Highlight
A rare SNP mutation in Brachytic2 underlies a major QTL affecting plant height, and moderately reduces plant height and increases yield potential in maize.
Key words: Brachytic2, major QTL, maize (Zea mays), mild mutation, plant height, rare allele.
Abstract
Plant height has long been an important agronomic trait in maize breeding. Many plant height QTLs have been reported, but few of these have been cloned. In this study, a major plant height QTL, qph1, was mapped to a 1.6kb interval in Brachytic2 (Br2) coding sequence on maize chromosome 1. A naturally occurring rare SNP in qph1, which resulted in an amino acid substitution, was validated as the causative mutation. QPH1 protein is located in the plasma membrane and polar auxin transport is impaired in the short near-isogenic line RIL88(qph1). Allelism testing showed that the SNP variant in qph1 reduces longitudinal cell number and decreases plant height by 20% in RIL88 (qph1) compared to RIL88 (QPH1), and is milder than known br2 mutant alleles. The effect of qph1 on plant height is significant and has no or a slight influence on yield in four F2 backgrounds and in six pairs of single-cross hybrids. Moreover, qph1 could reduce plant height when heterozygous, allowing it to be easily employed in maize breeding. Thus, a less-severe allele of a known dwarf mutant explains part of the quantitative variation for plant height and has great potential in maize improvement.
Introduction
Short stature, erect leaf angle, disease resistance, and high yield are traits that have been pursued by breeders for decades. Cereal production sharply increased in the 1960s as the ‘Green Revolution’ popularized the use of dwarf and semi-dwarf cultivars. Likewise, maize (Zea mays) production improved dramatically due to the adoption of hybrids and use of moderately short varieties that are more resistant to lodging and compatible with higher planting density (Duvick, 2005). Reduced ear height and an increased plant height/ear height ratio also have the potential to increase dry matter accumulation (Yang et al., 2010a). Many of the Green Revolution genes, such as sd-1 in rice (Sasaki et al., 2002) and rht1 in wheat (Peng et al., 1999), have been identified and utilized in crop improvement. These genes encode proteins that either regulate the synthesis of plant hormones or modulate their signalling pathways. Several genes that strongly influence plant height and are qualitatively inherited have also been cloned in maize (Winkler and Helentjaris, 1995; Thornsberry et al., 2001; Multani et al., 2003); however, these mutants have not found applications in maize breeding due to their adverse impact on grain yield. Identification of alleles moderately reducing plant height is highly desirable.
Plant height QTLs are favourable candidates as mild height modulators. A number of such QTLs have been reported in cereal crops but only a few of these have been cloned (Xue et al., 2008; Yan et al., 2011; Teng et al., 2012). The large genome size of maize makes QTL cloning a time-consuming task, although some progress has been made (Wang et al., 2005; Salvi et al., 2007; Zheng et al., 2008). Moreover, planting density affects the grain yield of maize more than other members of the grass family (Vega et al., 2001). In the past few decades, maize yield has been increased mainly by adopting modern hybrids that are more tolerant to high planting density and other biotic or abiotic stresses, and have higher efficiency in resource usage (Liang et al., 1992; Vafias et al., 2006). Due to competition for light, plant height increases with planting density. This tends to decrease stalk diameter and increase the potential of stalk lodging, leading to yield losses (Park et al., 1989; Hashemi et al., 2005). Modifying plant height and other architecture components, while not influencing grain yield, is among the key factors in developing cultivars for compact planting.
We previously identified a major QTL affecting multiple traits in bin7 on chromosome 1 using F2:3 (a series of F3 families derived from F2 individuals), immortalized F2, and recombinant inbred line (RIL) populations derived from Zong3 and 87-1, a hybrid known as ‘Yuyu22’ that has been widely planted in China for the past two decades. This QTL, designated qph1, was mapped near marker umc1122 (chromosome 1: 201677909- 201678070) and explains ~10% of the phenotypic variation for plant height and ear height in the RIL, immortal F2, and F2:3 population of Zong3/87-1. qph1 was significantly associated with plant height and marginally affects yield and yield components (Yan et al. 2003; Yan et al, 2006 Tang et al, 2007a, b; Ma et al, 2007; Tang et al, 2010). The objectives of this study were to fine map and clone qph1 and to evaluate its use in maize breeding.
Materials and methods
Mapping population of qph1 and near-isogenic lines
A RIL population of 294 lines was constructed previously from a Zong3×87-1 cross. Two RILs, RIL88(qph1) and RIL279(QPH1), which differed in plant height but shared a large portion of the same genetic background, were chosen to generate the near-isogenic line RIL88(QPH1), with the QPH1 allele in a RIL88 background (Supplementary Figure S1). RIL88(qph1) was used as the recurrent parent to backcross RIL279 and plants were selected based on phenotype (tall plants were kept) in each generation until BC4F1. Ninety-four BC4F1 individuals were screened by 101 SSR markers all over the maize genome and a tall line with the QPH1 allele, 05YH175-2, was selected on the basis of having the smallest segments introgressed from RIL279 on chromosome 1. 05YH175-2 was then selfed repeatedly to produce BC4F3 and its progeny, 08YB036-7, was selected on the basis of containing a single segment of RIL279 on chromosome1 exclusively in RIL88 background. 08YB036-7 was then used to generate BC5F2 and BC6F2 fine-mapping populations, and to generate the near-isogenic line of QPH1 (Supplementary Figure S12). 8030 BC6F2 kernels were chipped and genotyped with two flanking markers, umc2396 and MHC412 (Supplementary Table S5).
F2 populations and single-cross F1 hybrids
Four F2 populations, 4F1×81162, Ye107 × Zheng32, Ye107 × B73, and Zong3 × Chuan48-2, were generated. Two markers closely linked to qph1, umc2396 and MHC412, were used for genotyping (umc2396 for the Zong3/Chuan48-2 F2 population and MHC412 for the other three F2 populations). A small piece of each F2 kernel was chipped and genotyped using the soda boiling DNA extraction method (Gao et al., 2008) before planting; more than 50, 100, and 50 seeds in the QPH1/QPH1, QPH1/qph1, and qph1/qph1 genotype classes were planted and phenotyped, respectively. Zong3, 81162, Ye107, W138, B73, and Chang7-2 were crossed with RIL88(qph1) and RIL88(QPH1) to generate six pairs of hybrids; >50 seeds were planted and analysed in each genotype class. Two replicates of the F2 population and hybrids were planted to collect data for plant height, ear height, and yield components. Plots were designed with 50 rows per plot and 13 individuals per row.
br2 mutant-derived populations
The br2 mutants 117A, 114F, 114G, and 121B were provided by MGCSC (Maize Genetics Cooperation Stock Centre); 117A carries the Hahn6 allele of br2 (Leng and Vineyard, 1951). 114G and 114F are linkage stocks from two different sources that each carried an unspecified br2 mutant; these two lines also have hm1, hm2 mutations besides the br2 mutation (http://www.maizegdb.org). 121B was originally named mi8043 and was found to be an allele of the br2 gene (Marty Sachs, personal communication). 117A, 114F, 114G, and 121B were crossed with RIL88(qph1) and RIL88(QPH1) to develop four pairs of single-cross hybrids. 117A × RL88(qph1) was selfed to generate an F2 population; umc2396 was used for genotyping. Plant height and ear height data were obtained for 36, 89, and 54 individuals in br2-117A/br2-117A, qph1/br2-117A, and qph1/qph1 genotype classes, respectively. Plant height and ear height of individuals in these populations were measured and analysed. Plot design was the same as used for the four F2 populations and six pairs of single-cross hybrids.
Dwarf line N546 is derived from Mexican super dwarf (Johnson et al., 1998). Nine crosses, 93NEX501 × PHVRZ, 93NEX501 × PHHHN, 93NEX501 × PHVNV6, Y93NEX504 × PHHHN, YN546 × PHF0D, YN546 × PHCCW, YN546 × PHVRZ6, YN546 × PHVNV, and YN546 × PH128S were made between parents carrying the br2-bj allele (the former) and normal elite inbred lines (the latter). F2 individuals were genotyped with markers PZE-101155635 and PZE-100001759, which are adjacent to br2-bj, and phenotyped for plant height and ear height (Ganal et al., 2011).
DNA preparation and sequence analysis
DNA was extracted from young seedlings using the CTAB method (Dellaporta et al., 1983). For BC6F2 mapping population and the four F2 populations, DNA was extracted from chipped kernels using the soda boiling DNA extraction method (Gao et al., 2008). The genomic sequences of qph1 and QPH1 were amplified with H109F/H106R, H114F/R, H103F/R, H212F/233R, and H115F/116R. The Br2-bj allele was amplified with HF1F/R and HF2F/R. Primer sequences are listed in Supplementary Table S5. PCR was performed with Phusion High Fidelity Master Mix with HF buffer (Thermo Fisher Scientific, Pittsburgh, PA, USA) according to standard protocol. PCR products were ligated into the T-easy vector and colonies containing the desired PCR fragment were picked and sequenced; multiple colonies for each PCR product of each recombinant were sequenced and analysed to eliminate PCR errors.
Scanning electron microscopy
The second and sixth internodes at the mid-elongation stage (15 expanded leaves and 19 visible leaves) and the uppermost internodes at the adult stage (19 expanded leaves) of RIL88(qph1) and RIL88(QPH1) were subjected to scanning electron microscopic examination. Stem tissues from corresponding internodes of RIL88(qph1) and RIL88(QPH1) were cut into 1mm longitudinal and transverse sections and fixed in FAA (Formalin:acetic acid:70% ethanol, 1:1:18, v/v/v). Fixed samples then went through dehydration with a series of graded ethanol (15min in 70%, 80%, 90%, and 100% ethanol). Samples were then treated with isoamyl acetate for 15min twice to replace the remaining ethanol and subjected to critical point drying (HITACHI, HCP-2). Dried samples were mounted on a suitable working stage and coated with Pt using a high vacuum (Eiko IB.3, ION COATER). Scanning electron microscope HITACHI S-3400N was used for imaging.
Subcellular localization of QPH1
QPH1 coding sequence was amplified by PCR using primer pair H235/GFP-R and cloned into a GFP vector with the 35S promoter to express the QPH1–GFP fusion protein. Plasmid containing the QPH1–GFP construct was transformed into onion epidermal cells through gene gun bombardment (Bio-Rad PDS-1000). Transformed cells were then incubated in 1/2 MS media for 20h at 28°C and examined by confocal laser scanning microscopy (Nikon EZ-C1).
Expression analysis
Stem tissues of RIL88(qph1) and RIL88(QPH1) in three stages, at the beginning, middle, and end of elongation, was collected. In each developmental stage, 10 biological replicates of RIL88(qph1) and RIL88(QPH1) were sampled. Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and complementary DNA was synthesized using the AMV reverse transcription system (Promega, Madison, WI, USA) with Oligo (dT) primer. RT12F/R were used to amplify QPH1 (qph1) using SYBR Premix Ex TaqTM (TAKARA, Shuzo, Kyoto, Japan), with β–actin1 as the endogenous control. Real-time PCR was performed with Real Time PCR system 7500 (Applied Biosystems) using the 2-ΔΔCT method (Livak and Schmittren, 2001) according to the standard procedure.
Sequence analysis in diverse maize lines and teosinte accessions
The association panel with 527 inbred lines used in this study is described by Yang et al. (2010b). The 1.4kb qph1 target region containing all five SNPs was amplified with primers 5N3F and 211R. The 839bp SNP5259-containing region was amplified with 212F and 211R from 192 teosinte accessions. The CIMMYT genebank represented a major portion of the diversity present in the teosinte collection.
Gene transformation in Arabidopsis T-DNA insertional mutant atpgp1-2
The Arabidopsis mutant SAIL_716_H02 (Columbia ecotype), designated atpgp1-2 (At2g36910), was obtained from ABRC (Ohio State University, Columbus, OH) and genotyped with primers 716F, 716R, and LB2 for the homozygous mutant according to standard procedure (http://signal.salk.edu/tdnaprimers.2.html). Coding regions of maize qph1 and QPH1 alleles were amplified with primer pair 232F/233R. Site-directed mutagenesis was performed on qph1 coding sequence to change the T to G (wild-type genotype). QPH1, qph1, and mqph1 (the mutated qph1) were overexpressed using PBI121 under the CaMV 35S promoter in the atpgp1-2 mutant. Transgenic plants were selected with kanamycin (50mg l–1). Plant height was measured at 28 days (16/8 light/dark photoperiod, 21–23°C). Coleoptile length for 7-day-old plants grown on 1/2MS solid medium was measured with ImageJ software (National Institutes of Health, http://imagej.nih.gov/ij) with the pictures of the plants taken by digital camera.
Polar auxin transport measurements
Auxin transport assays were performed using the protocol described by Li et al. (2007) and Multani et al. (2003) with some modifications. Ten replicates of RIL88(qph1) and RIL88(QPH1) seeds were grown in sand for 5 days and harvested for coleoptiles in length of 3cm. Coleoptile segments were equilibrated in1/2MS (pH 5.8) liquid media for 2h and the apical portion was submerged in 1/2 MS solid medium containing 0.35% phytogel, 500nM unlabelled IAA (Sigma-Aldrich), and 500nM [3H]-labelled IAA (specific activity 20 Ci mmol–1; American Radiochemical, St Louis, MO, USA) and incubated in the dark for 5h at 25°C. Coleoptiles were then cut into 0.5cm sections and washed twice with 1/2 MS liquid medium. Washed coleoptile sections were incubated in 1ml scintillation fluid for 16h and counts were made with a scintillation counter (Perkin-Elmer MicroBetaTriLux1450).
Results
Effects of qph1 on plant height and other traits
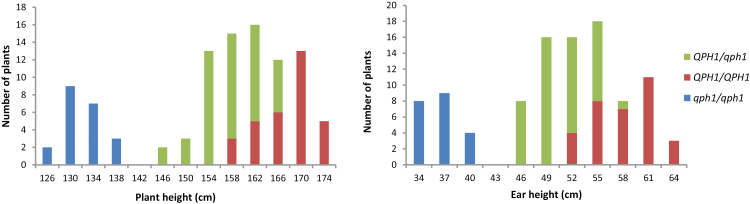
To identify the underlying gene for QTL qph1, RIL88(qph1), and RIL279(qph1), two lines carrying qph1 and QPH1 alleles, respectively, and sharing a large proportion of the same genetic backgrounds, were used to generate a BC4F2 fine-mapping population (Supplementary Figure S1). The previously identified plant height QTL qph1 was further mapped between markers umc1035 (chromosome 1: 195219753-195219900) and umc2236 (chromosome 1: 198266651-198266734) on chromosome 1 in BC4F2 (Supplementary Figure S2). In BC4F2:3, the numbers of qph1/qph1, qph1/PQH1, and QPH1/QPH1 individuals were 21, 42, and 32; segregation of tall to short plants was ~3:1 for both plant height and ear height (χ 2 = 0.196, P > 0.05), indicating the presence of a single recessive Mendelian factor (Fig. 1). In contrast, flowering time and yield components displayed continuous variation (Supplementary Figure S3), consistent with the hypothesis that qph1 has major effects on plant height and ear height and minor effects on other traits. Plant height and ear height differences were highly significant with P-values of 9.30E-48 and 3.40E-44 (t-test) with the dominant allele of QPH1 contributing 90% to plant height and 87% to ear height. Differences in days to silking and days to tasseling were also marginally significant (P = 0.0303 and 0.0002, respectively), revealing that the QPH1/QPH1 individuals flower slightly earlier. Although ear length, diameter, and weight were all significantly different (P = 0.0002, 9.50E-05 and 9.00E-08), the effects of qph1 on these yield components were relatively small (Table 1).
Fig. 1.
Plant height and ear height variation in BC4F2:3. Blue, green and red bars represent the number of qph1/qph1, QPH1/qph1, and QPH1/QPH1 individuals in the BC4F3 population developed from RIL88 and RIL279. Plant height (A) and ear height (B) segregations are consistent with the segregation of a single Mendelian factor.
Table 1.
Analysis of the agronomic traits in BC4F2:3 (2007, Beijing)
| qph1/qph1 | QPH1/QPH1 | QPH1/qph1 | A | D | D/A | R2 (%) | P-value | |
|---|---|---|---|---|---|---|---|---|
| PH | 130.5±3.3 | 165.9±4.7 | 156.2±4.8 | 17.7 | 7.8 | 0.4 | 90 | 9.30E-48 |
| EH | 35.4±1.9 | 57.2±3.4 | 49.6±3.1 | 10.9 | 3.3 | 0.3 | 87 | 3.40E-44 |
| ED | 4.3±0.1 | 4.4±0.1 | 4.4±0.1 | 0.1 | 0 | 0 | 17 | 9.50E-05 |
| EL | 13.8±0.4 | 14.4±0.6 | 14.2±0.5 | 0.3 | 0.2 | 0.5 | 16 | 0.0002 |
| EW | 58.9±9.3 | 74.9±9.8 | 67.5±8.8 | 8 | 0.6 | 0.1 | 28 | 9.00E-08 |
| DTS1 | 90.7±1.0 | 90.4±1.4 | 89.6±1.0 | –0.2 | –1 | 6.6 | 16 | 0.0002 |
| DTS2 | 93.2±0.8 | 92.9±1.4 | 92.5±0.9 | –0.2 | –0.6 | 3.5 | 7 | 0.0303 |
| N | 21 | 32 | 46 |
PH, plant height; EH, ear height; ED, ear diameter; EL, ear length; EW, ear weight; DTS1, days to shedding; DTS2, days to silking. Data are expressed as mean ± SD.
Morphological and cytological observations
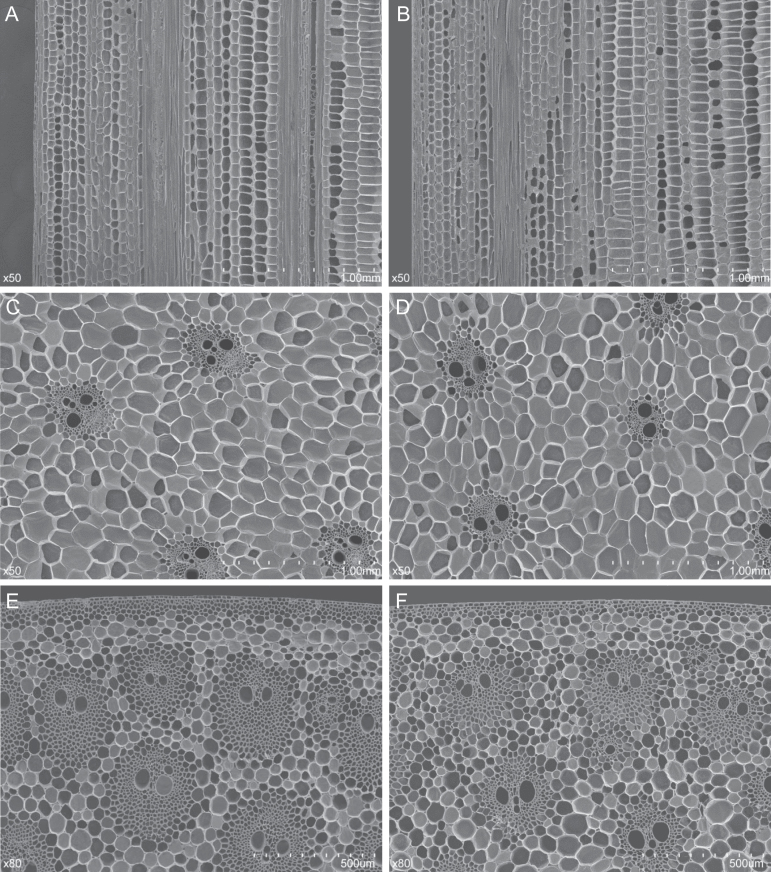
To study the effect of QPH1 and qph1 alleles on plant height and its components in the RIL88 background, RIL88(QPH1) (BC6F3), which only has a 5kb segment of RIL279 introgressed into RIL88 background, was generated (Supplementary Figure S1). Plant height exhibited an incomplete recessive (partial dominant) effect (Fig. 2a). Plant height variation between RIL88(QPH1) and RIL88(qph1) was affected by both average internode length and difference in internode number. RIL88(QPH1) had one more internode below the ear; however, plant height difference was contributed mostly by internode length difference, especially below the ear (Fig. 2BC). Moreover, a gradual increase in internode length difference from the top of the plant down was observed; the uppermost internode showed no difference in length between the near-isogenic lines while the second lowest internode of RIL88(qph1) was more than 50% shorter than wild type (Supplementary Figure S4). The short stature of qph1 was similar to other br2 mutants but with a milder phenotype (Scott et al., 1969). Cytological analysis was performed on the second, sixth, and top internodes of RIL88(QPH1) and RIL88(qph1) through scanning electron microscopy, statistical data on cell length, cell number per mm, and calculated longitudinal total cell number per internode is summarized in Supplementary Table S1. No significant difference was detected between RIL88(QPH1) and RIL88(qph1) for either cell length (P = 0.71, 0.43, and 0.48) or number of cells per mm (P = 0.33, 0.23, and 0.52) in the second (Fig. 3), sixth (Supplementary Figure S5) and top internodes (Supplementary Figure S6). This indicates that the length difference was caused by the different longitudinal cell number between near-isogenic lines rather than different cell size (Supplementary Table S1). Furthermore, altered structure of the vascular bundles in epidermal regions of the second and sixth internodes was observed between RIL88(QPH1) and RIL88(qph1) (Fig. 3E, F; Supplementary Figure S5); however, the uppermost internodes do not show cell number differences or differences in epidermal vascular structure (Supplementary Table S1; Supplementary Figure S6), indicating that the reduced longitudinal cell number is related to the short stature of RIL88(qph1).
Fig. 2.
Plant height and internode length of RIL88(QPH1) (BC6F3) and RIL88(qph1). (A) RIL88(qph1) (two rows on the left), RIL88(QPH1) (two rows on the right), and their F1 hybrid (two rows in the middle). Plant height of the hybrid is between the two parental lines and slightly shorter than RIL88(QPH1). (B) Plant height and ear height comparison between RIL88(QPH1) (left) and RIL88(qph1) (right). (C) Internode length comparison between RIL88(QPH1) (left) and RIL88(qph1) (right); a greater decrease in internode length from the top of the plant down is observed between near-isogenic lines.
Fig. 3.
Scanning electron microscopy examination of the second internodes from RIL88(qph1) and RIL88(QPH1). The second internodes of near-isogenic lines in mid-elongation stage were subjected to histological analysis. (A, B) Longitudinal view of the parenchyma cells of RIL88(QPH1) and RIL88(QPH1). (C, D) Transverse view of the parenchyma cells of RIL88(qph1) and RIL88(QPH1). (E, F) Transverse view of the epidermal region of RIL88(qph1) and RIL88(QPH1).
Map-based cloning of qph1 and validation through allelism testing
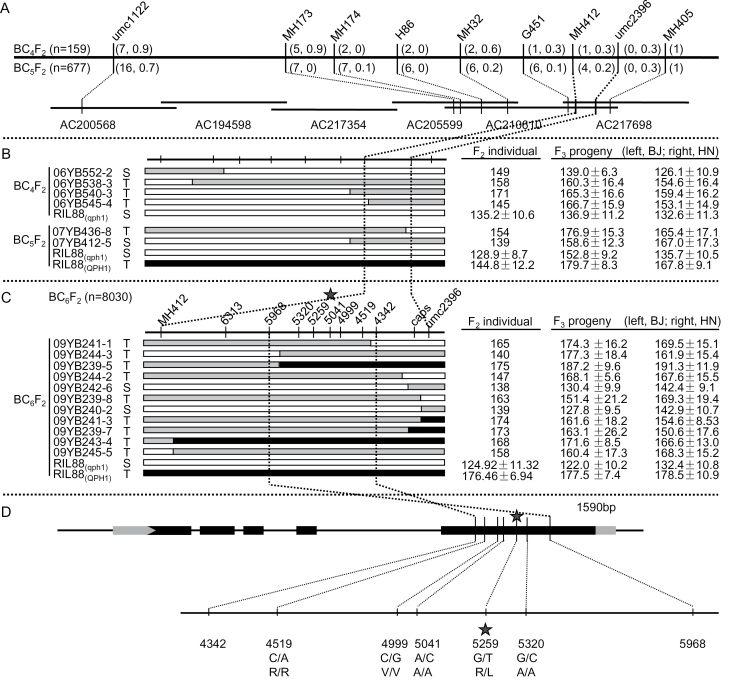
Two lines, RIL279 (the tall line and donor of allele QPH1) and RIL88 (the short line and recurrent parent, containing the qph1 allele), which shared 70% genetic background, were selected from the Zong3/87-1 RIL population to generate a fine-mapping population of qph1. The qph1 allele was first mapped to a 4.9kb region flanked by umc2396 (chromosome 1: 202337847-202337976) and MH412 (chromosome 1: 202342789-202342908) on BAC AC210610 in BC4F2 and BC5F2 populations (Fig. 4AB). To achieve higher map resolution, an expanded BC6F2 mapping population was developed and out of the 8030 individuals genotyped from kernel chips, 45 recombinants of 11 crossover types were identified by genotyping with flanking markers umc2396 and MH412 (Fig. 4C). Eleven recombinants were sequenced for the five SNPs within the target region, and the progeny plants were phenotyped in two different locations (Beijing and Hainan). Two key recombinants, 09YB241-1 and 09YB244-3, allowed the mapping interval to be narrowed down to 1590bp. The final interval has both borders falling in the fifth exon of the maize Brachytic2 gene (ZmPGP1, GRMZM2G31537), and among the five identified SNPs, only SNP5259 (G/T) caused an amino acid substitution from arginine (R) to leucine (L) (Fig. 4D).
Fig. 4.
Map-based cloning of qph1. (A) Locations of markers used in BC4F2 and BC5F2 populations. Number of recombinants and crossover rates between markers are shown before and after the comma. (B) Recombinants identified in BC4F2 and BC5F2. White, black, and grey indicate homozygous RIL88(qph1), homozygous RIL88(QPH1), and heterozygous genotypes. Plant height of recombinant individuals and their progenies in two locations are shown. T, tall; S, short. (C) Fine mapping of qph1 using the BC6F2 population. MHC412 and umc2396 were used as flanking markers to identify recombinants. Recombinants were sequenced to determine crossover break points, allowing the qph1 interval to be narrowed down to 4.9kb between SNPs 5968 and 4342. (D) Sequence analysis of the qph1 target region. The qph1 interval locates in exon 5 of the Br2 gene. Five SNPs were identified within it; only SNP5259 caused an amino acid substitution from arginine to leucine. Nucleotides and amino acids in RIL88(qph1) and RIL88(QPH1) for polymorphic loci are shown before and after the slash.
To evaluate the allelic effect between the natural variation present and the known br2 mutants, four br2 mutants (114F, 114G, 117A, and 121B) were obtained and crossed with RIL88(QPH1) and RIL88(qph1), respectively. Significant differences in plant height and ear height between the hybrids were observed; the phenotypic defects of the four br2 mutants were completely compensated by crossing with RIL88(QPH1) and only partially reversed by crossing with RIL88(qph1) (Supplementary Figure S7), implying that qph1 is allelic to the Br2 gene and is defective compared with QPH1. Moreover, in the F2 population of 117A and RIL88 (qph1), plant height and ear height segregated in a 3:1 ratio (qph1/qph1 and qph1/br2-117A plants to br2-117A/br2-117A plants), indicating that the qph1 allele is dominant to br2-117A (Supplementary Figure S7). Moreover, although RIL88(qph1) is not as short as the four br2 mutants, it has comparably low ear height (Supplementary Figure S7), which is a favourable feature directly related to lodging resistance.
Subcellular localization of QPH1
In order to detect the subcellular location of QPH1, the full-length QPH1 protein fused at its C-terminus with the GFP coding region protein was overexpressed in onion epidermal cells using the GFP construct not fused to a plant protein as the control. As revealed by transient expression results, the untargeted GFP was expressed throughout the whole transformed cell in cytoplasm, nucleus, and membrane. However, the QPH1–GFP fusion protein was expressed exclusively in membrane (Supplementary Figure S8); this result is consistent with the known localization of br2 homologues and cellular function of ABC transporters.
Expression analysis
The Br2 gene was previously shown to have high expression in stalk internodes, moderate expression in leaves, and very low expression in roots (Mutani et al., 2003). To investigate the underlying mechanism for the reduction in internode length of RIL88(qph1), stem tissues of RIL88(qph1) and RIL88(QPH1) from three developmental stages during elongation, when the phenotypic difference in internode length starts to be visible, were subjected to qRT-PCR analysis. The expression of qph1 was lower in period A at the beginning of elongation, reached a higher level in mid-elongation at period B, and began to decrease in period C before the end of elongation. T-test showed no significant differences in expression between RIL88(qph1) and RIL88(QPH1) in any of the three stages, indicating that the plant height and ear height difference between the near-isogenic lines is not caused by the difference in transcription level of qph1 (Supplementary Figure S9).
Association analysis using the qph1 SNPs
Based on the association result of plant and ear height in a population consisting of 527 inbred lines (Yang et al, 2010b), SNP5259 is a rare SNP that only exist in five lines and the four synonymous SNPs (SNP4519, 4999, 5041, and 5320) were shown not to be associated with phenotype. Due to the low number of inbred lines that have nucleotide T at position 5259, the non-synonymous SNP5259 could not be validated by association analysis (Supplementary Table S2). The five inbred lines that harbour SNP5259 (T), 81162, Ye107, Dan9046, W138, and Zong3 are all temperate lines that belong to the same heterotic group. All five lines have plant heights shorter than average: 113, 144, 133, 136, and 153cm for 81162, Ye107, Dan9046, W138, and Zong3, respectively. (Data was collected from >50 plants grown in five different locations; the average plant height of 527 inbred lines was 173cm.)
A total of 192 teosinte entries (data not shown) were also sequenced and analysed for the qph1 confidence interval. The four synonymous SNPs were found existing in either homozygous or heterozygous states in teosinte; however, only the homozygous G allele (wild-type genotype) was identified for SNP5259, suggesting that the causative mutation in qph1 most likely occurred as part of the temperate maize breeding program after the domestication of maize. The rare frequency of SNP5259 (T) also implies that the mutation occurred very recently and is not widely used in breeding programs.
Validation of qph1 and its functional site with Arabidopsis mutant atpgp1-2
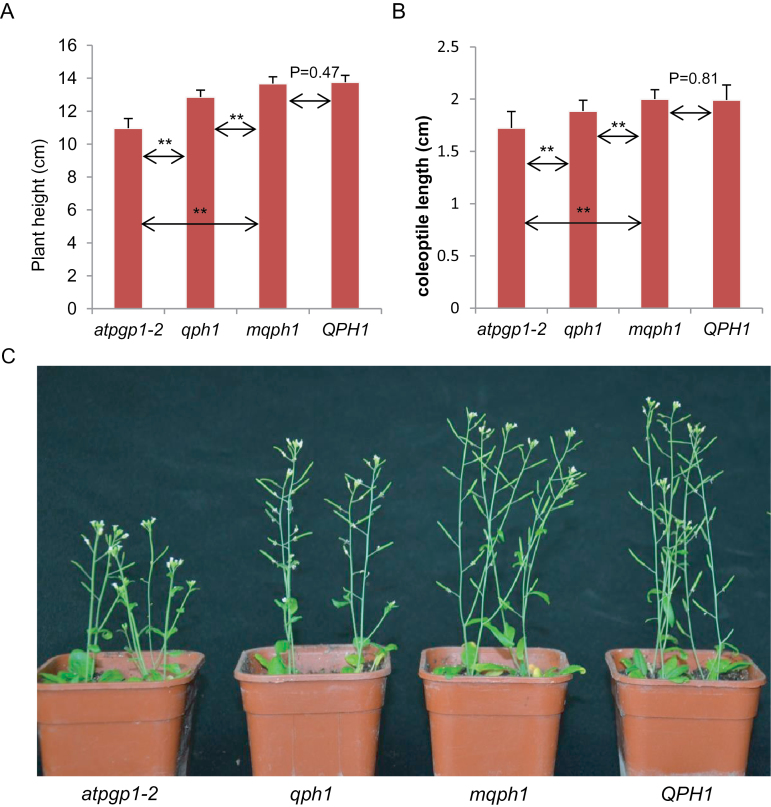
To validate the function of qph1 and its functional site SNP5259, site-directed mutagenesis was performed on the maize qph1 allele to mutate SNP5259 (T) to SNP5259 (G). QPH1, qph1, and mqph1 (mutagenized qph1) were cloned into vector PBI121 and overexpressed with the 35S promoter in Arabidopsis T-DNA insertional mutant atpgp1-2 (AtPGP1, At2g36910, is the homologue of the maize Br2 gene in Arabidopsis), which has reduced plant height and coleoptile length (Ye et al, 2013). QPH1 and mqph1 could restore the plant height and coleoptile length of the atpgp1-2 mutant to that of the wild type; in contrast, transformation of atpgp1-2 with the qph1 allele led to an intermediate level of rescue (Fig. 5). No significant difference was observed between QPH1 and mqph1 transgenic plants in plant height and coleoptile length, but a significant difference was detected between qph1 and mqph1 transgenic plants (P = 0.0007 for plant height and P = 0.005 for coleoptile length) (Fig. 5AB); the only difference between mqph1 and qph1 coding sequence is the G/T polymorphism at SNP5259; this result is consistent with the hypothesis that SNP5259 (T) is the underlying causative mutation in qph1. Moreover, statistical analysis showed that the difference in plant height and coleoptile length between qph1 transgenic plants and the atpgp1-2 control also reached significant levels (P = 0.00023 and 2.93E-11), suggesting that qph1 is not a complete loss of function allele (Fig. 5A, B).
Fig. 5.
Validation of qph1 and its functional site through transformation of Arabidopsis homologous mutant atpap1-2. (A) Plant height of the atpgp1-2 mutant transformed with pBI121 empty vector, qph1, mqph1 (mutated qph1), and QPH1. (B) Coleoptile length of the atpgp1-2 mutant transformed with pBI121 empty vector, qph1, mqph1 (mutated qph1), and QPH1. (C) Plant height of qph1, mqph1 (mutated qph1), and QPH1 transformation plants. **, significant difference (0.01 level). Data is expressed as mean ± SD.
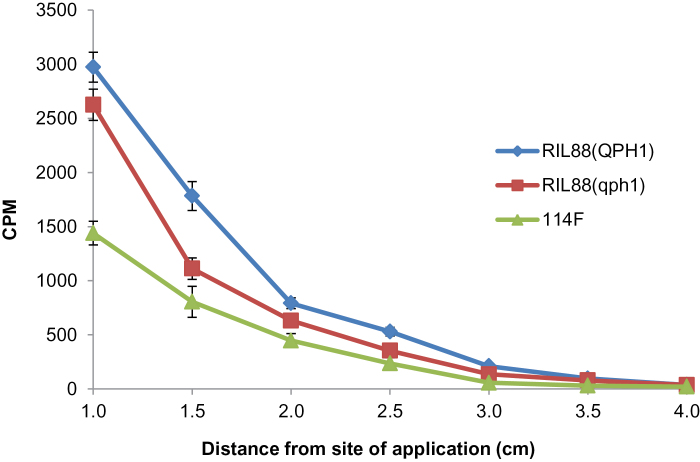
Polar auxin transport in RIL88(qph1), RIL88(QPH1) and the br2 mutant 114F
Because the maize Br2 gene is known to function in polar auxin transport (Multani et al, 2003), an assay of basipetal transport of [3H] IAA was carried out in the coleoptiles of RIL88(qph1), RIL88(QPH1), and the null br2 mutant 114F. Results showed that [3H] IAA translocation to the lower sections of the coleoptiles was significantly reduced in RIL88(qph1) and the br2 mutant 114F compared to RIL88(QPH1) (Fig. 6). As the br2 mutant consistently showed increased loading of auxin into the upper coleoptile near the site of application in former studies (Multani et al, 2003), the difference was probably caused by the impaired polar auxin transport in defective br2 plants. Consistent with the hypothesis that the qph1 allele is partially defective compared with the br2-117A allele, a significant difference in [3H] IAA translocation between RIL88(qph1) and 114F was detected, indicating that qph1 was not a complete loss of function allele.
Fig. 6.
Transport of [3H] IAA in coleoptiles of RIL88(QPH1), RIL88(qph1), and the br2 mutant 114F. CPM, counts per minute. Error bars indicate SD, n=8.
Functional assessments of qph1 in F2 populations of different genetic backgrounds
To evaluate the effect of qph1 under different genetic backgrounds, four F2 populations, 4F1×81162, Ye107 × Zheng32, Ye107 × B73, and Zong3 × Chuang48-2 were constructed by crossing the three lines containing the rare SNP (T) (81162, Ye107, and Zong3) with normal inbred lines (4F1, Zheng32, B73, and Chuan48-2). The four F2 populations were planted in two different locations, Beijing and Hainan (200 individuals per population in each location). In each replicate, 50 qph1/qph1, 100 qph1/QPH, and 50 QPH1/QPH1 individuals per population were phenotyped. In both locations, a significant difference in plant height and ear height was detected between QPH1/QPH1 and qph1/qph1 individuals and between QPH1/qph1 and qph1/qph1 individuals. Plant height difference between QPH1/QPH1 and qph1/qph1 individuals in the B73 × Ye107 F2 population is shown as an example (Supplementary Figure S10). Single-factor ANOVA analysis in 4F1/81162 F2 detected a strong dominant effect for plant height (D/A = 1.3 and 0.9, P = 6.06E-16 and 1.21E-18 in 09HN and 10BJ, respectively); similar results were also observed in the other three F2 populations. 15–49% and 4–37% phenotypic variation could be explained by qph1 for plant height and ear height, respectively, in different genetic backgrounds and environments (Supplementary Table S3 and Supplementary Figure 7A). These results show that qph1 has a consistent effect in reducing plant height and ear height in multiple genetic backgrounds.
A semi-dwarf mutant N546 (Johnson et al., 1998) has features very similar to the br2 recessive lines and a QTL peak for plant/ear height was detected at the Br2 region in the N546 × PHB00 F2 population. The N546 Br2 allele is designated br2-bj; it has a 3.5kb insertion of En/Spm-like transposon 660bp upstream of the predicted TATA box and a complete gag/pol retro-transposon insertion of 4.7kb in exon 5 which truncates the last 153 amino acids of the protein. Mean plant/ear height for br2-bj/br2-bj individuals are 88% and 50% of the heterozygous and wild-type plants, respectively. Consistent with the predicted reduction or loss of function of this complex allele, br2-bj behaves as a recessive allele in nine F2 populations constructed by crossing N546 and two of its conversion lines N501 and N504 (with the br2-bj allele) with normal lines (PHVRZ, PHHHN, PHVNV, PHF0D, OHCCW, and PH128S). In these segregating populations, heterozygous and wild-type plants showed no significant difference in plant height; ear height between wild-type and heterozygous plants showed a significant difference in some crosses depending on the background (Supplementary Figure S11). Results indicate that br2-bj has different effects on plant height and ear height in F2 segregation populations from qph1.
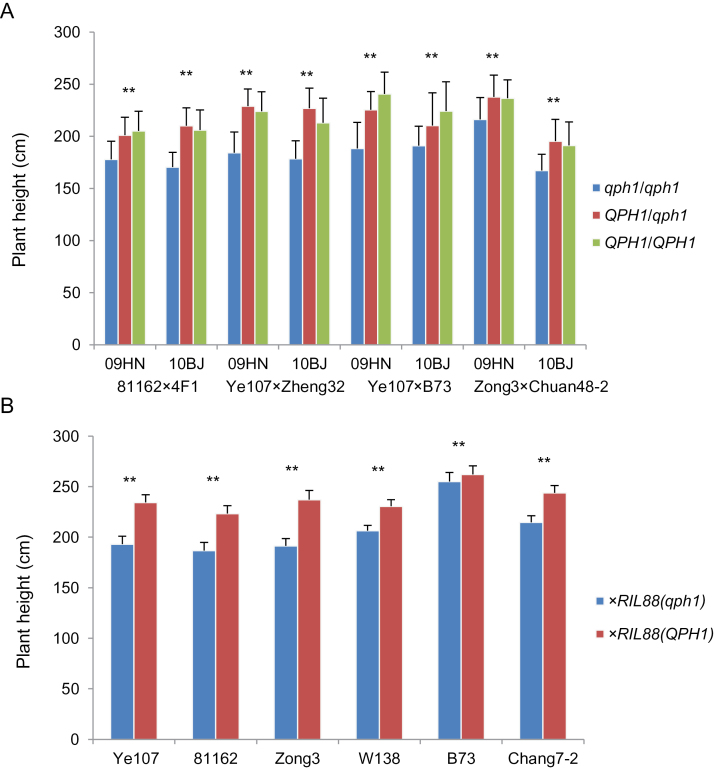
Functional assessments of qph1 in six pairs of single-cross hybrids
Because hybrid maize is used in agriculture, the effect of qph1 was also estimated in hybrid backgrounds. Six pairs of single-cross hybrids were generated by crossing RIL88(QPH1) and SNP5259 (T)-containing RIL88(qph1) with the inbred lines with or without the SNP5259 (T); effects of qph1/qph1 with qph1/QPH1 and QPH1/qph1 with QPH1/QPH1 under the same hybrid genetic backgrounds were evaluated. Plant and ear height showed significant differences between each pair of hybrids (Fig. 7B). In the hybrids derived by crossing Ye107, 81162, Zong3, and W138 with RIL88(qph1), plant heights between F1 individuals of qph1/qph1 were reduced by 10–24%, respectively, compared to their QPH1/qph1 counterparts derived from RIL88(QPH1). Reductions in ear height were also significant, ranging from 26 to 38%. These results indicate the strong effect of the homozygous recessive qph1/qph1 in reducing plant height and ear height. Moreover, in B73 and Chang7-2 F1 hybrids, due to the incomplete recessive feature of qph1, significant differences in plant height and ear height between QPH1/QPH1 and QPH1/qph1 hybrids were also detected; phenotypic variations in plant height and ear height range from 3 to 12% for plant height and 6 to 23% for ear height, indicating that qph1 is able to reduce plant height and ear height in a heterozygous state. qph1 is proved to have a significant effect on plant height and ear height in hybrids, and the incomplete recessive feature of qph1 allows the plant and ear height of a hybrid to be modified by introducing it into only one parent.
Fig. 7.
Plant height analysis of the four F2 populations and six pairs of single-cross hybrids. (A) Blue, red, and green represent the plant height of the qph1/qph1, QPH1/qph1, and QPH1/QPH1 individuals, respectively. Phenotype data were collected from two replicates in Hainan (2009, left) and Beijing (2010, right). 50, 100, and 50 individuals of qph1/qph1, QPH1/qph1, and QPH1/QPH1 were planted and analysed, respectively, for each population. (B) Plant height of RIL88(qph1) (blue) and RIL88(QPH1) (red) derived single-cross hybrids; 30–40 individuals were planted and analysed, respectively, in each genotype class, and data was collected from two replicates for each hybrid. **, significant difference (0.01 level). Data are expressed as mean ± SD.
Prospect evaluation of the impact of qph1 on grain yield
In our previous studies, a pleiotropic effect of qph1 causing a marginal influence on yield and yield components was detected in RIL, IF2, F2:3, and BC4F2 populations of Zong3/87-1; this was probably contributed by the genetic backgrounds (Tang et al., 2007a, b ). To evaluate the potential of qph1 in maize improvement in terms of increasing yield potential, a number of agronomic traits of the six pairs of F1 and four F2 populations derived from qph1-containing lines were measured and analysed. Most yield components exhibited no significant difference between qph1/qph1, QPH1/qph1, and QPH1/QPH1 individuals in the four F2 populations, and significant differences could be detected in only one biological repeat for a few traits in some F2 populations, indicating that qph1 has no or a slight influence on maize yield under different genetic backgrounds while maintaining a shorter plant height and ear height (Supplementary Figures S10 and S12). Likewise, except for ear kernel weight and 100-kernel weight in 81162 F1, and ear weight and days to shedding in Zong3 F1 and ear kernel weight in B73 F1, no additional significant difference was identified between each pair of the single-cross hybrids, suggesting that qph1 only has a minor impact on yield under hybrid backgrounds (Supplementary Table S4). These results suggested that qph1 could significantly reduce plant height and ear height with no or very little negative impact on yield under multiple genetic backgrounds. qph1 could affect plant height and ear height when heterozygous, making it very useful for hybrid maize breeding.
Discussion
Molecular mechanism underlying the major plant height QTL qph1
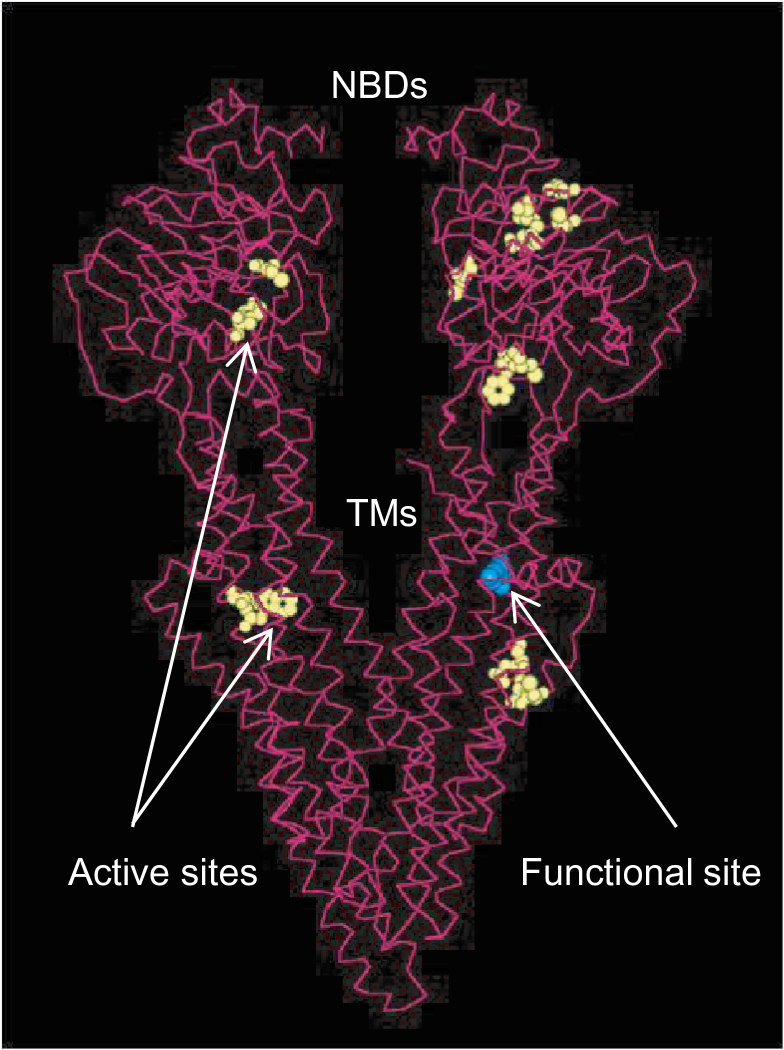
In this study, we report a rare SNP mutation in the maize Brachyric2 gene underlying the major plant height QTL qph1. The maize Br2 (ZmPGP1) gene is an ABC (ATP-binding cassette) transporter which belongs to the MDR (multi-drug resistant) class of P-glycoprotein (Noh et al., 2001; Multani et al., 2003) and functions in polar auxin transport as an efflux carrier. The protein molecule consists of two transmembrane domains (TMDs) that provide the translocation pathway of auxin and two cytoplasmic nucleotide-binding domains (NBDs) that hydrolyse ATP and drive the transport reaction (Chang and Roth, 2006; Aller et al., 2009). The two TMDs span the membrane through the 12 α-helices (six per domain) and enable membrane insertion and regulation. The predicted causative mutation of qph1, SNP5259 (T), which resulted in the arginine to leucine substitution, is located on the ninth α-helix in the TMD of Br2 and thus affects the efficiency of the transmembrane channel (Fig. 8). As an efflux carrier, amino acid residues along the transporter channel are strictly arranged (in this case, all residues are positively charged); Arg has a hydrophilic and positively charged side chain, while Leu is hydrophilic and neutral; the substantial change from Arg to Leu in qph1 is very likely to have affected its interaction with negatively charged IAA– inside the cell. Moreover, auxin is synthesized predominantly in the shoot apex, young leaves, and developing seeds (Ljung et al., 2001) then dispensed to other organs by multiple efflux and influx transporters (Friml et al, 2002; Zhao et al, 2010). The results are consistent with the hypothesis that the defective qph1 allele in RIL88(qph1) impaired basipetal auxin transport, which led to the auxin insufficiency in lower internodes and resulted in shortened internodes. Reduced cell division and changes in vascular bundle development were observed in RIL88(qph1) lower internodes, which is consistent with auxin deficiency (Galweiler et al., 1998). qph1 affects leaf number, leaf angle, and flowering time minimally compared with plant height and ear height, suggesting that it has potential for maize improvement.
Fig. 8.
Protein structure simulation of qph1 generated using Pymol. The six active sites of the protein are shown in yellow; the Arginine to Leucine amino acid substitution on the ninth α-helix in the transmembrane domain is indicated in blue. TMs, transmembrane domains; NBDs, nucleotide binding domains.
Potential application of qph1 in maize improvement
Cereal production went through a dramatic increase due to the adoption of short cultivars during the last century, known as the Green Revolution. Underlying genes were later isolated in rice, wheat, sorghum and several other crops (Galweiler et al., 1998). Obvious defects in major genes were found to be responsible for phenotypic variation in most cases, and thus made them easily manipulated for practical use. In maize, however, loss of function mutations of the major plant height genes often led to serious defects and very large yield loss (Winkler et al., 1995; Thornsberry et al., 2001), so moderate-effect QTLs were considered to be excellent alternatives. QTL mapping has long been conducted to localize maize plant height regulators with desirable effects, but has rarely resulted in candidate gene cloning. Several maize genes underlying quantitative traits have been cloned and validated based on linkage analysis; examples include Vgt1, Tga1, and DGAT1-2 (Wang et al., 2005; Salvi et al., 2007; Zheng et al., 2008), but plant height QTLs were rarely cloned (Teng et al, 2012). Among the plant height factors identified in maize so far, the recessive br2 gene is considered to have great potential and efforts have been made to use it practically (Anderson and Chow, 1960; Djisbar and Brewabaker, 1987). Introgression of br2 into normal varieties could reduce plant height and ear height by shortening each internode (Souza and Zinsly, 1985), but unfortunately all the recessive br2 alleles identified so far cause severe phenotypes and it has not been possible to use them in breeding. Here, we provide the detailed phenotypic and molecular characterization of the naturally occurring mild allele of br2, qph1; it is a very rare SNP mutation that might have occurred recently and hasn’t been widely used in breeding programs. The cloning of qph1 sheds more light on the molecular nature of natural variation at maize QTLs; it demonstrates that the naturally occurring allele at a QTL locus and a strong dwarf mutant are genetic variants of the same gene. Results of this study revealed qph1 as a major plant height QTL that has a moderate effect on plant height and no or minimal negative effects on grain yield under various genetic backgrounds tested, suggesting its potential in maize improvement by marker-assisted selection for reduced plant height and lodging resistance.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. Cytology analysis of the RIL88(qph1) and RIL88(QPH1) stem tissues.
Supplementary Table S2. Association analysis of the five SNPs within the target region of qph1 in a population of 500 inbred lines.
Supplementary Table S3. Plant height and ear height analysis of the four F2 populations.
Supplementary Table S4. Yield-related trait analysis of single-cross hybrids derived from RIL88(qph1) and RIL88(QPH1).
Supplementary Table S5. Primers used in this study.
Supplementary Figure S1. Construction of qph1 fine-mapping population BC6F2 and near-isogenic lines RIL88(QPH1) and RIL88(qph1).
Supplementary Figure S2. Fine mapping of qph1 in BC4F2 and BC4F2:3.
Supplementary Figure S3. Phenotypic variation and distribution of the yield-related traits in BC4F2:3.
Supplementary Figure S4. Stalk internode length variation between RIL88(qph1) and RIL88(QPH1).
Supplementary Figure S5. Scanning electron microscopy examination of the sixth internodes from RIL88(qph1) and RIL88(QPH1).
Supplementary Figure S6. Scanning electron microscopy examination of the uppermost internodes from RIL88(qph1) and RIL88(QPH1).
Supplementary Figure S7. Allelism test of qph1.
Supplementary Figure S8. Subcellular localization of QPH1.
Supplementary Figure S9. qph1 expression in RIL88(qph1) and RIL88(QPH1) internodes from three developmental stages during elongation.
Supplementary Figure S10. Plant height and yield performance comparison between individuals of different genotypes in the B73 × Ye107 F2 population.
Supplementary Figure S11. Plant height and ear height segregation in nine F2 populations of N546 conversion lines and normal inbred lines.
Supplementary Figure S12. Yield component analysis of the four F2 populations.
Funding
This work was supported by a grant from the National High Technology Research and Development Program of China (863 Program, No. 2012AA10A307)
Supplementary Material
Acknowledgments
We thank Dr Kevin Fengler for helping with BAC AC210610 and providing the physical locations of genetic markers; and Dr J. Antoni Rafalski and Dr Robert L. Last for editing the paper. We also thank Marty Sachs for providing the four br2 mutants.
References
- Aller SG, Yu J, Ward A, et al. 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Chow PN. 1960. Phenotypes and grain yield associated with br2 gene in single cross hybrids of dent corn. Crop Science 1, 335–337. [Google Scholar]
- Chang G, Roth CB. 2006. Structure of MsbA from E. coli: A homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293, 1793–1800. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter 1, 19–21. [Google Scholar]
- Djisbar A, Brewabaker JL. 1987. Effects of the brachytic-2 gene on maize yield and its components. Maydica 32, 107–123. [Google Scholar]
- Duvick DN. 2005. Genetic progress in yield of United States maize (Zea mays L.). Maydica 50, 193–202. [Google Scholar]
- Friml J, Palme K. 2002. Polar auxin transport – old questions and new concepts? Plant Molecular Biology 49, 273–284. [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Joseph D, Clarke JD, Graner E, Hansen M. 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE. 6, e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Martinez C, Skinner DJ, Krivanek AF, Crouch JH, Xu Y. 2008. Development of a seed DNA-based genotyping system for marker-assisted selection in maize. Molecular Breeding 22, 477–494. [Google Scholar]
- Hashemi A, Herbert SJ, Putnam DH. 2005. Yield response of corn to crowding stress. Agronomy Journal 97, 839–846. [Google Scholar]
- Johnson BE, Rodríguez-Herrera SA, Donald H. 1998. A parental line of maize. Crop Science 38, 574–574. [Google Scholar]
- Leng ER, Vineyard ML. 1951. Dwarf and short plants. Maize Genetics Cooperation Newsletter 25, 31–32. [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J. 2007. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Research. 17, 402–410. [DOI] [PubMed] [Google Scholar]
- Liang BC, Remillard M, Mackenzie HF. 1992. Effects of hybrids, population densities, fertilization and irrigation on grain corn (Zea mays L.) in Qubec. Canadian Journal of Plant Science 72, 1163–1170. [Google Scholar]
- Livak KJ, Schmittren TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ljung K, Ostin A, Lioussanne L, Sandberg G. 2001. Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiology. 125, 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Tang J, Teng W, Yan J, Meng Y, Li J. 2007. Epistatic interaction is an important genetic basis of grain yield and its components in maize. Molecular Breeding 20, 41–51. [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302, 81–84. [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. 2001. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 13, 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KY, Park SU, Moon HG. 1989. Effects of planting density and tiller removal on growth and yield of sweet corn hybrids. Korean Journal of Crop Science 34, 192–197. [Google Scholar]
- Peng JR, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F. 1999. “Green revolution” genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E. 2007. Conserved non-coding genomic sequences controlling flowering time differences in maize. Proceedings of the National Academy of Sciences, USA 104, 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. 2002. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Scott GE, Campbell CM. 1969. Internode length and brachytic-2 maize inbreds in single cross. Crop Science 9, 293–295. [Google Scholar]
- Souza CL, Jr, Zinsly JR. 1985. Relative genetic potential of brachytic maize (Zea mays L.) varieties as breeding populations. Brazilian Journal of Genetics 3, 523–533. [Google Scholar]
- Tang J, Teng W, Yan J, Ma X, Meng Y, Li J. 2007a. Genetic analysis of plant height using a set of recombinant inbred line population in maize. Euphytica 155, 117–124. [Google Scholar]
- Tang J, Ma X, Teng W, Yan J, Wu W, Dai J, Li J. 2007b. Detection of quantitative trait loci and heterotic loci for plant height using an immortalized F2 population in maize. Chinese Science Bulletin 52, 477–438. [Google Scholar]
- Tang J, Yan J, Ma X, Teng W, Wu W, Dai J, Dhillon BS, Melchinger AE, Li J. 2010. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theoretical and Applied Genetics 120, 333–340. [DOI] [PubMed] [Google Scholar]
- Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z. 2012. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. The Plant Journal 73, 405–416 [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES. 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics 28, 286–289. [DOI] [PubMed] [Google Scholar]
- Vafias B, Ispalandis CG, Goulas C, Deligeorgidis PN. 2006. An approach on yielding performance in maize under varying plant densities. Asian Journal of Plant Science 5, 690–694. [Google Scholar]
- Vega CRC, Andrade FH, Sadras VO. 2001. Reproductive partitioning and seed set efficiency in soybean, sunflower and maize. Field Crops Research 72, 165–173. [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF. 2005. The origin of the naked grains of maize. Nature 436, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Helentjaris T. 1995. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. The Plant Cell 7, 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yan J, Tang H, Huang Y, Shi Y, Li J, Zheng Y. 2003. QTL mapping for developmental behavior for plant height in maize. Chinese Science Bulletin 48, 2601–2607. [Google Scholar]
- Yan J, Tang H, Huang Y, Zheng Y, Li J. 2006. Quantitative trait loci mapping and epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica 149, 121–131. [Google Scholar]
- Yan WH, Wang P, Chen HX, et al. 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319–330. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang Y, Zhang J, Liu P, Li C, Zhu Y, Hao M, Li D, Dong S. 2010a. Dry matter production and photosynthesis characteristics of three hybrids of maize (Zea mays L.) with super-high-yielding potential. Acta Agronomica Sinica 37, 355–361. [Google Scholar]
- Yang X, Yan J, Shah T, Warbuton ML, Li Q, Li L, Gao Y, Chai Y, Fu Z, Zhou Y. 2010. Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theoretical and Applied Genetics 121, 417–431. [DOI] [PubMed] [Google Scholar]
- Ye L, Liu L, Xing A, Kang D. 2013. Characterization of a dwarf mutant allele of Arabidopsis MDR-like ABC transporter AtPGP1 gene. Molecular Cell Biology Research Communications 441, 782–786. [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2010. Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology 61, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W. 2008. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nature Genetics 40, 367–372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.