Highlight
MYB and bHLH bind to two identifiable syntaxes present in target gene promoters, and the anthocyanin pathway is regulated as one unit as in other branches of the flavonoid network.
Key words: Anthocyanin pathway, Arabidopsis, bHLH, cis element, Ipomoea, MBW complex, MYB, promoter activity, promoter architecture, regulatory module, trans factor, transcription initiation, WDR.
Abstract
Cellular activities such as compound synthesis often require the transcriptional activation of an entire pathway; however, the molecular mechanisms underlying pathway activation have rarely been explained. Here, the cis regulatory architecture of the anthocyanin pathway genes targeted by the transcription factor (TF) complex including MYB, bHLH, and WDR was systematically analysed in one species and the findings extended to others. In Ipomoea purpurea, the IpMYB1-IpbHLH2-IpWDR1 (IpMBW) complex was found to be orthologous to the PAP1-GL3-TTG1 (AtPGT) complex of Arabidopsis thaliana, and interacted with a 7-bp MYB-recognizing element (MRE) and a 6-bp bHLH-recognizing element (BRE) at the proximal promoter region of the pathway genes. There was little transcription of the gene in the absence of the MRE or BRE. The cis elements identified experimentally converged on two syntaxes, ANCNNCC for MREs and CACN(A/C/T)(G/T) for BREs, and our bioinformatic analysis showed that these were present within anthocyanin gene promoters in at least 35 species, including both gymnosperms and angiosperms. For the anthocyanin pathway, IpMBW and AtPGT recognized the interspecific promoters of both early and later genes. In A. thaliana, the seed-specific TF complex (TT2, TT8, and TTG1) may regulate all the anthocyanin pathway genes, in addition to the proanthocyanidin-specific BAN. When multiple TF complexes in the anthocyanin pathway were compared, the cis architecture played a role larger than the TF complex in determining the variation in promoter activity. Collectively, a cis logic common to the pathway gene promoters was found, and this logic is essential for the trans factors to regulate the pathway.
Introduction
Anthocyanins are widely synthesized in seed plants to provide colouration, protection under various circumstances, and components for cellular activities. The synthesis of anthocyanins is the end product of genetically well characterized enzymes (Winkel-Shirley, 2001). These enzymes also comprise the backbone of the flavonoid synthesis network, supporting the network to metabolize arrays of secondary compounds in different species (Vogt, 2010). Although the regulation of the anthocyanin pathway genes has been known to be under the control of a transcription factor (TF) complex that consists of MYB, bHLH, and WD-repeat (WDR) proteins (Broun, 2004; Koes et al., 2005; Ramsay and Glover, 2005; Xu et al., 2015), little consensus has been reached about the cis elements recognized by the complex at the pathway level. The MYB-bHLH-WDR (MBW) complex represents a mode of gene regulation not only for the anthocyanin pathway but also proanthocyanidin (PA) synthesis and epidermal cell differentiation (reviewed by Feller et al., 2011). Systematic examination of the cis structures involved in the regulation of pathway genes is important for understanding these biological processes.
The involvement of MYB and bHLH in flavonoid gene regulation was initially found in Zea mays, with mutants of c1 (Paz-Ares et al., 1987) and Lc (Ludwig et al., 1989), respectively; the necessity for both MYB and bHLH in the activation of the anthocyanin genes was soon established in this species (Goff et al., 1990; Bodeau and Walbot, 1992). Still in maize, details followed that the N-terminus of B (a bHLH) directly interacted with the R2R3 domain MYB C1 (Goff et al., 1992), and that the C-terminus of C1 was responsible for transcriptional activation (Sainz et al., 1997). Interestingly, C1 and Lc were both required for restoring the phenotype of an11 mutant in Petunia (Quattrocchio et al., 1993), and AN11 was later found to be a WDR regulating the anthocyanin pathway (deVetten et al., 1997). The participation of WDR in the pathway was further validated with Arabidopsis TTG1 (Walker et al., 1999) and Zea PAC1 (Carey et al., 2004).
In Arabidopsis, characterization of MBWs led to the identification of PAP1-GL3/EGL3-TTG1 transiently expressed in the seedlings for anthocyanin synthesis (Zhang et al., 2003; Gonzalez et al., 2008) and TT2-TT8-TTG1 in developing siliques for PA synthesis (Nesi et al., 2001; Baudry et al., 2004; Lepiniec et al., 2006). At the same time, anthocyanin regulatory genes MYB1, bHLH2, and WDR1 were reported from Ipomoea mutants (Chang et al., 2005; Morita et al., 2006; Park et al., 2007), complementing previously well characterized AN2-AN1-AN11 in the Petunia hybrid Vilmorin (Beld et al., 1989; Quattrocchio et al., 1993; deVetten et al., 1997). Nonetheless, the collaborative action of the Ipomoea TFs requires more evidence. Reported components of the anthocyanin MBW complexes appear to form their own clades, with MYBs from the subgroup 6 (Dubos et al., 2010), bHLHs (subfamily IIIf) (Pires and Dolan, 2010), and recently WDRs (subgroup 19) (Li et al., 2014).
Binding of MYB and bHLH to promoters of anthocyanin structural genes was first reported with maize C1 and B (Roth et al., 1991). Details showed that from –123 to –88bp of the A1 promoter was critical for C1/B activation (Tuerck and Fromm, 1994); a region within –224bp of the Bz2 promoter was adequately regulated by R (a B homologue) and C1 (Bodeau and Walbot, 1996). For the MYB part, C1 could bind to variable sites in the maize a1 gene promoter (Sainz et al., 1997), and a 16 bp-long consensus motif was identified from a2 and other C1-binding genes (Lesnick and Chandler, 1998); recently, the consensus was narrowed down to ANCNACC by site mutagenesis tests with anthocyanin MYB1s in coloured Ipomoea petals and magnolia tepels (Wang et al., 2013). For the bHLH part, after reported binding of CG-1 protein (Staiger et al., 1991) and human c-MYC (Blackwell et al., 1990) to CACGTG, the G-box was recently shown to bind to maize R (Kong et al., 2012), petunia AN1, and Ipomoea bHLH2 (Wang et al., 2013). EMSA tests on anthocyanin gene promoter fragments involving Ipomoea CHS-D, Zea 3GT, and Gerbera DFR led to a tentative consensus of bHLH-recognized elements in the form of CACNN(G/T) (Wang et al., 2013). All these analyses agreed in that the binding of MYB and bHLH occurred at locations within 200bp upstream of the translation start site. This feature of cis locations was also reported in bean (Loake et al., 1992), grape (Gollop et al., 2002), African daisy (Elomaa et al., 2003), and apple (Espley et al., 2009), indicating that the short functional promoters are common to the anthocyanin genes. The focus of these studies was usually on one or a few promoters. Analysis of TF–promoter interactions at the level of pathway has been limited to several Arabidopsis promoters activated by maize TFs (Hartmann et al., 2005). How conspecific pathway genes interact in the 5′-noncoding region remains to be explored. Obviously, conspecific TF interactions are most relevant to understanding pathway regulation.
In documented MBW complexes, specific residues were identified on maize C1 for its interaction with R (Grotewold et al., 2000), while protein interaction was also observed between Arabidopsis GL3 and TTG1 (Payne et al., 2000); however, little interaction was observed between MYB and WDR (Zhang et al., 2003). Meanwhile, transcription of Arabidopsis TT8 and petunia AN1 require the presence of both MYB and WDR (Baudry et al., 2006; Albert et al., 2014). For transcription of MYB and WDR, however, the necessity for the presence of MBW remains unclear, except in one case of a MYB mutant (Espley et al., 2009). With all that is known about MBW regulation, however, little can be said about the conspecific interactions between the trans and cis components. Although anthocyanin TFs have been found to be conserved in both monocots and dicots (e.g. Quattrocchio et al., 1993; Hartmann et al., 2005), the molecular mechanism for observed trans-specific regulation is unclear. While the lack of identified cis components and the range of a gene’s promoter previously prevented systematic and quantitative analysis of the regulatory mechanism, accumulating data indicate that transcription in metazoa typically occurs within the proximal promoter region (Lenhard et al., 2012), which appears true also for the anthocyanin genes. With a recently reported method (Wang et al., 2013), the cis element(s) may be pinpointed to an anthocyanin promoter through both bioinformatic and experimental approaches, and quantitative evaluation of contributions of TFs and cis elements to gene regulation is now achievable.
In order to elucidate the molecular mechanism(s) of anthocyanin pathway regulation in the proximal regions, we analysed interactions of TFs and cis motifs of conspecific genes at the pathway level in I. purpurea Roth (common morning glory) and made parallel examinations of the homologous genes in Arabidopsis thaliana (L.) Heynh. as a reference. The I. purpurea genes included those encoding chalcone synthase D (CHS-D), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol reductase B (DFR-B), anthocyanidin synthase (ANS), UDP-glucose:flavonoid 3-O-glucosyltransferase (3GT), UDP-glucose:anthocyanidin 3-O-glucoside-2′-O-glucosyltransferase (3GGT), and the trans factors MYB1, bHLH2, and WDR1. Transcriptional activities of MBW complexes were subsequently compared between I. purpurea and Arabidopsis on interspecific and conspecific promoters. A bioinformatic analysis of homologues in multiple species was used to explore the generality of cis patterns (initially identified in I. purpurea). These extensive analyses revealed the features of the MYB- and bHLH- recognizing elements (MREs and BREs, respectively), patterns of MREs and BREs on the proximal promoters, and one cis logic for pathway regulation.
Materials and methods
Plant materials
Seeds of I. purpurea taken from a nationwide collection (Lu et al., 2009) were grown in the botanical garden of IB-CAS, Beijing. These plants provided petal tissue for anthocyanin gene cloning and quantitative analysis of gene expression, and leaf tissue for promoter amplification and genotyping. Seeds of Arabidopsis (tt3 mutant and the Columbia accession) were obtained from the Arabidopsis information resource (TAIR) and those of Petunia hybrida (R27) were a kind gift from Dr Quattrocchio (Free University, The Netherlands). All seeds were germinated in the growth chamber.
qRT-PCRs
RNA samples were extracted in TRIzol (Life Technologies) from petals of I. purpurea (III6-4 and III6-9). Each plant was sampled at 2-h intervals starting 60h before flower opening. RNAs were reverse transcribed and quantifications of transcript copies followed the protocol reported by Lu et al. (2012). About 1–10ng cDNA was used in each qPCR reaction, and the running conditions were optimized for efficiency with each gene taken (Supplementary Table S1).
Gene cloning and sequencing
Standard PCRs were applied to leaf genomic DNAs to amplify five gene promoters (IpCHS-D, IpF3′H, IpDFR-B, IpbHLH2, and IpWDR1) based on primers (Supplementary Table S1) whose designs followed the available sequences of conspecific or closely related targets (Supplementary Table S2). The promoter sequence of IpF3H was obtained by inverse PCR (Ochman et al., 1988). Promoter sequences of IpMYB1, IpCHI, IpANS, Ip3GT, and Ip3GGT were isolated by tail-PCRs (Liu and Whittier, 1995). Homologue promoters from Arabidopsis were PCR-amplified via primers targeting the Columbia accession (Supplementary Table S1). Ipomoea TFs were reverse transcribed from RNAs extracted from fresh petals of I. purpurea. Arabidopsis TFs were similarly amplified from seedlings of the Columbia accession with the appropriate primers (Supplementary Table S1).
Dual luciferase assays
The promoters were ligated into vectors to prepare for dual luciferase assays as described by Wang et al. (2013). The complete coding regions of TFs were integrated into pJIT163 (Guerineau et al., 1992) in frame with appropriate restriction sites (Supplementary Table S1) to replace its reporter gene, resulting in effector vectors. The reporter vectors were adapted from pJIT163-eGFP with the CaMV 35S promoters replaced by the proximal promoters and GFP replaced by the firefly luciferase (LUC) gene. The reference vector was also based on pJIT163-eGFP, with its GFP replaced by the renilla luciferase (RUC) gene (Wang et al., 2013). The vectors were introduced into leaf cells of the Ipomoea nil wdr1 mutant (seeds available on request) by particle bombardment (BioRad PDS-1000/He equipment). A construct mixture (0.5 μg effector, 0.5 μg reporter, and 0.1 μg reference for each shot) was prepared for the plasmid cocktail, which was coated with 50mg ml–1 microparticles (Bio-Rad) at 2.0 μl per 1.0 μg plasmid DNA. The complex was then mixed with 2.5M CaCl2 and 0.1M spermidine in a ratio of 1:5:2. Other details followed Wang et al. (2013).
Yeast two-hybrid assays
The coding sequences of IpMYB1, IpbHLH2, and IpWDR1 were integrated in frame into vectors pAD-GAL4 2.1 and pBD-GAL4 Cam of HybriZAP-2.1 (Stratagene), respectively, according to the manufacturer’s instructions. Constructs were introduced in pairs into the YRG-2 yeast strain (Stratagene). The transformants were plated onto SD media minus histidine, leucine, and tryptophan for identifying positive interactions. The strengths of TF interactions were then measured by β-galactosidase activities according to the protocol of the Matchmaker system (Clontech).
Electrophoretic mobility shift assays
A complete coding sequence of MYB was integrated in frame into pMAL-c2G (New England Biolabs) to express a tagged protein consisting of maltose-binding protein (MBP) and MYB. A partial coding region of bHLH was similarly expressed. The vector-transformed Escherichia coli strain BL21 (DE3) was incubated at 37°C in LB solution and treated as described (Wang et al., 2013). Protein quantification followed the Bradford method using the bovine gamma globulin (BIO-RAD) as the standard. Probes were quantified by the Picogreen quantification module of Rotor Gene 3000 (Corbett Research) with λDNA (Life technologies) as the reference. Details for binding reactions and gel documentation followed Wang et al. (2013).
Transgenic experiments
Homozygous Arabidopsis tt3 mutants were grown in the growth chamber for transgenic tests of the IpDFR-B promoter series. The coding regions of IpDFR-B were first subcloned into the SN1301 binary vector for the construction of SN1301-35S::DFR-Bcoding. Varying promoter regions (IpDFRB-1559, IpDFRB-191, and IpDFRB-166) independently replaced the 35S promoter to generate series of SN130-proDFR::DFR-Bcoding vectors. These vectors were singly introduced into Agrobacterium strain GV3101 via the standard freeze-thaw protocol. Transformation of Arabidopsis followed a modified flower dip protocol (Logemann et al., 2006), and the selection of transformants was on MS medium containing 50mg l–1 hygromycin and 50mg l–1 carbenicillin. DNAs were subsequently extracted from these plants and scored for the presence of the transgene by PCRs and DNA sequencing.
Phenotypic complementation was observed in the T2 generation. Seeds were sterilized and cultivated on 1/2MS medium containing 5% sucrose, kept at 4°C in darkness for 2 days, and transferred to constant light conditions (100 μmol m–2 s–1, 24°C) for observations. For 4–5-week old seedlings, treatment by dehydration (up to a week) and a low temperature (10–16°C) also induced anthocyanin accumulation. Anthocyanins extracted from representative seedlings were further examined by high performance liquid chromatography using cyanidin-3-O-glucoside (Extrasynthese) as the standard.
Bioinformatic analysis
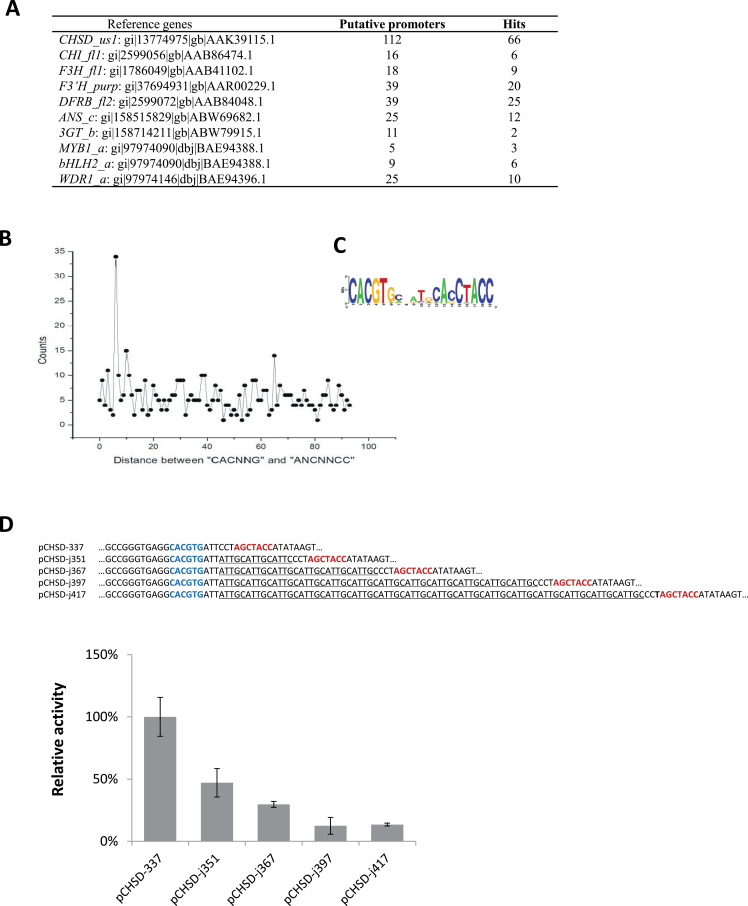
Multiple blastp searches in the NCBI database (ftp://ftp.ncbi.nih.gov/genbank/, up to 10 October 2012) identified protein homologues for each of the anthocyanin genes. The results were trimmed for redundancy following the protocol reported by Wang et al. (2013). For each of the promoters, a sliding window (6-bp width) was initially adapted to search for four or more bases matching CACNNG; if this condition was met, the second sliding window of 7bp was taken to examine whether or not there were four or more bases matching ANCNNCC in the next 100 bases. When both searches returned hits, the non-coding sequences were recorded and the candidate cis elements were uppercased by WxW_Align.gene, and the frequency of each case was counted by WxW_Align.PosD (Supplementary File 1). We ended up with 159 sequences satisfying the search criteria, which provided the basis for mapping the natural distribution of known cis elements on anthocyanin gene promoters. The sequences included in the peak of the distribution were further characterized by two motifs identified via MEME software following the parameters of Wang et al. (2013).
Statistical analysis
Standard t-tests were performed on comparisons between averages of experiments. Two-way ANOVAs were carried out after log transformation of the promoter activity. Pearson correlation coefficients were calculated with the significance levels corrected for multiple comparisons by the Dunn-Šidák method (taking the experiment-wise error rate α = 0.05). All tests were computed using SAS (ver. 9.0).
Results
IpMYB1, IpbHLH2, and IpWDR1 collaboratively regulate the anthocyanin pathway genes
Although the regulatory genes IpMYB1, IpbHLH2, and IpWDR1 are expressed in the corolla of I. purpurea (Chang et al., 2005; Morita et al., 2006; Park et al., 2007; Guan and Lu, 2013), whether or not they regulate the anthocyanin pathway as a complex remains to be confirmed. If they do, expression of the anthocyanin pathway genes is expected to be correlated. We quantified the pair-wise Pearson correlation coefficients among transcript levels of ten anthocyanin genes via reversely transcribed quantitative PCRs (RT-qPCRs) in developing petals of two wild-type I. purpurea individuals (Supplementary Table S3). The transcript levels of the enzyme-coding (structural) genes (IpCHS-D, IpCHI, IpF3H, IpDFR-B, and IpANS) were significantly correlated with those of regulatory genes (IpMYB1, IpbHLH2, and IpWDR1) and among themselves (except between IpDFR-B and Ip3GT). The expression of IpF3′H was highly correlated with that of IpbHLH2 and IpWDR1, but not with that of IpMYB1. The quantitative data largely agreed with the perception that IpMYB1, IpbHLH2, and IpWDR1 regulated pathway transcription collaboratively. The features of the TFs were examined in two further experiments. One showed the subcellular localization of the three TFs. The vectors with fluorescent-tagged TFs were introduced into the onion epidermis (Supplementary Figure S1A), and the in vivo images indicated that IpbHLH2 and IpWDR1 were localized in both the nucleus and the cytoplasm, whereas IpMYB1 was mostly in the nucleus. This pattern agrees with the documented subcellular sites of their homologues (Ness et al., 1989; Matus et al., 2010). The other experiment described protein interactions via yeast two-hybrids. Quantifications of β-galactosidase activities suggested significant interactions between IpMYB1 and IpbHLH2 as well as between IpbHLH2 and IpWDR1 (Supplementary Figure S1B), as expected.
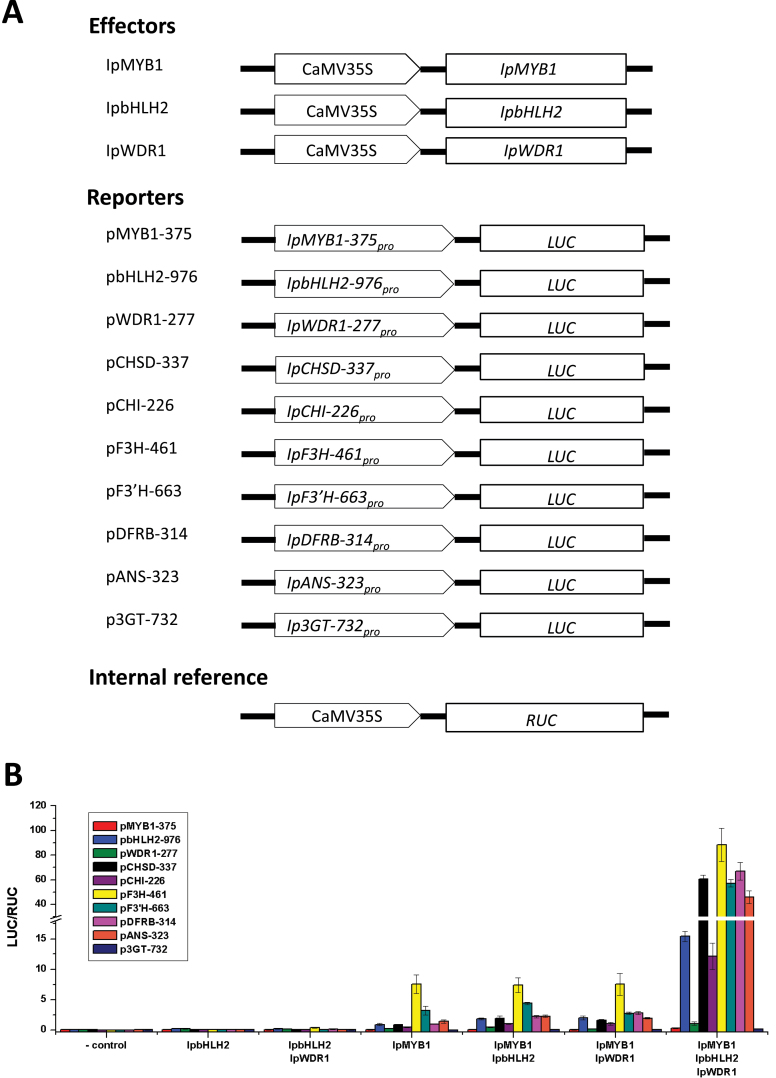
The TF validations paved the way for identification of the conspecific structural genes of the pathway. For I. purpurea, some pathway genes (IpCHS-D, IpF3′H, and IpDFR-B) were characterized through mutants or mutant complementation (Habu et al., 1998; Hoshino et al., 2003; Zufall and Rausher, 2004), while others, including IpCHI, IpF3H, IpANS, and Ip3GT, were largely identified via gene expression and sequence homology to genes identified in other model species (Tiffin et al., 1998; Durbin et al., 2000; Clegg and Durbin, 2003; Lu et al., 2009). Assuming that they were the bona fide genes of the anthocyanin pathway, we collected their 5′-noncoding sequences upstream of the translation start sites (Supplementary Table S2); similar information was also gathered for IpMYB1, IpbHLH2, and IpWDR1 to evaluate the TF regulatory capacity on the expression of each. By single and combinatory tests of the TFs as effectors in dual luciferase assays (Fig. 1A), we detected how IpMYB1, IpbHLH2, and IpWDR1 (IpMBW) regulated the reporter gene via the 5′-noncoding regions of the pathway genes. We found that seven (IpCHS-D, IpCHI, IpF3H, IpF3′H, IpDFR-B, IpANS, and IpbHLH2) of the ten promoter constructs could be activated strongly by IpMBW; however, the constructs hosting the IpWDR1 5′-noncoding regions responded only at low levels to IpMBW, and little promoter activity was found on those hosting various lengths of Ip3GT and IpMYB1 promoter sequences (Supplementary Figure S1C). The tests also showed that IpMYB1 could, alone, initiate a low level of transcription on the seven promoters (particularly F3H). By contrast, IpbHLH2 or IpWDR1 alone failed to activate observable promoter activities in the same setting. Only the collective presence of all three TF effectors generated considerable promoter activities (Fig. 1B), suggesting that the TFs acted as an IpMBW complex in transcriptional activation of the main anthocyanin pathway genes in I. purpurea. The activation of IpMBW on the promoter of IpbHLH2 resembles that of TT8 in Arabidopsis (Baudry et al., 2006).
Fig. 1.
Collaborative regulation of IpMYB1, IpbHLH2, and IpWDR1 on the conspecific genes of the anthocyanin pathway of I. purpurea. (A) Construct details for dual luciferase assays. Effectors and reporters were based on the same plasmid pJIT163 (black line). The promoter region of each construct is shown by an arrowed rectangle, while the coding region is shown by the plain rectangle. The firefly luciferase (LUC) was taken as the reporter gene while renilla luciferase (RUC) was the reference for internal control. (B) Results of transient expression assays expressed as the ratios of LUC and RUC for trials on different combinations of TFs. The 5′-noncoding regions are shown using bars for the ten genes tested. The negative controls were the reporters only. Effectors were added as shown on the x-axis for each reporter, with the error bars based on four replicates. This figure is available in colour at JXB online.
Two syntaxes emerge from identification of cis elements in pathways
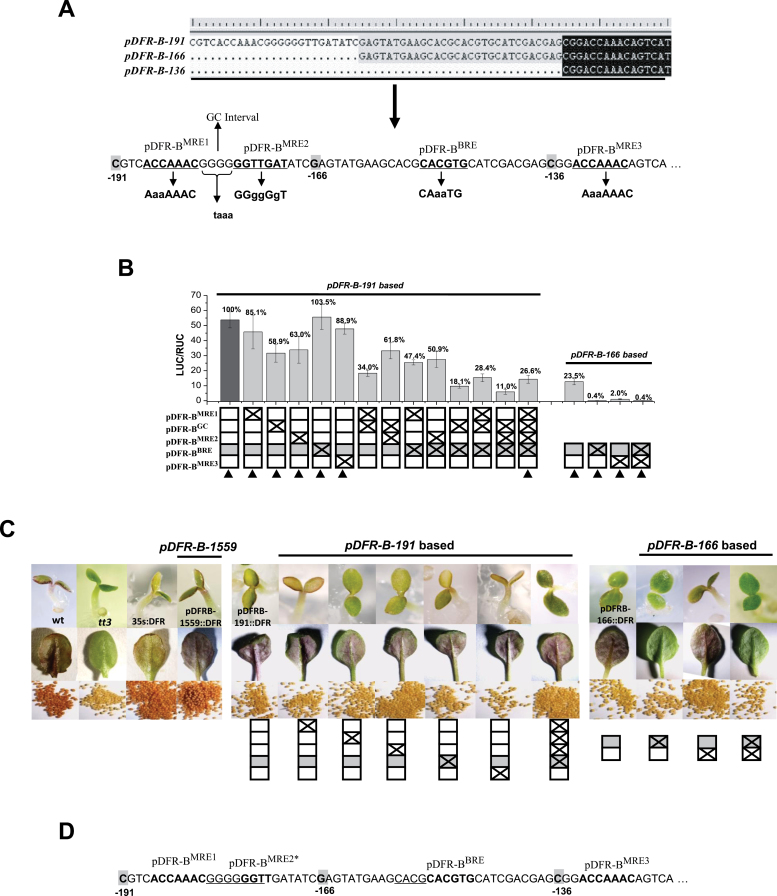
The central aim of our study was to understand how the TFs interacted with the cis elements at the target gene promoters. Our identification of cis elements was extended from IpCHS-D pro (Wang et al., 2013) to all other gene promoters including IpCHI pro, IpF3H pro, IpF3′H pro, IpDFR-B pro, IpANS pro, Ip3GGT pro, IpWDR1 pro, and IpbHLH2 pro. Both sequential deletion tests (Supplementary Figure S1C) on the cloned 5′-noncoding regions of the anthocyanin genes and site-by-site tests on the predicted motifs (concentrated on the proximal promoter regions) were adapted. In the initial case of IpDFR-B, three experimental approaches were taken for site-specific analysis, including dual luciferase assays performed on leaves of the wdr1 mutant of I. nil, gene transformations of the tt3 mutant of Arabidopsis, and electrophoretic mobility shift assays (EMSAs) with IpMYB1 and IpbHLH2. Starting by mutating predicted motifs (Fig. 2A), we examined the effects of the mutations in both dual luciferase assays (Fig. 2B) and transgenic plants (Fig. 2C). The presence of multiple MREs was evident for IpDFR-B-191 pro as mutations at one of the MREs did not much affect the transcriptional intensity, and only mutations at multiple motifs led to a significant reduction in promoter activity (Fig. 2B). The initially suspected effect of the ‘GC interval’ overlapped with that of pDFR-BMRE2, and the two were better explained by the modified pDFR-BMRE2* (Fig. 2D). The function of pDFR-BMRE2* was further supported by the EMSA assay with the probe (DFRBpro) hosting a suspected motif (GGGGGTT or GGGGGGT, a reverse complement of MRE) as mutations at the site (DFRB-M4) demolished the binding capacity of IpMYB1 (Fig. 3A). In the transgenic results, promoter activity as little as 2% could still result in anthocyanin accumulation in the seedlings of Arabidopsis (e.g. pDFR-B-166 MRE3; Fig. 2C); the promoter construct IpDFRB-1559 pro ::IpDFRB-coding caused a higher anthocyanin accumulation than that of 35S::DFRB-coding in the tt3 background (Fig. 2C), suggesting that the 35S promoter did not lead to excessive gene expression in vivo. This was relevant to the dual luciferase assay where the effectors were driven by 35S promoters.
Fig. 2.
Analysis of cis elements on IpDFR-B pro.(A) Sequence features of pDFR-B-191, pDFR-B-166, and pDFR-B-136. Predicted cis motifs on pDFR-B-191are in bold and underlined, and the mutated sites shown in lower case. The nucleotides that are shaded and in bold mark the beginnings of pDFR-B-191, pDFR-B-166, and pDFR-B-136. (B) Results of dual luciferase assays. Effectors were IpMYB1, IpbHLH2, and IpWDR1 (0.5 µg each). The black bar for pDFRB-191 was set as 100%. Mutation types (BRE in grey) are checked boxes. The error bars indicate the standard errors based on four independent trials. The tests highlighted by triangles were further examined by the following transgenic trials. (C) Phenotypes of wild type, dfr mutant (tt3), and transgenic seedlings of Arabidopsis. The upper panel shows 2–3 day old seedlings. Some mature leaves were induced by 10–18°C and dehydration. The seeds of each line are shown in the lower panel. The schematic boxes follow the same notation as in (B). (D) A summary of cis motifs on IpDFR-B-191 pro. The cis elements BRE and MRE3 were confirmed on IpDFR-B-166 pro in the mutation tests of transient expression shown in (B) and the additional transgenic in vivo test of BRE shown in (C). MRE1 and MRE3 are identical in sequence and weak in effect as seen in (B). Effects of the previous MRE2 and GC interval in (B) were similar whether alone or in combination, and are thus best explained by a unified MRE2*, which was also supported in the subsequent EMSA test with the IpDFRB-m4 probe. The underlined motifs are possibe additional targets of the TFs in vivo. This figure is available in colour at JXB online.
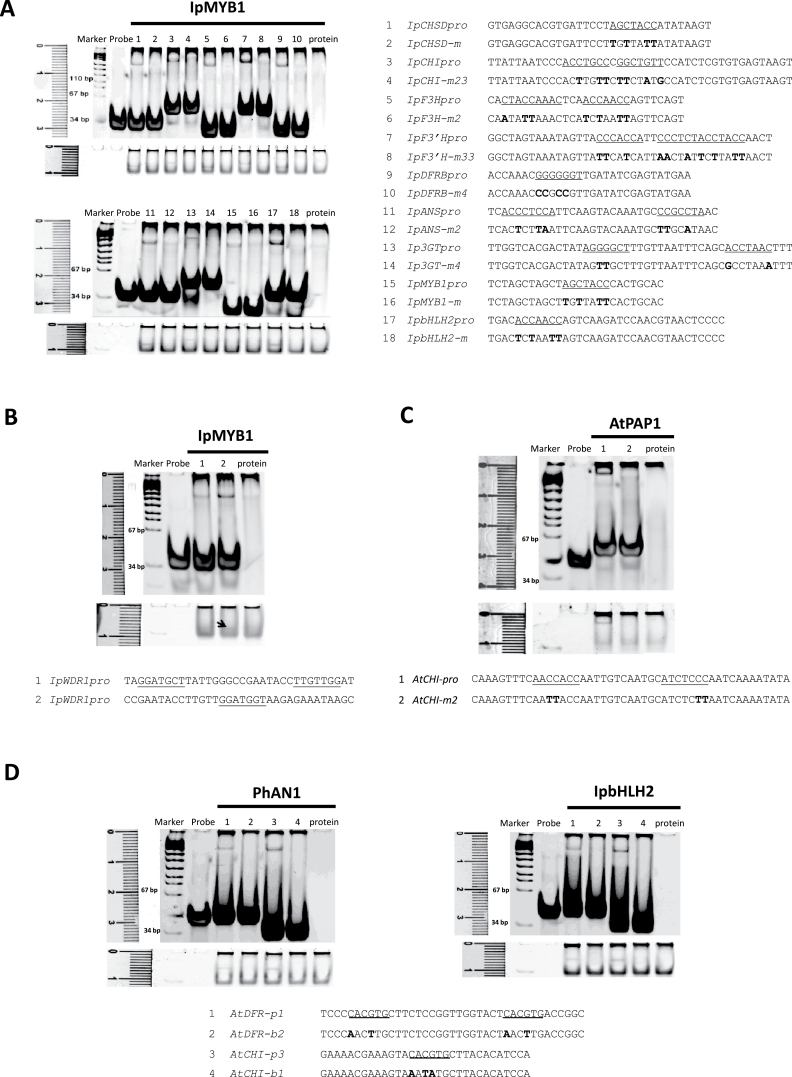
Fig. 3.
Results of EMSAs with MYBs and bHLHs. (A) I. purpurea probes based on the 5′-noncoding regions of the anthocyanin genes and their binding results with IpMYB1 in EMSAs. The probes harboured candidate cis motifs (underlined) and their mutations (in bold). The gels are shown in two parts, with the upper panel for the DNA binding and the lower panel for protein binding of the same gel photographed under a different light filter (Wang et al., 2013). The lane numbers correspond to those of probes used in the binding reactions; the probe lane contains probe 1 (20 pmol) only; the protein lane has MBP-IpMYB1 (2 µg) only. (B) Results of EMSAs with probes based on the 5′-noncoding region of IpWDR1. Three likely cis motifs (underlined) were tested with IpMYB1; only the second probe showed positive binding as indicated by the arrow. The arrangement shown follows that described in (A). (C) Arabidopsis probes based on AtCHI pro and their binding results with AtPAP1. The probes and EMSA results are shown as in (A). (D) EMSAs of bHLHs of I. purpurea (IpbHLH2) and Petunia (PhAN1) with probes based on the 5′-noncoding regions of Arabidopsis DFR and CHI genes. The experimental conditions followed (A), and the mutations to the G-box are indicated in bold.
Though effective in validating cis elements, the time commitment for the transgenic tests and common occurrence of multiple cis elements prevented the transgenic approach being used on a large scale. We identified cis elements for other anthocyanin genes mainly through mutation tests by means of EMSAs and transient expressions. Locating and testing cis elements on those genes were guided partially by the previous results on MREs and BREs (Wang et al., 2013) and partially by our ongoing accumulating data. The EMSA results were presented in both DNA- and protein-staining to reduce false positives. Effective cis-containing probes that showed positive binding to MBP-tagged IpMYB1 were identified for all tested anthocyanin genes including IpCHS-D, IpCHI, IpF3H, IpF3′H, IpANS, Ip3GT, IpMYB1, IpbHLH2, and IpWDR1 (Fig. 3A, B). To know whether or not TF orthologues could recognize similar cis elements, we expressed MBP-tagged PAP1 of Arabidopsis to detect its binding capacity to the predicted cis elements of AtCHI pro. The results suggested that the MYB orthologues recognized the cis elements of similar structure (Fig. 3C). Likewise, IpbHLH2 shared the same recognition pattern with PhAN1 for the G-box present at the promoters of AtDFR and AtCHI (Fig. 3D), as with the known binding capacity of AtGL3 to the G-box (Shangguan et al., 2008). Further EMSA tests with IpbHLH2 indicated that its BREs could change from canonical CACGTG to CACGTT (Supplementary Figure S2).
Complementing the EMSAs but in a cellular environment, dual luciferase assays showed that mutations correctly targeting MREs and BREs could reduce promoter activities (Supplementary Figure S3). Except in one case (IpbHLH2 pro), where accidental mutations to pbHLHMRE2 caused an exceedingly high promoter activity, all other introduced mutations were mostly on targets, judging from their reduced promoter activities. To make sure that the TFs were not overexpressed to bias the detection of effective motifs, we compared 10-fold levels of particle bombardment in the assays for two genes including IpCHSD pro and IpF3′H pro (Supplementary Figure S4A, B). Although lowered TF dosages led to reduced promoter activities (Supplementary Figure S4C), the patterns of the relative effects of mutated motifs were largely unchanged (Supplementary Figure S4D). To further make sure that the results of mutation tests were not biased by using leaves of I. purpurea, we compared the same promoters and effectors in the leaves of cultivated rice seedlings in transient assays. The results showed essentially the same patterns for the mutation effects (Supplementary Figure S4E, F) despite change in promoter strength. These experiments effectively validated applications of transient assays to assessing mutation effect of cis motifs.
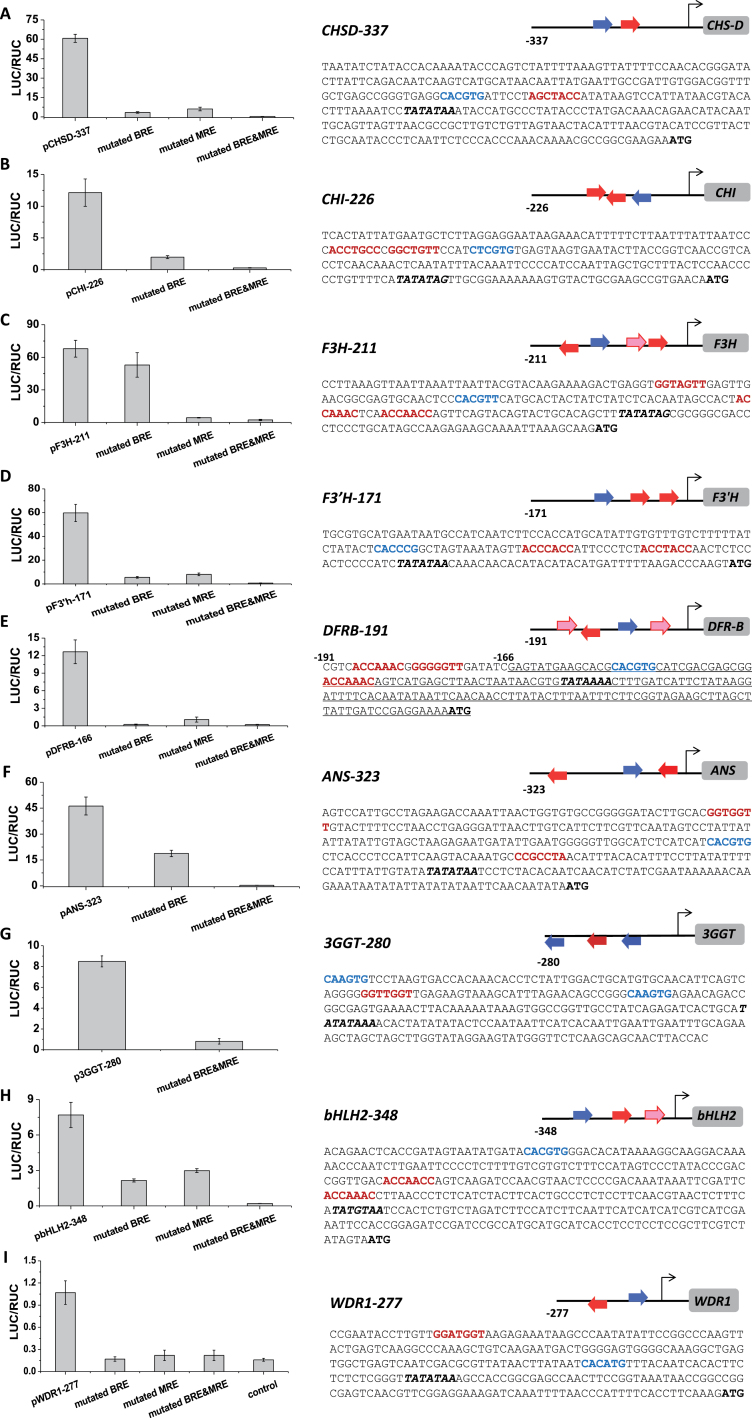
Collectively, transient and EMSA results led to identification of the major cis elements over the anthocyanin pathway (Fig. 4), including combined identification of MRE and BRE on the Ip3GGT promoter. These valid motifs indicated that the orientation of the cis elements could change, as could the numbers of MREs and BREs. The presence of both MRE and BRE is essential for the promoters to respond to IpMBW. In addition, variants of some of the cis elements identified were also functional, as tested on IpCHSD-337 pro (Supplementary Figure S5). Putting all the information on cis motifs together, we found they converged on ANCNNCC for MREs and CACN(A/T/C)(G/T) for BREs (Table 1). The two syntaxes representing nine anthocyanin pathway genes indicate a cis logic shared by the pathway. As the two syntaxes all fall in the proximal promoter regions typically within 350bp from the translation start site (Supplementary Figure S1C), the pathway genes appear to have a tight motif organization in the proximal region of a promoter in common.
Fig. 4.
Proximal promoter architectures of the I. purpurea anthocyanin pathway genes. (A–I) Test results of the 5′-noncoding regions of the anthocyanin genes from IpCHS-D to IpWDR1. Each gene is displayed by its dual luciferase results on the left and features of the proximal promoter sequence on the right (see Fig. 3 and Supplementary Figure S3 for mutation details). The proximal regions are marked at the sites of experimentally confirmed BREs (in blue and bold) and MREs (in red and bold, along with the expected TATA box (in italic and bold)). The mutated MREs and BREs shown on the x-axis on the left correspond to those confirmed cis elements on the right. The orientations of the cis elements are cartooned by the insert figure with arrows representing the cis motifs for each gene. The arrows are coloured in accordance with those of MREs and BREs; and those above the line and pointing to the translation start site are oriented following the syntax and the ones below the line are in the complementary layout. The pink arrows are for the weak MREs. The angled arrow shows the start site for the transcription of the gene. The box indicates the coding region. The numbers above the sequences show the relative positions from the translation start site, and those after the gene names the lengths of the regions.
Table 1.
Experimentally confirmed cis elements on anthocyanin gene promoters
| Method | Species | Gene | BRE | BRE location | MRE | MRE location |
|---|---|---|---|---|---|---|
| Luciferase assay and EMSA | I. purpurea | CHS-D | CACGTG | 205–200 | AGCTACC | 193–187 |
| ACCCACC | 321–315 | |||||
| CHI | CACGAG | 143–148 | ACCTGCC | 167–161 | ||
| AACAGCC | 153–159 | |||||
| F3H | CACGTT | 135–130 | AACTACC | 160–166 | ||
| ACCAAAC (weak) | 97–91 | |||||
| ACCAACC | 87–81 | |||||
| F3′H | CACCCG* | 106–101 | ACCCACC | 86–80 | ||
| ACCTACC | 70–64 | |||||
| DFR-B | CACGTG | 152–147 | ACCAAAC (weak) | 187–181 | ||
| ACCCCCC, ATCAACC | 170–180 | |||||
| ACCAAAC (weak) | 133–127 | |||||
| ANS | CACGTG | 155–150 | AACCACC | 265–271 | ||
| ATCCGCC | 117–123 | |||||
| 3GGT | CACTTG | 275–280, 177–182 | ACCAACC | 212–218 | ||
| bHLH2 | CACGTG | 321–316 | ACCAACC | 226–220 | ||
| WDR1 | ACCATCC | 257–263 | ||||
| EMSA | I. purpurea | 3GT | ACCTAAC | 105–99 | ||
| Z. mays | Bz (3GT) | ACCTGCC | Wang et al., 2013 | |||
| A1 (DFR) | ACCTACC | |||||
| Malus domestica | MYB10 | AGCTACC | ||||
| Gerbera hybrida | DFR2 | AACTTAC | ||||
| Luciferase assay | I. purpurea | CHSD-337 pro | cacgtt | agcaacc | mutation | |
| cacgcg | agcgacc | mutation | ||||
| Consensus | CACN(A/C/T)(G/T) |
ANCNNCC
(ANCNNAC weak) |
The numbers for location are relative to the translation start site of the gene. Sequences underlined are ones in the reverse or/and complementary form on the promoters. Those in lower case were tested on mutated IpCHSD-337 pro. *, Little binding to IpbHLH2 in EMSA.
Generality and layout pattern of cis elements on the anthocyanin gene promoters
We searched for the syntaxes in multiple species to test for their generality. Searches using the blastp command led to the identification of available coding sequences of anthocyanin genes in the NCBI database after taking those of Ipomoea genes as templates (Fig. 5A). A collection of 299 anthocyanin gene promoter sequences was retrieved. A fixed cis layout pattern (CACNNG (N1–100)ANCNNCC) was taken as the search criterion for these sequences, as the pathway-based identification of cis motifs suggested a common layout of BRE and MRE (5′→3′) on the anthocyanin genes (Fig. 4). From the collected promoter sequences, 159 promoters of 35 species (53%) were found exhibiting the pair of cis elements within 100bp of each other. The species included Ginkgo biloba, monocots and dicots (Supplementary Table S4). Interestingly, the distance between the cis elements showed a peak at 6-bp apart (Fig. 5B). The sequences giving rise to the peak showed two significant motifs agreeing with the syntaxes identified in Ipomoea and Arabidopsis (Fig. 5C). As IpCHSD pro hosts the BRE and MRE at this preferred distance of 6-bp apart (Fig. 4A), the biological significance of the distance can be evaluated in transient expression assays. By artificially increasing the between-cis distance, we found that the distance affected the promoter activity negatively. Lengthening the distance (up to 60bp) significantly weakened the promoter strength; nevertheless, about 12% of the promoter activity still remained even after the distance was increased further to 80-bp apart (Fig. 5D).
Fig. 5.
A common layout for the BRE and MRE on the anthocyanin gene promoters across species. (A) Results of a bioinformatic survey. The column for putative promoters lists the numbers of 5′-noncoding sequences with their coding regions sharing >40% amino acid identity with those of I. purpurea anthocyanin genes. Of these sequences, the number of promoter sequences having the specific arrangement of BREs and MREs [from 5′ to 3′ as CACNNG(N1–100)ANCNNCC] is listed as hits for each gene. (B) Frequency distribution of the distance (bp) between BRE and MRE candidates detected on the 159 promoter sequences. The cis candidates were within 100bp of each other (Supplementary Table S4). The sequences are 200–2000bp long, covering the 5′-noncoding region upstream of the translation start site. (C) The cis motifs reported by MEME for the peak in (B). The peak consists of 34 sequences. (D) Effect of between-cis distance on promoter activity. The distance alterations were carried out on IpCHSD-337 pro. All five constructs had the same cis elements (in bold, MRE to the right and BRE to the left) but different spacer lengths to separate the distances between the cis elements. The effect of changing the distance was analysed in the transient expression system driven by the IpMBW complex. The activity of the native promoter estimated in LUC/RUC was set as 100%. The error bars are from three replicates. This figure is available in colour at JXB online.
The regulatory specificities of MBW complexes
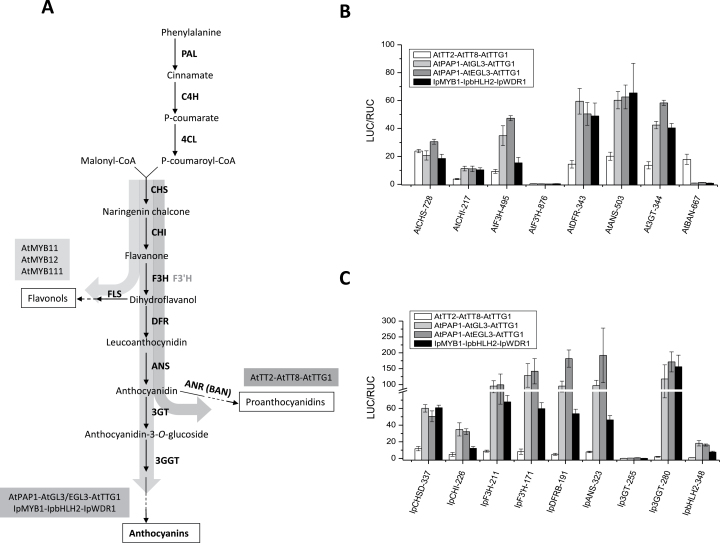
Since both trans and cis components are vital for the regulatory module formed at the 5′-noncoding region, we evaluated the specificities of the trans factors in governing the transcription of flavonoid genes. This analysis included proximal promoters of ten flavonoid genes (CHS, CHI, F3H, F3′H, DFR, ANS, 3GT, AtBAN, Ip3GGT, and IpbHLH2) and four sets of MBW complex from two species (Arabidopsis and I. purpurea). Three TF complexes [AtTT2-AtTT8-AtTTG1 (AtTTT), AtPAP1-AtGL3-AtTTG1 (AtPGT), and AtPAP1-AtEGL3-AtTTG1 (AtPET)] were from Arabidopsis, and one (IpMBW) from I. purpurea. We found that AtPGT, AtPET, and IpMBW could positively regulate all tested genes except AtF3′H, AtBAN and Ip3GT. In comparison, AtTTT also activated the same set of genes (except IpbHLH2), in addition to AtBAN (Fig. 6), congruent with its role in PA synthesis; the activation intensity was lower than AtPGT and AtPET for the same constructs. While all TF complexes were capable of regulating CHS, CHI, and F3H, none could activate AtF3′H or Ip3GT. A close search of the 876-bp long 5′-noncoding region of AtF3′H indicated that MREs and BREs were both absent within the region about 500-bp upstream of the translation start site (Supplementary Figure S6), despite typical BRE and MRE sequences being located at –659 and –584 from the translation start site, respectively. Both Ip3GT and IpMYB1 were found lacking BREs in their proximal promoters (Supplementary Figure S6), which explained their low promoter activities in the presence of IpMBW (Fig. 1B). A common pattern in the TF complexes such as IpMBW, AtPGT, and AtPET was that they all demonstrated strong power of activation towards the later enzyme genes of the anthocyanin pathway (DFR, ANS, and At3GT). In contrast, AtTTT could activate these genes at a relatively low level (Fig. 6).
Fig. 6.
Comparisons of the flavonoid network regulation between Arabidopsis and Ipomoea. (A) The anthocyanin pathway backbone and the major branches. The arrows and associated TFs indicate the known regulated segments of the network after this work and that of Stracke et al. (2007). Pathway regulation by four MBW complexes on the flavonoid genes of A. thaliana. (B) and I. purpurea (C) were measured in three replicates in dual luciferase assays. The 5′-noncoding regions of the genes are labelled along the x-axis. The normalized promoter activities (in LUC/RUC) are indicated by the y-axis.
After excluding AtF3′H and the more-or-less redundant AtPET (relative to AtPGT), we analysed the transient expression data in two-way ANOVAs. The analysis suggests that promoter identity is the leading factor for proximal promoter activity on the anthocyanin pathway, and the interactions between the TF complex and promoter (TF × promoter) plays a role as significant as the TF complex itself (Table 2). When the analysis was performed again with the transient expression data of only Arabidopsis or Ipomoea, the pattern remained the same. Proximal promoters are hence considered the major force governing gene transcription in the flavonoid network, irrespective of species.
Table 2.
Analysis by two-way ANOVA on pathway promoter activities of flavonoid genes
| Source | df | Mean square | F-value | Pr > F | Power |
|---|---|---|---|---|---|
| Promoter | 7 | 1.58 | 101.19 | < 0.0001 | 0.999 |
| TF complex | 2 | 0.43 | 27.20 | < 0.0001 | 0.999 |
| Promoter × TF complex | 14 | 0.46 | 29.47 | < 0.0001 | 0.999 |
Eight proximal promoters included AtBAN, At3GT, AtANS, AtDFR, AtF3H, AtCHI, AtCHS, and IpCHS-D. Three TF complexes were from Arabidopsis (TT2-TT8-TTG1, PAP1-GL3-TTG1) and I. purpurea (IpMYB1-bHLH2-WDR1). Each promoter activity (log transformed) was measured by three replicates of its transient expression (R 2 = 0.96, n = 72). The F-value was from F-test on the effect of each variable (source).
Discussion
Our analysis has provided a bilateral view of pathway regulation, with the molecular mechanism discussed in both promoter architecture and TF–cis interactions featuring the MBW complex. The promoter architecture was found following a common logic for the anthocyanin pathway, including the orientation of the BRE and MRE within the proximal promoter and the semi-conservative composition of cis elements for MYB and bHLH. The syntaxes for both BRE and MRE contain highly variable nucleotide sites, which may lead to segmented summaries for the binding sites of the complex. For instance, for 85 R2R3 MYBs of the Arabidopsis genome, three categories of MYB binding site (MBS) were suggested (Romero et al., 1998). MBSI (CNGTTR, or T/CAACNG) partially overlaps with MBSII [GTTWGTTR, or T/CAAC(T/A)A(C/A)C], while MBSIIG [GKTWGGTR, or C/TACC(T/A)A(C/A)C] differs from MBSII by one nucleotide. The latter two also agree largely with the AC-rich element [A/CCC(T/A)A(C/A)C/G] proposed by Hartmann et al. (2005). Our summary of MREs for MYB1- like homologues in the syntax of ANCNN(C/A)C appears to unify these categories in both length and content, providing a basis for future expanded inquiries on R2R3 MYBs in the plant genome. In this study, however, we mainly focused on the anthocyanin pathway, showing that a systematic approach could enable a deeper understanding of the regulatory mechanism in the context of the related flavonoid branches.
MBW regulates the pathway genes as one unit
Identification of cis elements in I. purpurea pathways led to a mechanistic understanding of how TFs regulate groups of genes. In both Arabidopsis and Ipomoea, transcription of the enzyme-coding genes of the anthocyanin pathway were activated as one unit, just like the pathway regulation found in maize (Carey et al., 2004). Since AtPGT and AtPET activate the same set of genes as IpMBW does, their orthology can be established in functioning as bona fide regulators of the anthocyanin pathway. Interestingly, the PA pathway in Arabidopsis appears to be regulated together with the anthocyanin pathway, as shown by the wide-ranging regulation capacity of AtTTT on all the anthocyanin pathway genes (with the exception of AtF3′H but including At3GT) and AtBAN. The result agrees with reported regulation of AtTTT on AtCHS (Appelhagen et al., 2011), AtDFR, AtANS, AtBAN (Baudry et al., 2004; Xu et al., 2014a), and additionally AtCHI and AtF3H (Xu et al., 2014b). Against this background, all tested MBWs failed to activate AtF3′H and Ip3GT in the transient expressions, consistent with their lack of the appropriate cis elements in the proximal promoters. These genes are either regulated independently of MBW or need extra regulatory components beyond the proximal regions. For instance, the grape UFGT was considered to be regulated independently of the other genes (Boss et al., 1996), and ZmA1 required the presence of R-interacting factor 1 to be activated by its conspecific MBW (Kong et al., 2012). Regardless, the cases of AtF3′H and Ip3GT suggest that both an MRE and a BRE are required for the MBW complex to activate the promoter of a target gene after the local chromatin structure is cleared. Most pathway genes identified so far do contain the cis regulatory region with the necessary footings for the MBW complex.
Parallel redundancy of cis elements and multiple TFs feature combinatory regulation
The pathway genes show multiple copies of each type of cis element (particularly those of MREs). At least three functional MREs were experimentally confirmed on each of IpCHI-226 pro, IpF3H-211 pro , IpF3′H-171 pro, and IpDFRB-191 pro (Fig. 3; Fig. 4; Supplementary Figure S3). For instance, if one of the MREs was mutated on IpDFRB-191 pro, no phenotypic change could be found on the seedlings transformed with the mutated construct. Although refractory in experimental validations of the cis elements involved, the redundancy in binding sites may provide dynamics as well as intricacy to the gene transcription. A full occupancy of the cis sites in a promoter region, for instance, may result in a higher level of gene transcription than is the case for partial occupancy of the cis sites.
The multiple cis elements also mirror functional redundancy of TFs. Noticeably, AtTTT activates the same set of anthocyanin pathway genes as AtPGT and AtPET do, but possibly at a lower capacity (Fig. 6). This may explain why the transgenic Arabidopsis plants showed accumulation of anthocyanins but no obvious PA accumulation in the cases of the short promoter sequences (Fig. 2C). The level of PA synthesis in these cases was probably too low to be visible. Given the restricted yet high expression of AtTT2 in developing seeds (Nesi et al., 2001) and a low expression of AtPAP1 in developing siliques (Matsui et al., 2004), the extensive control from AtCHS to AtBAN by the AtTTT complex on the flavonoid network allows independent PA synthesis in the seed coat. Likewise, flavonol synthesis appears to use the same principle, where AtCHS, AtCHI, and AtF3H are regulated by AtMYB11, AtMYB12, and AtMYB111, which also target the flavonol synthase 1 gene (Stracke et al., 2007). The regulatory redundancy of the same genes may offer insurance to different functional branches of the network. For TF redundancy, maize B alleles can substitute for R in inducing the expression of Zm3GT (Goff et al., 1990). Also, the triple mutant of AtMYB11, AtMYB12, and AtMYB111 still accumulates anthocyanin (Stracke et al., 2007), but AtPAP1has little to do with PA accumulation (Gonzalez et al., 2008). The redundancy in both cis and trans factors in pathways enables a cell-specific regulation of the same pathway by different arrays of regulatory proteins.
Promoter architecture is the most important factor for pathway regulation
Promoter architecture is believed to take a central position in transcription (Spitz and Furlong, 2012), but detailed studies have been scarce. A promoter’s architecture is characterized here by the distribution of all cis elements in the region, including the binding sites of TFs and the basal transcription apparatus (such as the TATA box), numbers of cis elements, spatial distance between cis elements, and the relative orientations of cis elements. These factors may significantly affect a promoter’s activity. For the anthocyanin pathway genes, the cis regulatory sites summarized by the syntax identified for MREs (ANCNNCC) overlap with several previously postulated motifs. For example, cctacc(n)7ct, described as an H-box (Loake et al., 1992), shows an overlap of NCNNCC, and the consensus (AC)ACC(TA)A(AC)C described by Sablowski et al. (1994) is also mostly congruent with the MRE syntax here; in the case of TTGACTGGnGGnTGCG for C1 binding (Lesnick and Chandler, 1998), GGnGGnT is clearly the reverse complement of ANCNNCC. The syntax also includes ANCNACC, shown in a recent consensus summary (Wang et al., 2013). We have shown here that both orientations of the syntaxes are valid on the anthocyanin genes (Table 1). The intermittently conserved sites within the MRE syntax have greatly increased the difficulty of cis element predictions. Even when all the existing data support the syntax for MREs, for instance, it provides still more of guidance than a total replacement of experimental validations since a fraction of cases that obey the structure of the syntax have been proven nonfunctional, at least to IpMBW (Supplementary Figure S5). The same is also true for the syntax of BREs. It matches well with the previous G-box (Staiger et al., 1991; Loake et al., 1992) but not the E-box (CANNTG). Certain cases satisfying the syntax such as CACTTG were proven invalid for IpbHLH2 (Supplementary Figure S2). The mutation tests of Bodeau and Walbot (1996) suggested that the R-motif in the form of CACGAC and CACGAG was effective on the Bz2 (GST) promoter, suggesting that the BRE syntax holds for both monocots and dicots.
The proximity of BRE and MRE in the 5′-noncoding region was revealed by both the promoter survey (Fig. 5) and identification of the cis motifs over the anthocyanin pathway (Fig. 4). The influence of cis spacing on promoter activity was previously indicated in the rbcS promoter (Lopez-Ochoa et al., 2007), and further analysed here to show its relevance to transcriptional intensity. Locations of most MREs on the anthocyanin genes were found close to the translation start site; BREs, on the other hand, are largely located 5′ to the first MRE (Fig. 4). This order of BRE and MRE from 5′ to 3′ in the noncoding region is not only broadly found on the anthocyanin gene promoters (> 53%) but also present in the promoter of Rd22, an abscisic acid-mediated dehydration-responsive gene (Abe et al., 2003). The occurrence of the cis orientation in different molecular systems driven by MYB and bHLH implies an antiquity of the cis arrangement for genes regulated by bHLH and MYB.
We have shown that promoter architecture varied greatly among genes, facilitating variations of gene expression across pathways that is regulated by the same set of TFs. The regulatory mechanism of the anthocyanin pathway is mostly about the cooperation of the cis-regulatory elements and the cognitive TF complex, taking the same logic of TF–DNA interactions across the pathway. The findings have led to greater understanding of the regulatory mechanism of the anthocyanin pathway and the opportunity for more quantitative analysis of TF–DNA interactions. It will be interesting to see how other TFs affect pathway gene regulation.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. List of primers used in the experiments in this study.
Supplementary Table S2. Accessions of genes and promoter sequences.
Supplementary Table S3. Correlation coefficients of transcript levels by RT-qPCRs.
Supplementary Table S4. List of 159 sequences of the anthocyanin genes at the 5′-noncoding region.
Supplementary Figure S1. Characterization of Ipomoea TFs and promoter activities of the target genes.
Supplementary Figure S2. Binding capacity of IpbHLH2 in EMSAs.
Supplementary Figure S3. Analysis of cis elements on the anthocyanin proximal promoters of I. purpurea.
Supplementary Figure S4. Comparisons of TF dosages in dual luciferase assays.
Supplementary Figure S5. Mutations on CHSD-337 pro showing DNA binding spectrums of IpMYB1 and IpbHLH2.
Supplementary Figure S6. 5′-noncoding sequences of flavonoid genes of two species (A. thaliana and I. purpurea).
Supplementary File S1. Bioinformatic survey with Perl scripts and instructions.
Funding
The study was supported by National Science Foundation of China (91331116, 31070263) and Chinese Academy of Sciences (KSCX2-YW-N-043).
Supplementary Material
Acknowledgements
We thank anonymous reviewers for constructive comments, Dr Yan Wang for technical assistance during the early phase of the study, the group of Dr Kang Chong for access to equipment, and members of the ecological and evolutionary research group for discussions.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang HB, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I, Lu GH, Huep G, Schmelzer E, Weisshaar B, Sagasser M. 2011. TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. The Plant Journal 67, 406–419. [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. 2006. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana . The Plant Journal 46, 768–779. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. 2004. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . The Plant Journal 39, 366–380. [DOI] [PubMed] [Google Scholar]
- Beld M, Martin C, Huits H, Stuitje AR, Gerats AGM. 1989. Flavonoid synthesis in Petunia hybrida - partial characterization of dihydroflavonol-4-reductase genes. Plant Molecular Biology 13, 491–502. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. 1990. Sequence-specific DNA-binding by the c-myc protein. Science 250, 1149–1151. [DOI] [PubMed] [Google Scholar]
- Bodeau JP, Walbot V. 1992. Regulated transcription of the maize Bronze 2 promoter in electroporated protoplasts requires the C1 and R gene-products. Molecular and General Genetics 233, 379–387. [DOI] [PubMed] [Google Scholar]
- Bodeau JP, Walbot V. 1996. Structure and regulation of the maize Bronze2 promoter. Plant Molecular Biology 32, 599–609. [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. 1996. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L cv Shiraz grape berries and the implications for pathway regulation. Plant Physiology 111, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. 2004. Transcription factors as tools for metabolic engineering in plants. Current Opinion in Plant Biology 7, 202–209. [DOI] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL. 2004. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana . The Plant Cell 16, 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SM, Lu YQ, Rausher MD. 2005. Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea . Genetics 170, 1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg MT, Durbin ML. 2003. Tracing floral adaptations from ecology to molecules. Nature Reviews Genetics 4, 206–215. [DOI] [PubMed] [Google Scholar]
- deVetten N, Quattrocchio F, Mol J, Koes R. 1997. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes and Development 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis . Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Durbin ML, McCaig B, Clegg MT. 2000. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Molecular Biology 42, 79–92. [PubMed] [Google Scholar]
- Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH. 2003. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiology 133, 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagne D, et al. 2009. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. The Plant Cell 21, 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. 1992. Functional-analysis of the transcriptional activator encoded by the maize B gene - evidence for a direct functional interaction between two classes of regulatory proteins. Genes and Development 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Goff SA, Klein TM, Roth BA, Fromm ME, Cone KC, Radicella JP, Chandler VL. 1990. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. The EMBO Journal 9, 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollop R, Even S, Colova-Tsolova V, Perl A. 2002. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. Journal of Experimental Botany 53, 1397–1409. [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. 2000. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proceedings of the National Academy of Sciences, USA 97, 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S, Lu YQ. 2013. Dissecting organ-specific transcriptomes through RNA-sequencing. Plant Methods 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau F, Lucy A, Mullineaux P. 1992. Effect of two consensus sequences preceding the translation initiator codon on gene-expression in plant-protoplasts. Plant Molecular Biology 18, 815–818. [DOI] [PubMed] [Google Scholar]
- Habu Y, Hisatomi Y, Lida S. 1998. Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. The Plant Journal 16, 371–376. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. 2005. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology 57, 155–171. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Morita Y, Choi JD, Saito N, Toki K, Tanaka Y, Iida S. 2003. Spontaneous mutations of the flavonoid 3’-hydroxylase gene conferring reddish flowers in the three morning glory species. Plant and Cell Physiology 44, 990–1001. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kong Q, Pattanaik S, Feller A, Werkman JR, Chai CL, Wang YQ, Grotewold E, Yuan L. 2012. Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proceedings of the National Academy of Sciences, USA 109, E2091–E2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P. 2012. REGULATORY ELEMENTS Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nature Reviews Genetics 13, 233–245. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. 2006. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology 57, 405–430. [DOI] [PubMed] [Google Scholar]
- Lesnick ML, Chandler VL. 1998. Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiology 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhao PP, Li J, Zhang CJ, Wang LN, Ren ZH. 2014. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis . Molecular Genetics and Genomics 289, 103–124. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR - automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA. 1992. Combination of H-box [CCTACC(N)7CT] and G-box (CACGTG) cis elements is necessary for feedforward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proceedings of the National Academy of Sciences, USA 89, 9230–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ulker B, Somssich IE. 2006. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ochoa L, Acevedo-Hernandez G, Martinez-Hernandez A, Arguello-Astorga G, Herrera-Estrella L. 2007. Structural relationships between diverse cis-acting elements are critical for the functional properties of a rbcS minimal light regulatory unit. Journal of Experimental Botany 58, 4397–4406. [DOI] [PubMed] [Google Scholar]
- Lu YQ, Du J, Tang JY, Wang F, Zhang J, Huang JX, Liang WF, Wang LS. 2009. Environmental regulation of floral anthocyanin synthesis in Ipomoea purpurea . Molecular Ecology 18, 3857–3871. [DOI] [PubMed] [Google Scholar]
- Lu YQ, Xie LL, Chen JN. 2012. A novel procedure for absolute real-time quantification of gene expression patterns. Plant Methods 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. 1989. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proceedings of the National Academy of Sciences, USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Tanaka H, Ohme-Takagi M. 2004. Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnology Journal 2, 487–493. [DOI] [PubMed] [Google Scholar]
- Matus JT, Poupin MJ, Canon P, Bordeu E, Alcalde JA, Arce-Johnson P. 2010. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Molecular Biology 72, 607–620. [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. 2006. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant and Cell Physiology 47, 457–470. [DOI] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. 2001. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness SA, Marknell A, Graf T. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. 1988. Genetic applications of an inverse polymerase chain-reaction. Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S. 2007. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. The Plant Journal 49, 641–654. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazares J, Ghosal D, Wienand U, Peterson PA, Saedler H. 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb protooncogene products and with structural similarities to transcriptional activators. The EMBO Journal 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N, Dolan L. 2010. Origin and diversification of basic-Helix-Loop-Helix proteins in plants. Molecular Biology and Evolution 27, 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. 1993. Regulatory genes-controlling anthocyanin pigmentation are functionally conserved among plant-species and have distinct sets of target genes. The Plant Cell 5, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 10, 63–70. [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. 1998. More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana . The Plant Journal 14, 273–284. [DOI] [PubMed] [Google Scholar]
- Roth BA, Goff SA, Klein TM, Fromm ME. 1991. C1 dependent and R dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding-sites. The Plant Cell 3, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianezmacia FA, Schuch W, Martin C, Bevan M. 1994. A flower-specific myb protein activates transcription of phenylpropanoid biosynthetic genes. The EMBO Journal 13, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz MB, Grotewold E, Chandler VL. 1997. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. The Plant Cell 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan XX, Xu B, Yu ZX, Wang LJ, Chen XY. 2008. Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. Journal of Experimental Botany 59, 3533–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Staiger D, Becker F, Schell J, Koncz C, Palme K. 1991. Purification of tobacco nuclear proteins binding to a CACGTG motif of the chalcone synthase promoter by DNA affinity-chromatography. European Journal of Biochemistry 199, 519–527. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Barsch GHA, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Miller RE, Rausher MD. 1998. Control of expression patterns of anthocyanin structural genes by two loci in the common morning glory. Genes and Genetic Systems 73, 105–110. [Google Scholar]
- Tuerck JA, Fromm ME. 1994. Elements of the maize a1 promoter required for transactivation by the anthocyanin b/c1 or phlobaphene p regulatory genes. The Plant Cell 6, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. 2010. Phenylpropanoid biosynthesis. Molecular Plant 3, 2–20. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell 11, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Guan S, Zhu ZX, Wang Y, Lu YQ. 2013. A valid strategy for precise identifications of transcription factor binding sites in combinatorial regulation using bioinformatic and experimental approaches. Plant Methods 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiology 127, 1399–1404. [PMC free article] [PubMed] [Google Scholar]
- Xu WJ, Grain D, Bobet S, Le Gourrierec J, Thevenin J, Kelemen Z, Lepiniec L, Dubos C. 2014a. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytologist 202, 132–144. [DOI] [PubMed] [Google Scholar]
- Xu WJ, Lepiniec L, Dubos C. 2014b. New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signaling and behavior 9, e28736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WJ, Lepiniec L, Dubos C. 2015. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends in Plant Science 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis . Development 130, 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zufall RA, Rausher MD. 2004. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature 428, 847–850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.