Highlight
The homeostasis of biologically active cytokinins was the predominant leaf-inherent factor for winter oilseed rape cultivar-differences in nitrogen starvation-induced leaf senescence and thus nitrogen efficiency.
Key words: Brassica napus, cytokinins, genotypic differences, leaf senescence, nitrogen efficiency, nitrogen starvation, reciprocal grafting, stay-green.
Abstract
Nitrogen (N) efficiency of winter oilseed rape (Brassica napus L.) line-cultivars (cvs.), defined as high grain yield under N limitation, has been primarily attributed to maintained N uptake during reproductive growth (N uptake efficiency) in combination with delayed senescence of the older leaves accompanied with maintained photosynthetic capacity (functional stay-green). However, it is not clear whether genotypic variation in N starvation-induced leaf senescence is due to leaf-inherent factors and/or governed by root-mediated signals. Therefore, the N-efficient and stay-green cvs. NPZ-1 and Apex were reciprocally grafted with the N-inefficient and early-senescing cvs. NPZ-2 and Capitol, respectively and grown in hydroponics. The senescence status of older leaves after 12 days of N starvation assessed by SPAD, photosynthesis and the expression of the senescence-specific cysteine protease gene SAG12-1 revealed that the stay-green phenotype of the cvs. NPZ-1 and Apex under N starvation was primarily under the control of leaf-inherent factors. The same four cultivars were submitted to N starvation for up to 12 days in a time-course experiment. The specific leaf contents of biologically active and inactive cytokinins (CKs) and the expression of genes involved in CK homeostasis revealed that under N starvation leaves of early-senescing cultivars were characterized by inactivation of biologically active CKs, whereas in stay-green cultivars synthesis, activation, binding of and response to biologically active CKs were favoured. These results suggest that the homeostasis of biologically active CKs was the predominant leaf-inherent factor for cultivar differences in N starvation-induced leaf senescence and thus N efficiency.
Introduction
Nitrogen efficiency of winter oilseed rape line-cultivars, defined as high grain yield under N limitation, has been primarily attributed to maintained N uptake during reproductive growth (N uptake efficiency) (Kamh et al., 2005; Schulte auf’m Erley et al., 2007, 2011; Berry et al. 2010). A characteristic of N-efficient cultivars is a functional stay-green phenotype during reproductive growth, expressed through delayed senescence of the older leaves accompanied with maintenance of photosynthetic capacity (Schulte auf’m Erley et al., 2007). In many crop species, stay-green phenotypes are superior in yield formation particularly under abiotic stress conditions when senescence is prematurely induced, for instance by drought or N limitation (Gregersen et al., 2013). Functional stay-green results from a delayed onset and/or progression of senescence (Thomas and Howarth, 2000). Thus, in contrast to cosmetic stay-green without any photosynthetic activity (Thomas and Howarth, 2000), functional stay-green requires more than the retention of green colour based on delayed or completely inhibited chlorophyll catabolism (Kusaba et al., 2013).
Functional stay-green is an inheritable, mostly polygene-regulated quantitative trait (Wang et al., 2012). But the underlying mechanisms are currently poorly understood. The onset of leaf senescence is accompanied by transcriptional regulation of many genes (Smart, 1994). Master-regulators of the gene-expression network during senescence are senescence-inducible transcription factors (TFs), which positively or negatively act on leaf senescence (Balazadeh et al., 2008; Parlitz et al., 2011). Many of these TFs directly or indirectly control the activation or repression of down-stream senescence-associated genes (SAGs) to fine-tune both the onset and rate of leaf senescence (Kusaba et al., 2013). A hallmark of leaf senescence is the termination of photoassimilation. For functional stay-green, the maintenance of functional chloroplasts and thus photoassimilation is a prerequisite, a process that is under the control of nuclear as well as chloroplast encoded genes (Sakuraba et al., 2012a, b; Rauf et al., 2013).
Additional key regulators of leaf senescence are phytohormones (Lim et al., 2007). Alterations in phytohormone, particularly cytokinin (CK), metabolism and signalling lead to functional stay-green phenotypes (Thomas and Ougham, 2014). Cytokinins are the most potent general antagonist of senescence (Wingler and Roitsch, 2008; Zwack and Rashotte, 2013). Although currently nothing is known about the underlying mechanisms for genotypic variation in functional stay-green under N starvation in winter oilseed rape, leaf-inherent and/or root-mediated CKs might be important factors. For functional stay-green of mature leaves in cotton plants root-derived CKs are important (Dong et al., 2008; Dai and Dong, 2011), while leaf-inherent CK factors lead to functional stay-green in other plant species, exemplified by modification of leaf-inherent CK homeostasis in mature tobacco leaves (Gan and Amasino, 1995; del Mar Rubio-Wilhelmi et al., 2014) as well as CK perception and signalling in mature Arabidopsis thaliana leaves (Kim et al., 2006) or downstream targets in tomato (Albacete et al., 2014) and tobacco leaves (Balibrea et al., 2004).
An important aspect in addressing CK function is that CKs can be divided into biologically active CKs and biologically inactive CKs. The biologically inactive CKs are conjugated forms of the biologically active CKs and are used for storage and transport. In addition, the conjugation into biologically inactive CKs allows a fine-tuned regulation of the appropriate biologically active CKs levels. A subset of these conjugation reactions is reversible, enabling a rapid release of biologically active CKs from biologically inactive CK storage pools (Schmülling, 2004; Sakakibara, 2006).
To elucidate whether cultivar differences in N starvation-induced leaf senescence in winter oilseed rape are due to leaf-inherent factors and/or governed by root-mediated signals, a reciprocal grafting approach was applied in the present study using two cultivar pairs differing in N efficiency and N starvation-induced leaf senescence. To clarify the role of phytohormones for genotypic variation in functional stay-green, phytohormone levels were determined in roots, xylem sap and individual leaves in a complementary time-course experiment comprising the same four cultivars used for the grafting approach. In addition, the expression of selected genes involved in CK homeostasis, perception and signalling were analysed in the leaf tissue.
Material and Methods
Grafting experiment
Plant material
Based on a previous experiment comparing N starvation and leaf detaching as inducers of leaf senescence in 10 line-cultivars (cvs.), two cultivar pairs were selected for the reciprocal grafting approach. The cvs. NPZ-1 & NPZ-2 are breeding lines with a similar genetic background and reacted differently to N starvation and detaching, indicating root-derived factors. The commercial cvs. Apex and Capitol were selected since they did not differ in leaf senescence irrespective of the senescence inducer, suggesting leaf-inherent factors (Supplementary Fig. S1). Plants of both cultivar pairs were reciprocally-grafted or not-grafted and self-grafted as controls.
Grafting procedure
The seeds were germinated and the seedlings were grafted in a climate chamber (day/night 16/8h; temperature day/night 22/20°C; PAR 350 µmol m-2 s-1). The seeds were sown in substrate consisting of white peat, sand and perlite in the ratio of 3:1:2 (w:w:w). The substrate was limed to pH 6 using 4g l-1 limestone (85% calcium carbonate, Otterbeinkalk) and macro- and micronutrients were added using 0.5g l-1 Flory 3 (Euflor, Schrembeck, Germany) and 0.1g l-1 Flory 10 (Euflor, Schrembeck, Germany). Seven days after germination the hypocotyl reached a diameter of 1mm and the plants were grafted following a modified procedure described by Moroni (1997). The hypocotyls were cut horizontally, and roots and shoots fixed to each other via a well-fitting PVC-tube (PVC-Standard diameters: 1.020; 1.143; 1.295; 1.422mm, Spetec, Erding, Germany). The grafted plants were kept for 5 d under a wet tent. The humidification was gradually reduced to the ambient conditions in the climate chamber in daily steps. Afterwards the plants were acclimatized in the climate chamber for one day before they were transferred to hydroponics.
Hydroponic growing conditions
The plants were cultured in a greenhouse [assimilation light (16 klm) 16h; heating/ventilation temperature 22/20°C; shading at 15 klx solar radiation; relative humidity 80%] from 24 February to 4 April 2012. The roots of the plants were washed out of the substrate using deionized water. Two plants were transferred to a 6 l plastic pot and pre-cultured for 28 d at optimal N supply (2.0mM). The roots of the plants were separated by hand daily, to avoid intermingling of the root systems. The roots remained submerged during the separation to minimize root damage. The composition of the nutrient solution was 500 µM K2S04, 250 µM KH2PO4, 325 µM MgSO4, 50 µM NaCl, 8 µM H3BO3, 0.4 µM MnSO4, 0.4 µM ZnSO4, 0.4 µM CuSO4, 0.1 µM Na2MoO4 and 40 µM Fe-EDDHA. During pre-culture Ca(NO3)2 and (NH4)2SO4 were used as N sources in the ratio of 9:1, and 10 µM C2H4N4 (dicyandiamide) was added to prevent nitrification. Prior to treatment one plant per pot was discarded, which allowed selection for homogeneity of the experimental plants. For N starvation the plants were grown at 0.1mM N [Ca(NO3)2] and 1.0mM CaSO4 to allow for optimum Ca nutrition. For optimal N nutrition the plants were cultured at 4.0mM N doubling the concentration used for pre-culture. After the start of treatment the nutrient solution was changed every second day. The experiment was completely randomized with four biological replications.
Non-destructive measurements, plant harvest and analysis
During the treatment the senescence status of the third and the fourth leaf, counted from the bottom to the top of the plant, was measured using non-destructive methods. The fifth leaf from the bottom of the plant was the youngest fully expanded leaf at the end of pre-culture. The chlorophyll content of the third and the fourth leaf was assessed by a portable chlorophyll meter (SPAD-502, Konica Minolta, Tokyo, Japan). Photosynthesis rate of the fourth leaf was measured using a portable gas exchange system (LI-6400, LI-COR, Lincoln, USA) at a photon flux density of 1000 µmol m-2 s-1 and an incoming CO2 concentration of 400 µmol m-2 s-1 to assess functional stay-green. The plants were harvested 12 d after start of treatment (DAT) and were separated into shoot and root, while the third and the fourth leaf were harvested separately. The leaf area was measured by a portable leaf area meter (LI-3100, LI-COR, Lincoln, USA). The leaf was divided along the midrib with a razor blade. One half was immediately frozen in liquid N2 and the other half was dried at 70°C until constant weight for dry weight determination. N concentrations of the dried and ground root, shoot and leaf material were determined using an elemental analyser (Vario EL, Elementar Analysensysteme, Hanau, Germany). For RNA extraction, the frozen material was ground using a mixer mill (MM 400, Retsch, Haan, Germany).
Kinetics of N starvation-induced leaf senescence
Plant material and growing conditions
The kinetics of the development of N starvation-induced leaf senescence was investigated for the same four cvs.: NPZ-1, NPZ-2, Apex and Capitol. The seeds were germinated in the climate chamber as described above using a ‘sandwich’ method arranging the seeds between filter paper sandwiched between sponges and PVC-plates on both sides. The ‘sandwiches’ were placed into a box containing tap water. Seven days after germination the seedlings were cultured from 23 September to 3 November 2010 in the greenhouse in hydroponics as described above. The experiment was completely randomized with 3–4 biological replications.
Plant harvest and analysis
Plants were harvested after 0, 3, 5, 7, 10 and 12 d of N starvation and after 7 and 12 DAT of optimal N nutrition and divided into root, shoot and three previously marked individual leaves, which were the youngest fully expanded leaf at the end of pre-culture and the two older leaves. In most cases, the youngest fully expanded leaf at the end of pre-culture was the fifth leaf from the bottom of the plant. But sometimes it was the fourth leaf due to differences in development between the cultivars. During N starvation, the senescence status of the three leaves was measured using non-destructive methods as describe above, while photosynthesis rate was measured at a photon flux density of 400 µmol m-2 s-1. The harvest procedure, the sample preparation and the N analysis were performed as described above.
Xylem sap collection and analysis
Xylem exudates were collected at each harvest. The nutrient solution was changed 1h before starting xylem sap collection according to the N variants and additionally KCl (2.0mM) was added to enhance exudation. The plants were horizontally cut using a razor blade at the root collar and a well-fitting silicone tube was attached. After 5min the exudates were discarded and then collected for 1h. The collected exudates were kept on ice, protected from light and after determination of the exudate volume for the calculation of the metabolite xylem transport-rate, the exudates were stored at −20°C. For analysis the exudates were gently defrosted on ice and protected from light.
Phytohormone analysis
The levels of abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA) and of the cytokinins (CKs) trans-Zeatin (tZ), cis-Zeatin (cZ), dihydrozeatin (DHZ), isopentenyladenin (iP), trans-Zeatin-O-glucoside (tZOG) and trans-Zeatin-O-glucoside-riboside (tZOGR) were analysed in the root, the xylem sap and two individual leaves per plant (the second and third oldest harvested mature leaves at the end of pre-culture). Extractions were performed using 250mg of frozen and ground leaf and root tissue, or 100 µl xylem sap, after adding 4 μl of internal standard mix composed of deuterium labelled hormones. Phytohormone detection was performed on a UHPLC–MS/MS system consisting of a Thermo ACCELA pump (Thermo Scientific, Waltham/USA) coupled to a tempered HTC-PAL autosampler (CTC Analytics, Zwingen/Switzerland), and connected to a Thermo TSQ Quantum Access Max Mass Spectrometer (Thermo Scientific) with a heated electrospray ionization (HESI) interface, using a Nucleoshell-PFP column (2.7 μm, 100×2mm; Macherey-Nagel) according to Großkinsky et al. (2014).
RNA isolation and cDNA synthesis
RNA was extracted from 100mg of frozen and ground leaf material with TRIsure™ (Bioline, London, UK) reagent according to the instructions of the manufacturer. RNA integrity was tested on 1% agarose gel electrophoresis and photometrically quantified (NanoPhotometer™, Implen, München, Germany). Two micrograms of RNA were applied to synthesize cDNA with the RevertAid™ H Minus First Strand kit (Fermentas, Waltham, USA). For the reaction the supplied random hexamer primer were used to synthesize the first strand cDNA of the mRNA according to the instructions of the manufacturer. Quality and quantity of cDNA was determined as for RNA.
Primer design and qRT-PCR
The relative expression of the gene SAG12-1 encoding a senescence-specific cysteine protease and selected genes involved in CK homeostasis and biological impact were analysed using qRT-PCR. Among these were genes related to synthesis (isopentenyltransferase—IPT), to the reversible and irreversible glycosylation (uridine diphosphate glycosyltransferase—UGT) converting biologically active CKs to biologically inactive but activatable CKs or biologically inactive CK forms, respectively, to the release of highly biologically active CK nucleobases from their ribosides with lower biological activity (cytokinin riboside 5′-monophosphate phosphoribohydrolase—LOG), to the breakdown of biologically active CKs (cytokinin oxidase/dehydrogenase—CKX), and to the perception of and response to biologically active CKs (histidine kinase 3—AHK3; response regulator 2—ARR2). The gene EF1-alpha encoding an elongation factor was used as reference gene (Desclos et al., 2008). The initial selection of the analysed genes related to cytokinin homeostasis was based on the availability of sequence information in A. thaliana and B. napus. The final selection of the respective isogenes was based on the detectability of the respective gene expression in leaf tissues. A gene was detected when the fluorescence signal of its amplicon exceeded the threshold within 50 PCR cycles. For the design of the primer pairs B. napus sequences were used. If no suitable primer pair could be designed on the B. napus sequence the primer pair was designed on the sequence of the A. thaliana homologue gene. The B. napus sequences were retrieved from the NCBI public database and The Gene Index Project at the Dana-Farber Cancer Institute (DFCI; http://compbio.dfci.harvard.edu/tgi/plant.html). The A. thaliana sequences were obtained from the Arabidopsis Information Resource Version 10 (TAIR; http://www.arabidopsis.org). Primer pairs were designed using PrimerQuest (Integrated DNA Technologies, Coralville, USA) for genes having NCBI accession numbers, or using QuantPrime (Arvidsson et al., 2008) for genes having DFCI or TAIR accession numbers (Supplementary Table S1). PCR reactions for qRT-PCR were performed as described by Koeslin-Findeklee et al. (2015). The mean relative expression and the standard error of the qRT-PCR results were calculated based on the 2-ΔΔCt-method by Livak and Schmittgen (2001).
Statistical analysis
The statistical analysis was performed using the statistic software SAS version 9.2 (SAS Institute, Cary, USA). For the comparison of the two grafting experiments the data were ranked using the PROC RANK procedure followed by an analysis of variance (ANOVA) using the PROC GLM procedure. For analysis of data from the grafting experiment and the experiment investigating the kinetics of the development of N starvation-induced leaf senescence the ANOVA was calculated using the PROC GLM procedure. For the ANOVA the Type III sum of squares was applied, if an unequal number of replicates occurred. Multiple comparisons of means were calculated by the MEANS statement of the PROC GLM procedure. For all tests of significance a P-value of 0.05 was used and the P-values were Bonferroni-Holm adjusted. Curves were fitted using the graphic software SIGMA PLOT version 11 (Systat software, San Jose, USA). The relative qRT-PCR data were analysed using the %QPCR MIXED macro after Steibel et al. (2009) based on the PROC MIXED procedure.
Results
Grafting experiment
To investigate whether cultivar differences in N starvation-induced leaf senescence are due to leaf-inherent factors and/or governed by root-mediated signals, a reciprocal grafting experiment was performed using two cultivar pairs differing in N efficiency and N starvation-induced senescence described under Plant material above. The experiment was performed twice. Since these experiments did not differ in N starvation-induced leaf senescence (Supplementary Table S2), the results of only one experiment are shown.
Chlorophyll content and photosynthesis rate in senescing leaves
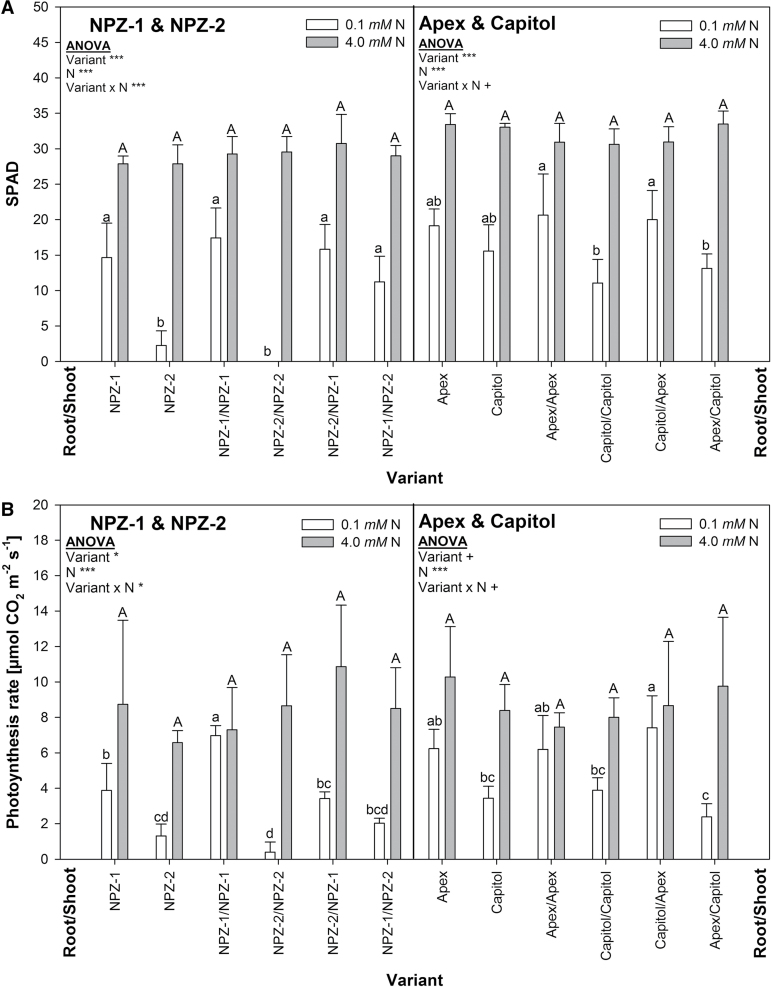
Since for both cultivar pairs the overall statistical analysis of cholorophyll content (represented by SPAD values) revealed that for all variants the fourth leaf (‘leaf 4’) reacted to N starvation, although delayed (significant leaf effect), in exactly the same way as the third leaf (no significant variant × leaf × N interaction, Supplementary Table S3), only the results for leaf 4 are presented. At optimal N supply the SPAD values of all variants were high and did not differ (Fig. 1A). The SPAD values declined after 12 days of N starvation, reflecting senescence induction, and highly significant differences existed between the variants. The non-grafted controls and the self-grafted plants showed the expected differences in senescence within the cultivar pairs: cvs. Apex and NPZ-1 remained greener (higher SPAD values) than the cvs. Capitol and NPZ-2, although the controls of cvs. Apex and Capitol differed only in tendency. The leaf SPAD value depended primarily on the origin of the shoot for the reciprocal grafts of cvs. Apex and Capitol, and cvs. NPZ-1 and NPZ-2: the cv. Apex shoot (significantly) and the cv. NPZ-1 shoot (only in tendency), conferred delayed leaf senescence when grafted on the cv. Capitol and cv. NPZ-2 roots, respectively. The lack of significance was due to an unusually high SPAD value of the cv. NPZ-2 shoot on the cv. NPZ-1 root in this experiment which might be interpreted as a positive effect of the cv. NPZ-1 root. However, this result could not be reproduced in the second repetition of the experiment (not shown). As an additional indicator of the leaf senescence status photosynthesis was measured (Fig. 1B). Photosynthesis reacted more sensitively to N starvation than SPAD with a significant decline. No differences existed between the variants at high N supply. Under N starvation leaf photosynthesis of non-grafted and self-grafted plants of cv. Capitol (in tendency) and cv. NPZ-2 (significantly) was lower than of cv. Apex and cv. NPZ-1, respectively. As for SPAD, photosynthesis of the grafted plants was determined mainly by the origin of the shoot: the cv. Apex shoot conferred less decline of photosynthesis than the cv. Capitol shoot when reciprocally grafted on the respective roots. For the cultivar pair NPZ-1 and NPZ-2 a comparative shoot effect was visible only in tendency for the reasons mentioned above for SPAD.
Fig. 1.
(A) SPAD values and (B) photosynthesis rate of leaf 4 in non-grafted, self-grafted and reciprocally-grafted plants of the winter oilseed rape cultivars NPZ-1 and NPZ-2 (left), and Apex and Capitol (right), grown in hydroponics after 12 d of N starvation (0.1mM N) or optimal N supply (4.0mM N). The plants were pre-cultured for 28 d at 2.0mM N. Different letters on top of the columns indicate differences between the variants (P<0.05). ANOVA: +, *, *** indicate significant differences at P<0.10, P<0.05, P<0.001, respectively. The error bars represent the standard deviations of the means (n=3–4).
Expression of senescence-specific cysteine protease gene SAG12-1 in senescing leaves
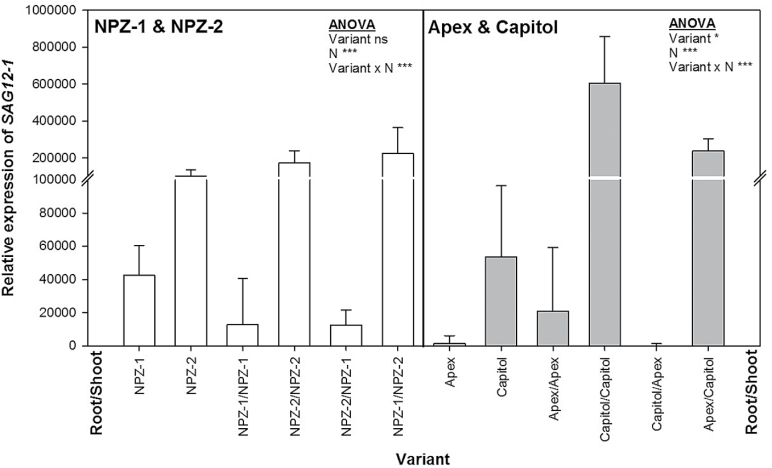
As a further senescence marker, the relative expression of SAG12-1 coding for a senescence-specific cysteine protease was determined. SAG12-1 expression proved to be a much more sensitive indicator than SPAD and photosynthesis (compare Figs 1 and 2). The lower upregulation by N starvation of SAG12-1 in the non-grafted and self-grafted cvs. Apex and NPZ-1 confirmed a delayed leaf senescence as compared to cvs. Capitol and NPZ-2. The dominant role of the shoot origin in N starvation-induced leaf senescence was well reflected by SAG12-1 expression in leaves of the grafted plants, which was only significantly increased when cv. Capitol and cv. NPZ-2 were used as shoots.
Fig. 2.
Relative expression (2-ΔΔCt) of SAG12-1 in leaf 4 in non-grafted, self-grafted and reciprocal-grafted plants of the winter oilseed rape cultivars NPZ-1 and NPZ-2 (left), and Apex and Capitol (right), grown in hydroponics after 12 d of N starvation (0.1mM N). The plants were pre-cultured for 28 d at 2.0mM N. The data are shown relative to the control (4.0mM N) harvested at the same day. ANOVA: *** indicate significant differences at P<0.001; ns, non-significant. The error bars represent the standard errors of the means (n=3–4).
Specific leaf N content in senescing leaves
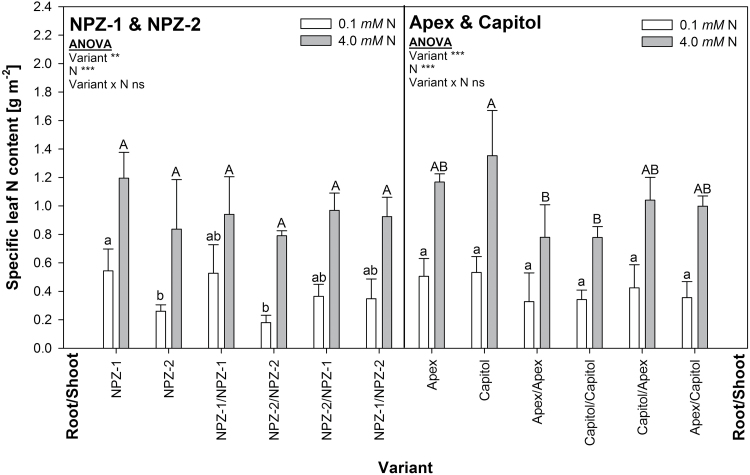
The cultivar differences in N starvation-induced leaf senescence could be due to differences in the depletion of the leaf N content. The specific leaf N content of leaf 4 significantly declined after 12 days of N starvation as compared to continuous high N supply for all cultivars and variants (Fig. 3). The non-grafted (significantly) and the self-grafted (tendentially) stay-green cv. NPZ-1 showed a higher specific leaf N content under N starvation as compared to the early-senescing counterpart cv. NPZ-2. However, no significant differences existed between the cvs. Apex and Capitol.
Fig. 3.
Specific leaf N content of leaf 4 in non-grafted, self-grafted and reciprocally-grafted plants of the winter oilseed rape cultivars NPZ-1 and NPZ-2 (left), and Apex and Capitol (right), grown in hydroponics after 12 d of N starvation (0.1mM N) or optimal N supply (4.0mM N). The plants were pre-cultured for 28 days at 2.0mM N. Different letters on top of the columns indicate differences between the variants (P<0.05). ANOVA: **, *** indicate significant differences at P<0.01 and P<0.001, respectively; ns, non-significant. The error bars represent the standard deviations of the means (n=3–4).
Under maintained high N supply, the cvs. NPZ-1 and NPZ-2 did not differ in the specific leaf N contents between the variants. In contrast, for the cultivar pair Apex and Capitol the specific leaf N content of the non-grafted was generally higher than that of the grafted plants. This difference was even significant between the non- and self-grafted plants of cv. Capitol.
Development of N starvation-induced leaf senescence
To further clarify whether root-to-shoot communication and which leaf-inherent factors are decisive for the cultivar differences in stay-green, phytohormone levels were analysed in roots, xylem sap and individual leaves in a complementary time-course experiment with the same four cultivars used for the grafting approach. In this experiment, the cultivars also showed the established cultivar-specific responses to N starvation characterizing the cvs. NPZ-1 and Apex as stay-green and the cvs. NPZ-2 and Capitol as early-senescing (Koeslin-Findeklee et al., 2015).
Plant phytohormone status
Phytohormones play a major role in root-to-shoot communication and in the control of leaf senescence. Therefore, the levels for the phytohormones SA, JA, ABA and CKs were analysed in roots, xylem exudates and leaves as affected by N starvation and cultivar. Only the results of the second oldest harvested leaf are presented since the statistical analysis did not reveal a consistent difference between the two analysed individual leaves (Supplementary Tables S4, S5). To identify a possible relationship between the root pool and the specific content of mature leaves showing leaf senescence during the treatment period, the root content, the xylem transport rate and the specific leaf contents were determined. Despite significant differences for some phytohormones between cultivars (Supplementary Tables S4, S5), only the measured biologically inactive but activatable CKs (iCKs) tZOG and tZOGR responded in a consistent way in relation to the cultivar-specific differences in N starvation-induced leaf senescence. Therefore, the following results showing the effects of treatment duration and N supply are means over the four cultivars.
The root SA content increased under both N supplies, reflecting the gain and differences in root biomass during the N treatment period (Supplementary Fig. S2A). The SA xylem transport rates did not differ either between the N supplies or the treatment times. The leaf specific SA content increased during the treatment time from 7 until 12 DAT independent of the N supply (no significant N effect). Also the JA root content increased until 12 DAT under both N supplies (Supplementary Fig. S2B). In contrast to SA the JA root content was higher under N starvation than with sufficient N supply at 12 DAT. The JA xylem transport rate did not show significant N supply and treatment-time effects. Up to 7 DAT the specific JA leaf content was neither affected by treatment duration nor by N supply. But after 7 DAT the JA content significantly increased under N starvation while at sufficient N supply it decreased (significant N × DAT interaction). The ABA root contents apparently increased more with treatment duration under high N supply than under N starvation, but this difference was not significant (Supplementary Fig. S2C). The ABA transport rate in the xylem increased significantly during the time-course of the treatment under sufficient N supply, but not under N starvation. The specific ABA content in the leaf tissue increased until 7 DAT for both N supplies. At 12 DAT, the ABA content was significantly higher in N-starved leaves, because it remained at high level, whereas it deceased under sufficient N.
Among the phytohormones, CKs play a major role in both root-to-shoot communication and leaf senescence. The individually measured CKs were grouped according to their biological activity (Schmitz et al., 1972; Schmülling, 2004) into biologically active (aCKs) and biologically inactive but activatable CKs (iCKs). The aCKs (tZ, iP, DHZ, cZ, tZR) are of particular importance in relation to stay-green. The overall statistical analysis (Supplementary Table S5) revealed that in spite of significant differences between the individual aCKs neither the cultivar nor the N supply affected the aCKs. Moreover, no systematic pattern was apparent in relation to cultivar, treatment duration (DAT) and N supply (no significant cultivar × DAT × N × CKs interaction). Therefore, the individual aCK data were pooled for the root, xylem sap and leaf tissue.
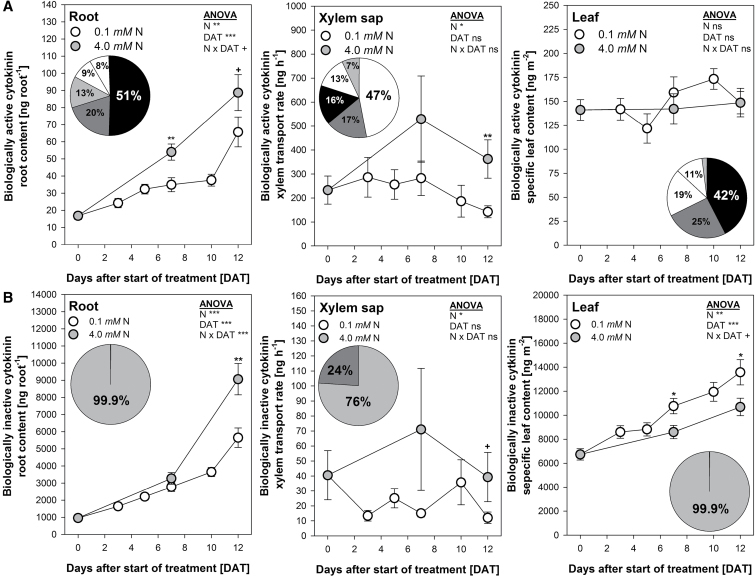
In the root and leaf tissue, tZ was the most abundant aCK (51% and 42%, respectively; Fig. 4A). However, in the xylem sap iP was the most abundant aCK (47%). In the root the aCK content increased until 12 DAT under both N supplies, but significantly more under sufficient N supply primarily due to increasing root biomass. The xylem transport-rate of aCKs remained stable over the treatment time (no significant DAT effect), but was significantly enhanced under high N supply. The specific leaf content of the aCKs was affected neither by the duration of N starvation (DAT) nor by N supply and remained at a constant level.
Fig. 4.
(A) Biologically active cytokinins (tZ, iP, DHZ, cZ, tZR) and (B) biologically inactive but activatable cytokinins (tZOR, tZORG) in the root, the xylem sap and the second oldest harvested mature leaf of the winter oilseed rape cultivars NPZ-1, NPZ-2, Apex and Capitol grown in hydroponics during 12 d N starvation (0.1mM) or optimal N supply (4.0mM). The plants were pre-cultured for 28 d at 2.0mM N. Pie chart in A: tZ (black), DHZ (dark grey), tZR (grey), cZ (white), iP (hatched). Pie chart in B: tZOG (grey), tZROG (dark grey). ANOVA: +, *, **, *** indicate significant differences at P<0.10, P<0.05, P<0.01, P<0.001, respectively; ns, non-significant. At 7 and 12 DAT: +, *, ** indicate differences between the N supplies at P<0.10, P<0.05, P<0.01, respectively. The error bars (visible only when greater than the symbols) represent the standard errors of the means across the four cultivars (n=3–4).
The glycosylation of biologically active CKs (aCKs) to O-glycosides (iCKs) leads to their reversible inactivation. In the leaf tissue this inactivation of the aCKs might be of importance for cultivar differences in N starvation-induced leaf senescence. The iCKs constitute the majority of the total CKs in root and leaf tissues (Fig. 4B). Among the iCKs, tZOG represented more than 99% of that fraction in root and leaf tissues independent of N supply and duration of N starvation (DAT, not shown). In the xylem, iCKs were also mostly transported as tZOG (76%). The highly significant influence of the duration of N starvation (DAT) on the root content of iCKs was due to root growth during the treatment period rather than root concentrations (not shown). The xylem transport rate of iCKs increased during the treatment period only at high N supply. The lack of a significant DAT effect (ANOVA) may be explained by the high variation of the mean at 7 DAT at high N supply. In the leaf tissue, the iCKs significantly increased during the treatment period. The increase was steeper under N starvation than at optimum N supply (significant N × DAT interaction).
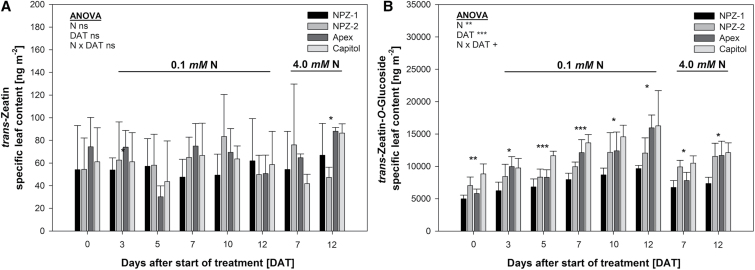
The overall statistical evaluation revealed that the iCKs but not the aCKs showed a consistent difference between the cultivars and N supplies in the leaf tissue (significant cultivar × DAT × N interaction, Supplementary Table S5), evident by the cultivar-specific specific leaf contents of the dominating individual aCKs (tZ) and iCKs (tZOG) (Fig. 5). The specific leaf content of tZ was not affected by N supply and the duration of N starvation (ANOVA). Differences between cultivars were only significant in high N leaves at 12 DAT. In contrast, the tZOG specific leaf content highly significantly increased during treatment duration to a larger extent in N-starved than in N-sufficient leaves (significant N × DAT interaction). The early-senescing cvs. NPZ-2 and Capitol mostly showed higher tZOG contents than their stay-green counterpart cvs. NPZ-1 and Apex, both under N starvation and at high N supply.
Fig. 5.
(A) trans-Zeatin and (B) trans-Zeatin-O-Glucoside specific leaf content of the second oldest harvested mature leaf of the winter oilseed rape cultivars NPZ-1, NPZ-2, Apex and Capitol grown in hydroponics during 12 d N starvation (0.1mM) or optimal N supply (4.0mM). The plants were pre-cultured for 28 d at 2.0mM N. ANOVA: +, *, **, *** indicate significant differences at P<0.10, P<0.05, P<0.01, <0.001, respectively; ns, non-significant. *, **, *** above the columns indicate significant differences between the cultivars at P<0.05, P<0.01, P<0.001, respectively. The error bars represent the standard deviations of the means (n=3–4).
Regulation of cytokinin homeostasis-related genes in senescing leaves
To investigate the importance of de novo synthesis, metabolic inter-conversion and breakdown of biologically active CKs for cultivar differences in functional stay-green during N starvation, the regulation of genes assigned to these processes were analysed.
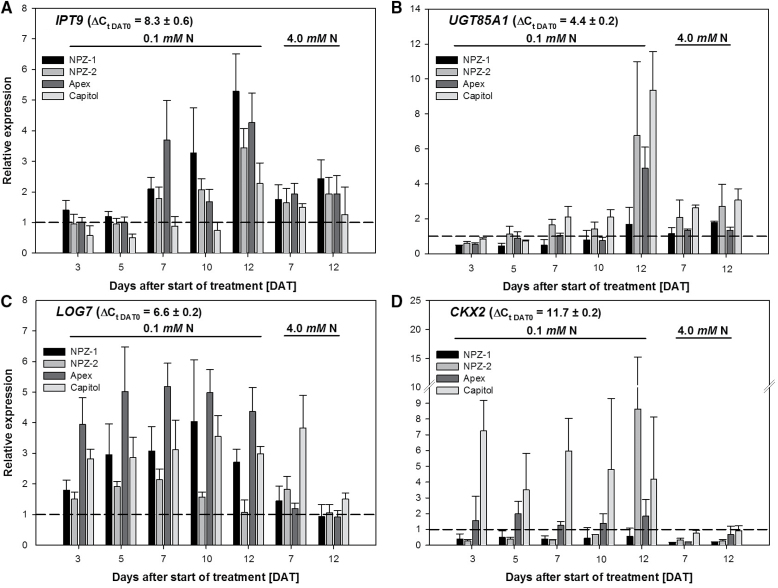
The first step of CK synthesis is catalysed by isopentenyltransferases (IPTs). Transcripts of the three IPT genes IPT2, IPT5 and IPT9 were detected in the leaf tissue under both N supplies. The cultivars generally showed a similar basal expression (DAT 0), but the basal expression levels (ΔCt) differed greatly between the three IPT genes, in ascending order: IPT2 < IPT9 < IPT5 (Supplementary Fig. S3). Among the three genes IPT9 was most clearly upregulated (up to 5-fold) under N starvation after 7 until 12 DAT and at most of the harvest days, at least in tendency, in a cultivar-specific way (Fig. 6A). IPT9 showed higher upregulation in the stay-green cvs. NPZ-1 and Apex as compared to their respective early-senescing counterpart cvs. NPZ-2 and Capitol. N starvation also generally enhanced IPT2 upregulation and at most of the harvest days, at least in tendency, a cultivar-specific higher upregulation occurred in the stay-green cultivars as compared to their respective early-senescing counterparts (Supplementary Fig. S3).
Fig. 6.
Relative expression (2-ΔΔCt) of (A) the isopentenyltransferase (IPT) gene IPT9, (B) the uridine diphosphate glycosyltransferases (UGT) gene UGT85A1, (C) the cytokinin ribosid 5′ monophosphate phosphoribohydrolase gene LOG7 and (D) the cytokinin oxidase (CKX) gene CKX2, in the second oldest harvested mature leaf of the winter oilseed rape cultivars NPZ-1, NPZ-2, Apex and Capitol grown in hydroponics during 12 d N starvation (0.1mM) or optimal N supply (4.0mM). The plants were pre-cultured for 28 days at 2.0mM N. The data are shown relative to the cultivar-specific control at DAT 0 (dashed line). The error bars represent the standard errors of the means (n=3–4). ± indicates the standard deviation of the mean for the ΔCt across the four cultivars.
The O-glycosylation of biologically active CKs leading to their reversible inactivation is catalysed by the uridine diphosphate glycosyltransferases (UGTs). In the leaf tissue under both N supplies transcripts were detected of the UGT genes UGT73C1, UGT73C5 and UGT85A1, which catalyse, and UGT73C4, which possibly catalyses, the formation of CK O-glycosides. The cultivars generally showed a similar basal expression (DAT 0), but the basal expression level (ΔCt) differed greatly between the four UGT genes, in ascending order: UGT73C1 < UGT73C4 < UGT73C5 < UGT85A1 (Supplementary Fig. S4). UGT85A1 with the highest basal expression level clearly responded to N starvation and showed higher upregulation in the early-senescing cvs. NPZ-2 and Capitol than in their respective stay-green counterpart cvs. NPZ-1 and Apex (Fig. 6B). The UGT genes UGT73C4 and UGT73C5 also responded to N starvation even more sensitively and showed a consistent cultivar-specific regulation pattern corresponding to the respective senescence phenotype (Supplementary Fig. S4B, C).
The release of the bioactive CK nucleobases from their CK riboside 5′ monophosphates is catalysed in a single step by the enzyme cytokinin riboside 5′-monophosphate phosphoribohydrolase, which is encoded by LONELY GUY (LOG). The cultivars generally showed a similar basal expression (DAT 0), but the basal expression level (ΔCt) differed greatly depending on the gene, in decreasing order LOG1 > LOG7 > LOG4 > LOG5 (Supplementary Fig. S5). Among the detected LOG genes LOG7 showed a mostly consistent cultivar-specific response to N starvation across the treatment duration (Fig. 6C). LOG7 showed higher levels of upregulation in the stay-green cvs. NPZ-1 and Apex as compared to their early-senescing counterpart cvs. NPZ-2 and Capitol (Fig. 6C). Considering the high levels of upregulation of LOG7 in cv. Capitol after 7 days of high N treatment, the upregulation by N starvation is even less than suggested by the ΔΔCt values shown in Fig. 6C, increasing the difference in comparison to cv. Apex. Also LOG4 and LOG5 clearly responded to N starvation (Supplementary Fig. S5). But only LOG4 showed a consistent cultivar-specific regulation pattern comparable to LOG7.
The catabolism of biologically active CKs is tightly regulated by cytokinin oxidase/dehydrogenases (CKXs). The five analysed CKX genes generally showed a similar basal expression between the cultivars (DAT 0; Supplementary Fig. S6). But the basal expression level (ΔCt) differed depending on the gene in ascending order CKX6 < CKX7 < CKX1 < CKX3 < CKX2. CKX1, CKX2 and CKX3 were generally upregulated by N starvation. Among all CKX genes, CKX2 with the lowest basal expression level was most strongly expressed by N starvation in a highly cultivar-specific way (Fig. 6D). The early-senescing cv. Capitol showed the highest upregulation independent of the duration of N starvation, whereas stay-green cv. Apex only weakly responded to N starvation. CKX2 was downregulated in the cvs. NPZ-1 and NPZ-2 until DAT 12 when the early-senescing cv. NPZ-2 strongly upregulated CKX2 expression.
Regulation of cytokinin perception and signalling-related genes in senescing leaves
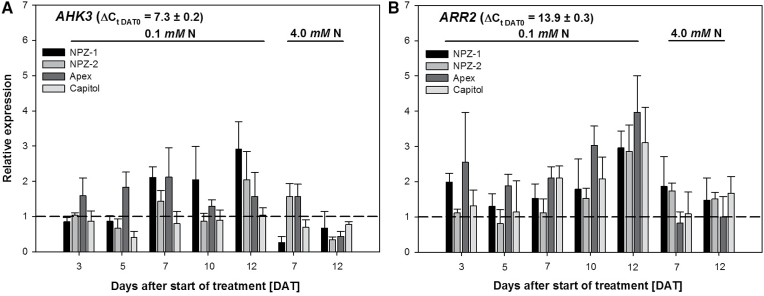
For the action of CKs on plant processes the local perception of and response to biologically active CKs, the binding to specific receptors and the activation of response regulators are necessary. The CK receptor encoded by the histidine kinase receptor AHK3 and the downstream response regulator encoded by ARR2 are involved in the CK-regulated leaf longevity. Basal expression levels did not differ between the cultivars for both genes (DAT 0; Fig. 7). AKH3 upregulation increased with the duration of N starvation in a cultivar-specific way (Fig. 7A). Considering the downregulation during the treatment period at high N supply, AHK3 was most strongly upregulated by N starvation in the stay-green cv. NPZ-1, particularly in comparison to its early-senescing counterpart cv. NPZ-2. A comparable higher expression was observed in the stay-green cv. Apex as compared to cv. Capitol, which did not show any upregulation. ARR2 upregulation generally increased with the duration of N starvation in all cultivars (Fig. 7B). As for AHK3, ARR2 was on the whole consistently more upregulated under N starvation in the leaves of the stay-green cvs. NPZ-1 and Apex than in the leaves of cvs. NPZ-2 and Capitol.
Fig. 7.
Relative expression (2-ΔΔCt) of (A) the histidine kinase gene AHK3 and (B) the response regulator gene ARR2 in the second oldest harvested mature leaf of the winter oilseed rape cultivars NPZ-1, NPZ-2, Apex and Capitol grown in hydroponics during 12 d N starvation (0.1mM) or optimal N supply (4.0mM). The plants were pre-cultured for 28 d at 2.0mM N. The data are shown relative to the cultivar-specific control at DAT 0 (dashed line). The error bars represent the standard errors of the means (n=3–4). ± indicates the standard deviation of the mean for the ΔCt across the four cultivars.
Discussion
Nitrogen efficiency of line cultivars has been primarily attributed to maintained N uptake during reproductive growth (N uptake efficiency) in combination with delayed senescence of the older leaves accompanied with maintained photosynthetic capacity (functional stay-green) (Schulte auf’m Erley et al., 2007). But currently it is not clear whether higher root growth and N uptake during early reproductive growth of N-efficient cultivars are causally related, and when related, if delayed leaf senescence is the cause or the consequence of maintained root growth.
The experimental approaches were based on experimental evidence showing that the cvs. NPZ-1 and NPZ-2 reacted differently in leaf senescence to N starvation and leaf detaching, indicating root-derived factors. In contrast, the cvs. Apex and Capitol reacted in the same way to the senescence inducers, indicating leaf-inherent factors (Supplementary Fig. S1). In order to unequivocally clarify whether the cultivar differences in N starvation-induced leaf senescence are root-mediated and/or leaf-inherent a reciprocal grafting approach was applied.
The senescence status of older leaves of the reciprocal grafts after 12 days of N starvation, assessed by SPAD (Fig. 1A), photosynthesis (Fig. 1B) and, most prominently, by SAG12-1 expression (Fig. 2) suggest that the cultivar differences between the stay-green cvs. NPZ-1 and Apex and their respective early-senescing counterpart cvs. NPZ-2 and Capitol were based primarily on leaf-inherent factors rather than on root-derived signals. SAG12-1 has been used as a suitable molecular leaf-senescence marker in winter oilseed rape (Etienne et al., 2007) and proved to be a highly sensitive marker for the detection of cultivar differences in N starvation-induced leaf senescence (Koeslin-Findeklee et al., 2014, 2015). The encoded cysteine protease is involved in protein breakdown for N remobilization during senescence and is controlled on the transcription level (Grbić, 2003). No cultivar differences existed under N starvation at 12 DAT in shoot and root N uptake (Supplementary Fig. S7) as well as N concentrations (Supplementary Fig. S8). Therefore, it is unlikely that the genotypic variation in functional stay-green was based on differences in N uptake or N utilization efficiency.
Since N content and photosynthetic activity of leaves are closely related (Evans, 1989), the specific leaf N content might be an important leaf-inherent factor for genotypic variation in functional stay-green. Particularly under N starvation, maintained photosynthetic activity could be related to the remaining specific leaf N content (Schulte auf’m Erley et al., 2007; Koeslin-Findeklee et al., 2014). But in the present study, cultivar differences in N starvation-induced leaf senescence at early stages with declined but still substantial photosynthesis rates, were not reflected by the specific leaf N content (Fig. 3). Solely the specific leaf N contents of the severely senescent non- and self-grafted cv. NPZ-2 at 12 DAT were either significantly or by trend lower as compared to the respective variants of cv. NPZ-1. This may explain the generally significant positive correlation between the specific leaf N content and photosynthesis rate (r2 = 0.20**; Supplementary Fig. S9). But only 20% of the variation in photosynthesis rate could be attributed to the specific leaf N content. Consequently, the development of cultivar differences in functional stay-green under N starvation depended primarily on other leaf-inherent factors than the specific leaf N content. This is in agreement with the results of Schulte auf’m Erley et al. (2010) for cultivar differences in N starvation-induced leaf senescence in maize.
Phytohormones are master-regulators of the leaf senescence process, in which SA, JA and ABA promote senescence whereas CKs delay it (Lim et al., 2007). Supporting the leaf-inherent control of N starvation-induced leaf senescence, the root contents of SA, JA, ABA, aCKs and iCKs and their transport rates in the xylem in the complementary time-course experiment did not suggest a role of root-derived phytohormone signals for cultivar differences in functional stay-green (Fig. 4A, B, Supplementary Fig. S2A–C). Under N starvation SA as well as JA accumulated in the leaf, but only at later stages of N starvation from 7 until 12 DAT (Supplementary Fig. S2A, B), suggesting that SA and JA were involved in the regulation of N starvation-induced leaf senescence only at advanced stages. Thus a major role of SA and JA as leaf-inherent factors for the development of genotypic variation in functional stay-green under N starvation is unlikely.
In the complementary time-course experiment leaf-photosynthesis rapidly declined after exposure to N starvation as shown previously (Koeslin-Findeklee et al., 2015). Abscisic acid regulates the stomatal conductance, which influences the photosynthesis rate (He et al., 2005). Under N starvation, ABA accumulation in the leaf was accompanied with a decrease of the photosynthesis rate (not shown). Thus, ABA accumulation in the leaf under N starvation might be an important leaf-inherent factor for the termination of photoassimilation and thus the final stage of the leaf senescence process. Nevertheless, the cultivar differences in functional stay-green could not be explained by differences in the specific ABA leaf content (Supplementary Table S4).
With regard to functional stay-green CKs are of particular importance (Balibrea et al., 2004, del Mar Rubi-Wilhelmi et al., 2014). In the present study, the measured aCKs remained at a constant level in the complementary time-course experiment, independent of N supply and cultivar (Fig. 4A). Only the iCKs responded in a N starvation and cultivar-specific way: the stay-green cvs. NPZ-1 and Apex showed a lower accumulation of iCKs in senescing leaf tissue as compared to their respective early-senescing counterpart cvs. NPZ-2 and Capitol (Fig. 4B). This suggests that CK homeostasis was a decisive leaf-inherent factor for genotypic variation in functional stay-green under N starvation. Hence the regulation of genes assigned to CK de novo synthesis, inter-conversion and breakdown were analysed in the same experiment. The rate-limiting step of CK biosynthesis is catalysed by the enzyme isopentenyltransferase (IPT) (Schmülling, 2004; Sakakibara, 2006). Among the detected IPT genes in the leaf, particularly the tRNA IPTs IPT9 (Fig. 6A) and IPT2 (Supplementary Fig. S3A) were upregulated by N starvation and in a cultivar-specific way. Both IPT isoforms catalyse the formation of cZ derived from tRNA degradation (Schmülling, 2004; Sakakibara, 2006). Although the cZ content in the leaf did not change, the IPT gene expression patterns indicated a higher leaf-inherent cZ biosynthesis in the stay-green cvs. NPZ-1 and Apex as compared to their respective early senescing counterpart cvs. NPZ-2 and Capitol under N starvation. Despite the fact, that cZ is less biologically active (Schmitz et al., 1972) and its biosynthesis requires a high tRNA turnover (Sakakibara, 2006), it might have significance for genotypic variation in functional stay-green, because during senescence a high tRNA turnover occurs (Buchanan-Wollaston, 1997) and cZ is less susceptible to degradation (Sakakibara, 2006). Moreover, the results of Miyawaki et al. (2004) support the view that in senescing leaves tRNA may be an important source for the leaf-inherent synthesis of aCKs.
In addition to the biosynthesis of biologically active CKs the spatial and temporal fine-regulated metabolic inter-conversion of CKs decisively affects the impact of CKs on plant processes (Schmülling, 2004; Sakakibara, 2006). The glycosylation of biologically active CKs lead to their inactivation. This is mediated by specific CK uridine diphosphate (UDP) glycosyltransferases (UGTs) forming either reversible O-glycosides or irreversible N-glycosides (Schmülling, 2004; Sakakibara, 2006). The UGTs encoded by UGT73C1, UGT73C5 and UGT85A1 particularly catalyse the formation of tZOG and DHZOG, whereas UGT76C1 is involved in the formation of irreversibly inactivated N-glycosides of tZ, DHZ, iP and cZ (Hou et al., 2004; Wang et al., 2013). Among the CK-specific UGTs, UGT85A1 has been identified as the key enzyme for the formation of tZOG in A. thaliana. The transcript amount correlated with the accumulation of tZOG in plant tissues (Hou et al., 2004; Jin et al., 2013). Transcripts of UGT85A1 were most abundant among the detectable UGT genes in winter oilseed rape leaves (Supplementary Fig. S4). In the complementary time-course experiment the expression pattern of UGT85A1 reflected the cultivar-specific tZOG accumulation in the leaf under both N supplies (Fig. 6B): UGT85A1 was more highly expressed in the leaf tissue of cv. NPZ-1 as compared to cv. NPZ-2, as well as in that of cv. Apex as compared to cv. Capitol, under both N starvation and optimal N supply. In addition the cultivar-specific higher upregulation of UGT73C4 and UGT73C5, particularly under N starvation, corresponded to the greater accumulation of iCKs in the leaf of the early-senescing cvs. NPZ-2 and Capitol as compared to their respective stay-green counterpart cvs. NPZ-1 and Apex (Supplementary Fig. S4B, C). In contrast, UGT73C1 was generally downregulated (Supplementary Fig. S4A). Although no N-glycoside CKs were determined, the expression pattern of UGT76C1 (Supplementary Fig. S4D) suggests that N-glycosylation did not play a role for leaf-inherent CK inter-conversion under N starvation.
An alternative pathway for limiting the biological activity of CKs is the attachment of ribose or ribose-5′-monophosphate to CK bases (Schmülling, 2004). The release of CK nucleobases from their respective CK ribose 5′-monophosphate is catalysed inter alia in a single-step by cytokinin-specific riboside 5′-monophosphate phosphoribohydrolase encoded by LONELY GUY (LOG) (Kuroha et al., 2009). Among the four LOGs detected in the leaf, particularly LOG7 (Fig. 6C) and also LOG4 (Supplementary Fig. S5B) were generally higher expressed under N starvation in the stay-green cvs. NPZ-1 and Apex as compared to their respective early-senescing counterpart cvs. NPZ-2 and Capitol. Particularly LOG7 has been identified as the key enzyme on the whole plant level for the release of highly biological active CKs from their corresponding ribosides (Tokunaga et al., 2012). But despite the cultivar-specific enhanced LOG7 (and LOG4) expression in the stay-green cultivars, the levels of the aCKs were not increased. In contrast to LOG4 and LOG7, LOG1 expression neither responded to the N supply nor the duration of the treatment (Supplementary Fig. S5A). Although occasionally cultivar differences occurred, no systematic expression pattern existed. However, the generally low change of LOG1 regulation is most likely the result of the comparable high basal transcript level (ΔCt=3.7±0.2). Although LOG5 showed the highest relative induction with the progression of N starvation-induced leaf senescence (Supplementary Fig. S5C) the initial (DAT0) and final (DAT12) expression levels (ΔCt) under both N supplies were comparable low. Thus it is most likely that LOG5 did not play a decisive role in leaf CK activation. This is in agreement with results in A. thaliana where the main sites of LOG5 transcription were the inflorescences (Kuroha et al., 2009).
The level of biologically active CKs is also determined by their rate of degradation (Schmülling, 2004; Sakakibara, 2006). The CK bases as well as their ribosides are irreversibly degraded by CK-specific oxidase/dehydrogenase (CKX) in a single-step by oxidative cleavage of the side chain (Schmülling et al., 2003). CKX activity is regulated on the transcriptional level, in which significant differences exist between the seven known isoenzymes in the ability to target specific CKs (Schmülling et al., 2003; Galuszka et al., 2007; Kowalska et al., 2010; Gajdošová et al., 2011; Trifunović et al., 2013; Köllmer et al., 2014). The regulation patterns of CKX1, CKX2, CKX3, CKX6 and CKX7 during N starvation-induced leaf senescence (Supplementary Fig. S6) suggest that CK degradation most likely did not play a major role for cultivar differences in functional stay-green.
The pool of biologically active CKs in the leaf may be insufficient to fully explain differences in CK-mediated leaf longevity. Of equal or even more importance is the binding of biologically active CKs to the CK-specific receptor histidine kinase AHK3, as well as the subsequent activation of the response regulator ARR2 (Kim et al., 2006). The response regulator ARR2 induces downstream CK-responsive genes and, directly or indirectly, induces or represses a set of target genes responsible for the regulation of leaf senescence (Kim et al., 2006). According to the cultivar differences in N starvation-induced leaf senescence AHK3 tended to be more highly upregulated in the stay-green cvs. NPZ-1 and Apex as compared to their respective early-senescing counterpart cvs. NPZ-2 and Capitol (Fig. 7A). In accordance to the regulation of AHK3, ARR2 too tended toward higher upregulation in the stay-green cvs. NPZ-1 and Apex as compared to their respective early-senescing counterpart cvs. NPZ-2 and Capitol (Fig. 7B). Thus, the expression patterns suggest that the binding of and the response to biologically active CKs mediated by AHK3 and ARR2 are additional leaf-inherent factors responsible for cultivar differences in N starvation-induced leaf senescence.
In conclusion, the present study revealed that cultivar differences in fuctional stay-green under N starvation were primarily governed by leaf-inherent factors. The specific leaf contents of CKs differing in biological activity and the expression of genes involved in CK homeostasis suggest that leaves of early-senescing cultivars were characterized by inactivation of biologically active CKs, whereas in fuctional stay-green cultivar synthesis, activation, binding of and response to biologically active CKs were favoured. Thus, the homeostasis of biologically active CKs was the predominant leaf-inherent factor for cultivar differences in N starvation-induced leaf senescence. The better understanding of the functional stay-green phenotype under N starvation could facilitate the breeding of N-efficient B. napus cultivars and thus contribute to reduce the large N balance surplus characteristic of this crop.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Comparison of cultivars for detaching and N starvation-induced leaf senescence.
Supplementary Fig. S2. SA, JA and ABA in root, the xylem sap and leaf of four cultivars.
Supplementary Fig. S3. Relative Expression of IPT2, IPT5 and IP9.
Supplementary Fig. S4. Relative Expression of UGT73C1, UGT73C4, UGT73C5, UGT76C1 and UGT85A1.
Supplementary Fig. S5. Relative Expression of LOG1, LOG4, LOG5 and LOG7.
Supplementary Fig. S6. Relative Expression of CKX1, CKX2, CKX3, CKX6 and CKX7.
Supplementary Fig. S7. N uptake of non-grafted, self-grafted and reciprocally-grafted plants.
Supplementary Fig. S8. N concentrations of non-grafted, self-grafted and reciprocally-grafted plants.
Supplementary Fig. S9. Correlation between specific leaf N content and photosynthesis rate.
Supplementary Table S1. Primer sequences of the genes of interest and the reference gene.
Supplementary Table S2. Statistical comparison of the two grafting experiments.
Supplementary Table S3. F test for the senescence status of the 3rd and 4th leaf.
Supplementary Table S4. F test for salicylic acid, jasmonic acid and abscisic acid.
Supplementary Table S5. F test for cytokinins.
Acknowledgments
Funding of the Forschergruppe FOR948 by the Deutsche Forschungs gemeinschaft (DFG) is gratefully acknowledged.
References
- Albacete Moreno A, Cantero AE, Grosskinsky D, et al. 2014. Ectopic overexpression of the cell wall invertase gene CIN1 increases water use efficiency in tomato (Solanum lycopersicum L.) and confers extreme drought tolerance. Journal of Experimental Botany 66, 863–878.25392479 [Google Scholar]
- Arvidsson S, Kwasniewski M, Diego M, Riaño-Pachón DM, Mueller-Roeber B. 2008. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. 2008. Transcription factors regulating leaf senescence in Arabidopsis thaliana . Plant Biology 10, 63–75. [DOI] [PubMed] [Google Scholar]
- Balibrea Lara ME, Gonzalez MCG, Fatima T, Ehneß T, Lee TK, Roels R, Tanner W, Roitsch T. 2004. Extracellular invertase is an essential component of cytokinin mediated delay of senescence. Plant Cell 16, 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry PM, Spink J, Foulkes MJ, White PJ. 2010. The physiological basis of genotypic differences in nitrogen use efficiency in oilseed rape (Brassica napus L.). Field Crops Research 119, 365–373. [Google Scholar]
- Buchanan-Wollaston V. 1997. The molecular biology of leaf senescence. Journal of Experimental Botany 48, 181–199. [Google Scholar]
- Dai J, Dong H. 2011. Stem girdling influences concentration of endogenous cytokinins and abscisic acid in relation to leaf senescence in cotton. Acta Physiologiae Plantarum 33, 1697–1705. [Google Scholar]
- del Mar Rubio-Wilhelmi M, Reguera M, Sanchez-Rodriguez E, Romero L, Blumwald E, Ruiz JM. 2014. PSARK::IPT expression causes protection of photosynthesis in tobacco plants during N deficiency. Environmental and Experimental Botany 98, 40–46. [Google Scholar]
- Desclos M, Dubousset L, Etienne P, Le Caherec F, Satoh H, Bonnefoy J, Ourry A, Avice JC. 2008. A proteomic approach to reveal a novel role of Brassica napus drought 22 kD/water-soluble chlorophyll-binding protein in young leaves during nitrogen remobilization induced by stressful conditions. Plant Physiology 147, 1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Niu Y, Li W, Zhang D. 2008. Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. Journal of Experimental Botany 59, 1295–1304. [DOI] [PubMed] [Google Scholar]
- Etienne P, Desclos M, Gou LL, Gombert J, Bonnefoy J, Maurel K, Le Dily F, Ourry A, Avice JC. 2007. N-protein mobilisation associated with the leaf senescence process in oilseed rape is concomitant with the disappearance of trypsin inhibitor activity. Functional Plant Biology 34, 895–906. [DOI] [PubMed] [Google Scholar]
- Evans JR. 1989. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78, 9–19. [DOI] [PubMed] [Google Scholar]
- Gajdošová S, Spíchal L, Kamínek M. 2011. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. Journal of Experimental Botany 62, 2827–2840. [DOI] [PubMed] [Google Scholar]
- Galuszka P, Popelková H, Werner T, Frébortová J, Pospísilová H, Mik V, Köllmer I, Schmülling T, Frébort I. 2007. Biochemical characterization of cytokinins oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. Journal of Plant Growth Regulation 26, 255–267. [Google Scholar]
- Gan S, Amasino RM. 1995. Inhibition of leaf senescence by autoregulated production of cytokinins. Science 270, 1986–1988. [DOI] [PubMed] [Google Scholar]
- Grbić V. 2003. SAG2 and SAG12 protein expression in senescing Arabidopsis plants. Physiologia Plantarum 119, 263–269. [Google Scholar]
- Gregersen PL, Culetic A, Boschian L, Krupinska K. 2013. Plant senescence and crop productivity. Plant Molecular Biology 82, 603–622. [DOI] [PubMed] [Google Scholar]
- Großkinsky DK, Albacete A, Jammer A, Krbez P, van der Graaff E, Pfeifhofer H, Roitsch T. 2014. A rapid phytohormone and phytoalexin screening method for physiological phenotyping. Molecular Plant 7, 1053–1056. [DOI] [PubMed] [Google Scholar]
- He P, Osaki M, Takebe M, Shinano T, Wasaki J. 2005. Endogenous hormones and expression of senescence-related genes in different senescent types of maize. Journal of Experimental Botany 56, 1117–1128 [DOI] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. 2004. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana . Journal of Biological Chemistry 279, 47822–47832. [DOI] [PubMed] [Google Scholar]
- Jin SH, Ma XM, Kojima M, Sakakibara H, Wang YW, Hou BK. 2013. Overexpression of glucosyltransferase UGT85A1 influences trans-Zeatin homeostasis and trans-zeatin responses likely through O-glucosylation. Planta 237, 991–999. [DOI] [PubMed] [Google Scholar]
- Kamh M, Wiesler F, Ulas A, Horst WJ. 2005. Root growth and N-uptake of oilseed rape (Brassica napus L.) cultivars differing in nitrogen efficiency. Journal of Plant Nutrition and Soil Science 168, 130–137. [Google Scholar]
- Kim HJ, Hojin R, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. 2006. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America 103, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeslin-Findeklee F, Meyer A, Girke A, Beckmann K, Horst WJ. 2014. The superior nitrogen efficiency of winter oilseed rape (Brassica napus L.) hybrids is not related to delayed nitrogen starvation-induced leaf senescence. Plant and Soil 384, 347–362. [Google Scholar]
- Koeslin-Findeklee F, Safavi Rizi V, Becker MA, Parra-Londono S, Arif M, Balazadeh S, Mueller-Roeber M, Kunze R, Horst WJ. 2015. Transcriptomic analysis of nitrogen starvation- and cultivar-specific leaf senescence in winter oilseed rape (Brassica napus L.). Plant Science 233, 174–185. [DOI] [PubMed] [Google Scholar]
- Kowalska M, Galuszka P, Frébortová J, Ŝebela M, Béres T, Hluska T, Ŝmehilová M, Bilyeu KD, Frébort I. 2010. Vacuolar and cytosolic cytokinin dehydrogenases of Arabidopsis thaliana . Phytochemistry 71, 1970–1978. [DOI] [PubMed] [Google Scholar]
- Köllmer I, Novák O, Strnad M, Schmülling T, Werner T. 2014. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant Journal 78, 359–371. [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokungaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. 2009. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis . Plant Cell 21, 3152–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Tanaka A, Tanaka R. 2013. Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynthesis Research 117, 221–234. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. 2004. Expression of cytokinin biosynthetic isopentenyltransferaes genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant Journal 37, 128–138. [DOI] [PubMed] [Google Scholar]
- Moroni JS. 1997. Studies on the efficiency of rapeseed (Brassica napus L.) germplasm for the acquisition and the utilization of inorganic nitrogen. PhD thesis, University of Alberta, Canada. [Google Scholar]
- Parlitz S, Kunze R, Mueller-Roeber B, Balazadeh S. 2011. Regulation of photosynthesis and transcription factor expression by leaf shading and re-illumination in Arabidopsis leaves. Journal of Plant Physiology 168, 1311–1319. [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Dortay H, Matallana-Ramírez LP, Waters MT, Nam HG, Lim PO, Mueller-Roeber B, Balazadeh S. 2013. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Reports 14, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A. 2012a. Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis . Plant Cell Physiology 53, 505–517. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paeck NC. 2012b. Stay-green and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis . Plant Cell 24, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RY, Skoog F, Playtis AJ, Leonard NJ. 1972. Cytokinins: synthesis and biological activity of geometric and position isomers of zeatin. Plant Physiology 50, 702–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T, Werner T, Riefler M, Krupková E, Bartina y Manns B. 2003. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. Journal of Plant Research 116, 241–252. [DOI] [PubMed] [Google Scholar]
- Schmülling T. 2004. Cytokinin. In: Lennarz W and Lane MD, eds. Encyclopedia of Biological Chemistry. Academic Press/Elsevier Science. [Google Scholar]
- Schulte auf’m Erley G, Wijaya KA, Ulas A, Becker H, Wiesler F, Horst WJ. 2007. Leaf senescence and N uptake parameters as selection traits for nitrogen efficiency of oilseed rape cultivars. Physiologia Plantarum 130, 519–531. [Google Scholar]
- Schulte auf’m Erley G, Ambebe TF, Worku M, Bänzinger M, Horst WJ. 2010. Photosynthesis and leaf-nitrogen dynamics during leaf senescence of tropical maize cultivars in hydroponics in relation to N efficiency in the field. Plant and Soil 330, 313–328. [Google Scholar]
- Schulte auf’m Erley G, Behrens T, Ulas A, Wiesler F, Horst WJ. 2011. Agronomic traits contributing to nitrogen efficiency of winter oilseed rape cultivars. Field Crops Research 124, 114–123. [Google Scholar]
- Smart CM. 1994. Gene expression during leaf senescence. New Phytologist 126, 419–448. [DOI] [PubMed] [Google Scholar]
- Steibel JP, Poletto R, Coussens PM, Rosa GJM. 2009. A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94, 146–152. [DOI] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ. 2000. Five ways to stay green. Journal of Experimental Botany 51, 329–337. [DOI] [PubMed] [Google Scholar]
- Thomas H, Ougham H. 2014. The stay-green trait. Journal of Experimental Botany 65, 3889–3900. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H. 2012. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant Journal 69, 355–365. [DOI] [PubMed] [Google Scholar]
- Trifunović M, Cingel A, Simonović A, Jevremović S, Petrić M, Dragićević IC, Motyka V, Bobrev PI, Zahajská L, Subotić A. 2013. Overexpression of Arabidopsis cytokinin oxidase/dehydrogenase genes AtCKX1 and AtCKX2 in transgenic Centaurium erythraea Rafn. Plant Cell, Tissue and Organ Culture 115, 139–150. [Google Scholar]
- Wang A, Li Y, Zhang C. 2012. QTL mapping for stay-green in maize (Zea mays). Canadian Journal of Plant Science 92, 249–256. [Google Scholar]
- Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. 2013. Glucosyltransgerase UGT76C1 finely modulates cytokinin responses via cytokinin N-glucosylation in Arabidopsis thaliana . Plant Physiology and Biochemistry 65, 9–16. [DOI] [PubMed] [Google Scholar]
- Wingler A, Roitsch T. 2008. Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biology 10, 50–62. [DOI] [PubMed] [Google Scholar]
- Zwack PJ, Rashotte AM. 2013. Cytokinin inhibition on leaf senescence. Plant Signal and Behavior 8, e24737; http://dx.doi.org/10.4161/psb.24737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.