugahara T, Kotake T, Nakagawa N, Sakurai N. 2010. Sugar treatment inhibits IAA-induced expression of endo-1,3:1,4-beta-glucanase EI transcripts in barley coleoptile segments. Physiologia Plantarum 139, 413–420. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar C. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G. 2011. First off the mark: early seed germination. Journal of Experimental Botany 62, 3289–3309. [DOI] [PubMed] [Google Scholar]
- Yan D, Duermeyer L, Leoveanu C, Nambara E. 2014. The functions of the endosperm during seed germination. Plant Cell and Physiology 55, 1521–1533. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Yang X, Lai J, Lin H, Cheng Z-M, Nonogaki H, Chen F. 2007. The endo-β mannanase gene families in Arabidopsis, rice, and poplar. Functional and Integrative Genomics 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Song D, Sun J, Li L. 2013. Populus endo-beta-mannanase PtrMAN6 plays a role in coordinating cell wall remodeling with suppression of secondary wall thickening through generation of oligosaccharide signals. The Plant Journal 74, 473–485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.