Highlight
Survival of A. tequilana, under arid conditions depends on a failsafe mechanism for asexual reproduction based on changes in auxin mobility controlled by two functionally distinct PIN1-related proteins.
Key words: Agave tequilana, auxin flux, bulbil, development, gene expression, immunolocalization, PIN proteins.
Abstract
In Agave tequilana, reproductive failure or inadequate flower development stimulates the formation of vegetative bulbils at the bracteoles, ensuring survival in a hostile environment. Little is known about the signals that trigger this probably unique phenomenon in agave species. Here we report that auxin plays a central role in bulbil development and show that the localization of PIN1-related proteins is consistent with altered auxin transport during this process. Analysis of agave transcriptome data led to the identification of the A. tequilana orthologue of PIN1 (denoted AtqPIN1) and a second closely related gene from a distinct clade reported as ‘Sister of PIN1’ (denoted AtqSoPIN1). Quantitative real-time reverse transcription–PCR (RT-qPCR) analysis showed different patterns of expression for each gene during bulbil formation, and heterologous expression of the A. tequilana PIN1 and SoPIN1 genes in Arabidopsis thaliana confirmed functional differences between these genes. Although no free auxin was detected in induced pedicel samples, changes in the levels of auxin precursors were observed. Taken as a whole, the data support the model that AtqPIN1 and AtqSoPIN1 have co-ordinated but distinct functions in relation to auxin transport during the initial stages of bulbil formation.
Introduction
Agaves are perennial monocarpic plants with life cycles ranging from 5 to 7 years for Agave tequilana (Valenzuela, 2003, 2011) to an estimated 53 years for A. deserti (Nobel, 1976), during which carbohydrates accumulate in stem tissue in the form of oligofructans (Lopez et al., 2003; Mancilla-Margalli and Lopez, 2006) which are exploited in A. tequilana to produce tequila under a controlled denomination of origin and in other species to produce mescal. Currently much interest has been shown in the potential of agave plants as sources for biofuel production since they are not exploited as food sources and can be grown on land not suitable for production of many traditional crop plants (Borland et al., 2009).
Many species of the Agave genus have evolved to survive under arid conditions (Nobel, 1976; Gentry, 1982; Arizaga et al., 2000a, b), and an important adaptation is a failsafe strategy involving the production of thousands of aerial vegetative plantlets or bulbils on the inflorescence (Arizaga and Ezcurra, 1995, 2002). This strategy comes into play when sexual reproduction is inefficient or unsuccessful, and ensures asexual reproduction even in the absence of other flowering individuals and pollinators or in the event of adverse environmental conditions (Arizaga and Ezcurra, 1995, 2002).
The formation of aerial bulbils has been documented in several groups of plant species; however, the mechanisms for inducing and carrying out bulbil development in different species are very distinct. In Titanotrichum oldhamii, vegetative meristems develop in the vicinity of the floral meristem when this tissue apparently loses its identity, leading to the formation of vegetative bulbils within the original floral structure (Wang and Cronk, 2003). In Kalanchoë species such as K. diagremontiana, a somatic embryogenesis programme similar to zygotic embryogenesis is undergone where the transcription factor Leafy Cotyledon1 plays an essential role (Garces et al., 2014).
Bulbil formation is also common in other members of the Liliopsida class (which includes the Agave genus), such as Allium sativum (Ebi et al., 2000; Mathew et al., 2005) and Dioscorea japonica where miniature bulbs are formed in leaf axils or on flower heads. These bulbils are often dormant and will germinate the following season (Kim et al., 2005; Walck et al., 2010). In contrast, agave species directly produce numerous small plantlets at the bracteoles of the inflorescence that eventually fall to the ground and grow independently (Abraham-Juarez et al., 2010). The formation of new meristems following bulbil induction implies that cells in the pedicel tissue are reprogrammed actively to undergo cell division to form vegetative structures similar to the activation of dormant axillary meristems following removal of the shoot apical meristem (SAM) (Leyser, 2005). Thousands of bulbils are produced on a single inflorescence in a few months in comparison with 1–2 offsets produced from the rhizomes of the mother plant each year, and reports have shown a slightly higher level of somaclonal variation in bulbils in comparison with offsets (Gonzalez et al., 2003; Diaz-Martinez et al., 2012).
A better understanding of the process of bulbil development will provide valuable information regarding the developmental and molecular mechanisms associated with this process in comparison with that of pre-formed, dormant axillary meristem development in other species where the number of meristems and new branches is strictly controlled. The plant hormone auxin plays an essential role in orchestrating the developmental changes that occur throughout a plant’s life cycle (Bohn-Courseau, 2010; Peer, 2013) and, when auxin gradients are altered, developmental and growth patterns are also modified (Mai et al., 2011; Goh et al., 2012; Rademacher et al., 2012). In particular, removal of the SAM leads to activation of dormant axillary meristems by a mechanism thought to involve localized changes in the balance of auxin and cytokinin levels (Muller and Leyser, 2011). Expression of several transcription factors is also affected by auxin including the KNOX, MADS, and LEAFY genes which have previously been implicated in bulbil induction in A. tequilana or T. oldhamii (Wang et al., 2004; Abraham-Juarez et al., 2010; Delgado Sandoval et al., 2012). These observations suggest that the modulation of auxin gradients following removal of flower buds in A. tequilana might also determine the initiation of bulbil formation.
The synthesis, degradation, conjugation, or transport of auxin molecules regulates the presence of auxin within specific plant tissues. In particular, auxin mobility mediated by polarized auxin efflux transporters known as PIN-formed (PIN) proteins plays an important role in determining developmental responses to environmental or physical cues. PIN proteins are composed of two highly conserved transmembrane domains that flank a more variable hydrophyllic loop. PIN proteins with long loop domains are located at the cell membrane in a polarized fashion where they mediate the directional flow of auxin within plant tissues. A detailed analysis of PIN1-related proteins from grass species identified a new clade ‘Sister of PIN1’ (SoPIN1) and, based on inmunolocalization and computer simulation, a model for the distinct roles of PIN1 and SoPIN1 during organ initiation was proposed (O’Connor et al., 2014). In addition, different combinations of motifs within the hydrophilic loop region are consistent with the functional divergence of the PIN proteins (Wang et al., 2014).
The aim of this work was to determine the role of auxin in the stimulation of bulbil development in A. tequilana and to explore the role of auxin transport mediated by PIN1-related proteins during this process. The characterization of two distinct A. tequilana PIN1 genes from different clades and their contrasting patterns of expression during bulbil development are described. By analysis of PIN1 protein localization, the formation of basipetal auxin gradients during the formation of vegetative propagules in A. tequilana is shown when the apical floral meristem is cut, demonstrating overlap between apical dominance and bulbil formation. By means of exogenous application of auxin, the role of auxin in the suppression of bulbil formation is also demonstrated for the first time and, based on the results, a preliminary model for the control of this process at the molecular level is proposed.
Materials and methods
Plant material
Bulbil samples were obtained at the same stages of development as described previously (Abraham-Juarez et al., 2010): S0, immediately before bud excision; S1, 7 d after bud removal (no visible change observed); S2, swelling; S3, necrosis; S4, bulbil eruption; and S5, nascent bulbils present (Supplementary Fig. S1A, B available at JXB online). These tissues were used to obtain mRNA and perform real-time PCR assays and immunolocalization assays.
Application of exogenous auxin
Three plants of A. tequilana with a 1 m long inflorescence were transferred from commercial fields to a greenhouse. They were monitored until flowering, and, when floral buds were 5cm long, bulbil induction was carried out by excision of the buds. Auxin or control treatments were applied to the pedicel daily beginning immediately after bud excision. Treatments were: (i) lanolin only as a control; (ii) lanolin containing 100 μM indole acetic acid (IAA); and (iii) lanolin containing 100 μM naphthylphthalamic acid (NPA). Treatments were carried out in summer (July–August), with a daylength of 14h light and 10h darkness. Tissue collection and treatment application were carried out in the morning. Each plant was divided into three sections and all treatments were applied to each individual plant. Pedicel tissue was collected for quantitative real-time reverse transcription–PCR (RT-qPCR) analysis and immunolocalization, and stored at –70 °C or fixed in FAA until needed. All samples were collected from the three individual plants as biological replicates.
Immunolocalization
Tissues were fixed in FAA [50% ethanol, 3.7% formaldehyde, 5% acetic acid, 0.5% Triton X-100, and 1% dimethylsulphoxide (DMSO)] overnight and were then dehydrated through an ethanol series and infiltrated with polyester wax (Steedman, Electron Microscopy Sciences). Following embedding and sectioning, 10 μm sections were placed onto Probe on plus® slides (Fisherbrand®). For antibody reactions, polyester wax was removed in 100% ethanol. The sections were then rehydrated in an ethanol series and blocked by incubating in phosphate-buffered saline (PBS) buffer (130mM NaCl and 10mM PO4) with 1% bovine serum albumin (BSA). The slides were incubated with an affinity-purified anti-Zea mays PIN1a antibody (Hake Lab; 1:200 dilution) in PBS with 1% BSA overnight at 4 °C. After three washes in PBS with 1% fish gelatin, the slides were incubated with a Cy3-conjugated anti-guinea pig secondary antibody (1:100 dilution; Jackson Immuno Research Laboratory). Slides were washed five times in PBS with 1% fish gelatin, mounted in 1% DABCO in 90% phosphate-buffered glycerol, and observed by fluorescence microscopy at 540nm using the same exposure time for all samples. Photographs were taken using an Olympus BX60 microscope fitted with an Olympus DP71 camera.
Sequence analysis
To identify PIN1 cDNA sequences within the A. tequilana transcriptome, two types of search were carried out: by key word and by using the BLAST tool to search specifically for PIN1-domain sequences. A threshold of E=10–6 was used to select sequences of interest. Alignments of amino acid sequences were performed using the Clustal W software (Larkin et al., 2007). A phylogenetic tree was constructed using the Neighbor–Joining (NJ) and maximum likelihood method in the MEGA 5.0 software suite (http://www.megasoftware.net/), and robustness was assessed by bootstrap analysis (Felsenstein, 1986) with 1000 replicates. Previously reported Arabidopsis, rice, and maize PIN proteins were used to classify the A. tequilana PIN sequences and name them accordingly. AtqPIN protein transmembrane helices were predicted using TMHMM2 (Krogh et al., 2001). Amino acid motifs were identified and annotated using Geneious software (http://www.geneious.com). AtqPIN1-1 and AtqSoPIN1 sequences have been deposited in GenBank under accession numbers: BankIt1715396 AtqPIN1 KJ676663 and BankIt1715396 AtqSoPIN1 KJ676664.
Quantitative real-time reverse transcription–PCR analysis
RT-qPCR was carried out for all tissues sampled. Total RNA was isolated using the PureLink Micro-to-Midi Total RNA Purification System (AmbionR) according to the manufacturer’s instructions. cDNA was synthesized using a reverse primer mix consisting of 1 μg of RNA, and RevertAid™ Reverse Transcriptase (Fermentas) on three independent replicates. All samples were run in duplicate, and qPCR was carried out for each cDNA replicate. The specific primers used for RT-qPCR are shown in Supplementary Table S1 at JXB online. PCRs were performed under the following conditions: 95 °C for 10min, 40 cycles of 95 °C for 15 s, 60 °C for 1min, and a final melt curve stage from 60 °C to 95 °C. Control reactions were carried out using specific primers for the ACT2 agave gene. For qPCR, the Step One Plus Real Time PCR System (Applied Biosystems) was used. SYBR Green PCR Master Mix (Applied Biosystems) was used for all reactions according to the manufacturer’s protocol. Gene expression was normalized by subtracting the CT value of the control gene from the CT value of the gene of interest. Average expression ratios were obtained from the equation 2–ΔΔCT, where ΔΔCT represents ΔCT (gene of interest in stage evaluated)–ΔCT (gene of interest at T0), according to the protocol reported by Czechowski et al. (2004).
Analysis of auxin, precursors, and conjugates
For quantification of IAA and related metabolites, extraction was carried out as described in Novak et al. (2012), using 10mg of dry tissue. To validate the quantification of endogenous auxin metabolites, the following stable isotope-labelled internal standards were added as tracers: [2H5]IAA, [2H5]IAAsp, [2H2]TRA, and [2H5]TRP (100 pmol per sample). Samples were analysed by UHPLC-Q-TOF MS (ultra high-pressure liquid chromatography/quadrupole time-of-flight mass spectrometry; Agilent Technologies, http://www.home.agilent.com), operating in positive ESI (electrospray ionization) mode. A 5 μl aliquot of each sample was injected onto a reversed-phase column [Waters Acquity CSH™ C18 1.7 μm, 2.1×50mm column, eluted using a 20min gradient comprising 0.1% acetic acid in water (A) and 0.1% acetic acid in methanol (B) at a flow rate of 0.2ml min–1], column temperature 30 °C. The following binary linear gradient was used: 0min, 90:10 A:B; 10.0min, 50:50 A:B; 12.5min, 40:60 A:B. At the end of the gradient, the column was washed with 100% methanol (2.5min), and re-equilibrated to initial conditions (5min). The eluent was introduced into the Agilent jet stream dual ESI source with the optimal settings as follows: drying gas temperature, 250 °C; drying gas flow, 10 l min–1; nebulizer pressure, 60 psi; sheath gas temperature, 400 °C; sheath gas flow, 12 l min–1; capillary voltage, 2250V; nozzle voltage, 500V. Peak integration was carried out under full scan conditions by using the extracted ion chromatogram (EIC) of the protonated molecule from the total ion chromatogram (TIC) with a 20 ppm mass extraction window. The accurate mass spectrum and retention time for all compounds were used for both confirmation and quantification purposes, with peak areas of EICs being used for quantification. Chromatograms were analysed using MASSHUNTER software version B.04.00 (Agilent Technologies).
Results
Removal of A. tequilana flower buds results in the formation of vegetative bulbils and/or new flower buds
In A. tequilana, bulbils are naturally produced close to the bracteoles when sexual reproduction fails and no fertilized seeds are produced. These structures can also be induced by removal of flower buds (Fig. 1A, B). Under field conditions, the majority of vegetative bulbils which form after floral bud removal produce viable plantlets that can eventually be removed from the mother plant and grown individually; however, removal of flower buds under greenhouse conditions leads to the formation of a majority of structures morphologically similar to flower buds or with mixed identity, where both vegetative leaves and reproductive organs are present (Fig. 1C). Detailed analysis of flower-like structures showed that all floral tissues including ovaries, anthers, carpels, pistils, and tepals have a distorted morphology (Fig. 1D–K). However, germination in vitro of pollen obtained from these secondary flower structures shows similar efficiency (42%) to that obtained from normal flowers (36%), suggesting that the distorted morphology of the maternal organs (ovaries and carpels) could explain why successful fertilization and formation of capsules and seeds is never observed in secondary flower structures. Overall these results show that induction of bulbil formation by removal of flower buds under normal field conditions in general leads to the formation of indeterminate vegetative meristems that produce new plantlets, whereas under greenhouse conditions, possibly due to changes in light intensity, determinate floral meristems are predominantly produced.
Fig. 1.
Removal of flower buds leads to formation of vegetative or floral structures at bracteoles in A. tequilana. (A) Unopened floral buds. (B) Pedicel tissue following removal of buds. (C) Vegetative bulbils and floral structures formed at bracteoles. (D–G) Normal flower morphology. (D) Completely normal flower; (E) normal carpelar organization shown in cross-section; (F) normal anther; (G) normal pistil. (H–K) Morphology of floral structures formed at bracteoles. (H) Completely distorted flower; (I) distorted carpelar organization; (J) distorted anther; (K) stigma-like structure. Scales are 2.0mm. (This figure is available in colour at JXB online.)
Expression patterns of agave KNOX and MADS box genes during bulbil formation were previously shown to be consistent with the expression patterns expected for vegetative growth; AtqKNOX1, AtqKNOX2, and AtqMADS5 transcripts increased and AtqMADS1 transcription decreased (Abraham-Juarez et al., 2010; Delgado Sandoval et al., 2012). In order to determine whether the expression patterns of these genes were reflected in the type of secondary structure formed (vegetative or floral), the expression patterns of the agave KNOX genes AtqKNOX1 and AtqKNOX2 and the agave MADS box genes AtqMADS1 and AtqMADS5 were determined during the formation of predominantly floral structures in greenhouse conditions. As shown in Supplementary Figure S2A–D at JXB online, the expression patterns correlate with the formation of floral rather than vegetative structures. Both AtqKNOX1 and AtqKNOX2 genes and AtqMADS5 (associated with vegetative growth) show a decrease in expression as determinate floral meristems are formed, whereas AtqMADS1, associated with flower development, increases in expression as floral structures develop and decreases to a minimum level of expression in stage S5 when determinate floral structures are observed.
Auxin as a major regulator of the cellular switch in bulbil formation in A. tequilana
It was hypothesized that removal of flower buds or ineffective fertilization in agave leads to modification of auxin levels and/or auxin transport in the underlying pedicel tissue which in turn leads to the induction of new meristems at the bracteoles. Since polar auxin transporters of the PIN family have been reported to play a central role in the formation of auxin gradients during organ formation (Bohn-Courseau, 2010), it was decided to examine patterns of PIN protein localization during bulbil formation. Figure 2a shows an example of a bisected floral bud and the underlying pedicel tissue, and Fig. 2b an example of pedicel tissue from which the bud has been removed. Figure 2c and d shows a light microscopy image and PIN immunolocalization image, respectively, at T0 (immediately after bud removal). PIN protein is observed in vascular tissue and at the lower periphery of both vascular and surrounding cells, suggesting auxin transport basipetally from the flower bud region. When vegetative bulbils begin to form, a localized concentration of PIN in a polar configuration typical of apical meristems is observed at the S5 stage, suggesting that a basipetal flux of auxin from the developing SAMs is established as the new vegetative structures develop (Fig. 2e, f).
Fig. 2.
Immunolocalization of agave PIN protein in pedicel and S5 bulbil tissue. (A) Bisected bud before sectioning. (B) Example of pedicel tissue with floral bud removed; the box indicates the region surrounding the bracteoles on which immunolocalization of PIN was carried out; brackets indicate pedicel tissue. (C) Immunolocalization of PIN protein using an anti-PIN1a antibody against maize PIN1a protein, in pedicel tissue where bud has been removed as shown in (B). An asterisk (*) indicates the cut surface. (D) Enlarged image showing PIN location in the lower periphery of cells; circles indicate individual cells outside vascular tissue; arrows indicate PIN localization in individual cells within the vascular tissue. Scale bar in (C) is 100 μm. (E) Light microscopy and (F) PIN immunolocalization images of untreated pedicel tissue (box in B), underlying the region where a floral bud was removed at the S5 stage of bulbil induction, ×20 magnification, scale bar=50 μm. (This figure is available in colour at JXB online.)
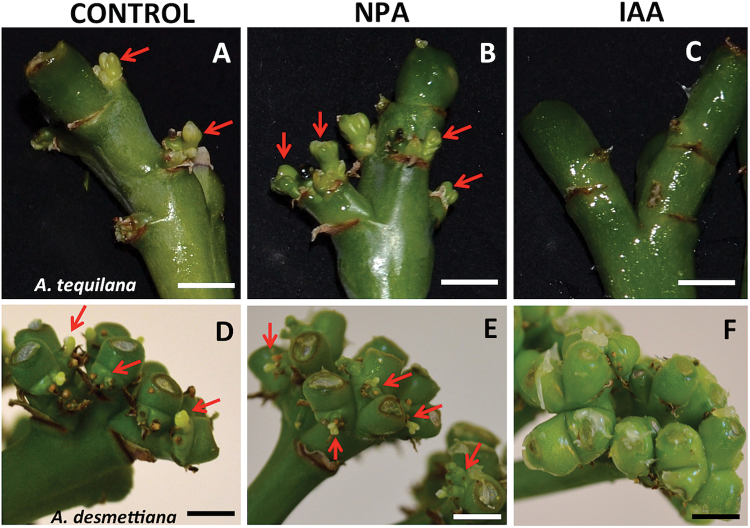
To test the hypothesis that auxin plays a central role in regulating bulbil formation, three different treatments were applied to pedicel tips from which floral buds had been removed for both A. tequilana and A. desmettiana: (i) exogenous application of IAA; (ii) exogenous application of the auxin transport inhibitor NPA; and (iii) application of a lanolin-only control (see the Materials and methods). As shown in Fig. 3A–F, application of exogenous auxin on cut pedicel tissue suppressed bulbil formation at the bracteoles, whereas the application of NPA or a lanolin-only control led to the formation of new meristems and bulbil structures. Bulbil auxin treatments were repeated in three consecutive years for A. tequilana and in two consecutive years for A. desmettiana. Similar results were obtained in all cases. These results demonstrate that pedicel tissue is only capable of developing active meristems in the absence of apical auxin, confirming that apical auxin determines the induction of reprogramming of the necessary cellular processes. Exogenous application of NPA did not inhibit the formation of new meristems.
Fig. 3.
Exogenous application of NPA and IAA to the cut surface of pedicel tissue following removal of floral buds. (A–C) Agarve tequilana 10 d after bud removal. (A) Lanolin-only control; (B) NPA; (C) IAA. (D–F) Agarve desmettiana 10 d after bud removal. (D) Lanolin-only control; (E) NPA; (F) IAA. Meristems are indicated with arrows. Bars are 0.5cm. (This figure is available in colour at JXB online.)
Identification and classification of A. tequilana PIN1-related cDNAs
Fourteen expressed sequence tags (ESTs) with highly significant homology to PIN1 sequences, most of which were expressed in SAM tissue or bulbil-induced pedicel tissue, were identified in an A. tequilana transcriptome database (Avila de Dios and Simpson, unpublished). Alignment of overlapping PIN ESTs produced the complete open reading frames of two putative A. tequilana PIN genes (Supplementary Fig. S3 at JXB online). Comparison with PIN amino acid sequences from a variety of plant species using both NJ and maximum likelihood methods allowed the identification of two distinct A. tequilana PIN-like genes: AtqPIN1 which is orthologous to other PIN1 genes and AtqSoPIN1 which groups within the SoPIN1 clade which is absent in Brassicacea species (O’Connor et al., 2014) as shown in Fig. 4. The percentage identity between the agave protein sequences and ZmPIN1a/ZMSoPIN1 also supports this classification (Supplementary Table S2). AtqPIN1 is 69 amino acids longer than AtqSoPIN1 in the hydrophilic domain, and comparison of the presence of conserved motifs in this region based on the report of Bennett et al. (2014) produced a complex pattern. Fifteen different motifs of the 32 motifs analysed (highly variable motifs were not included) were found in each agave PIN protein; however individual motifs differed between AtqPIN1 and AtqSoPIN1. A list of motifs found in the A. tequilana PIN1 and SoPIN1 clades, A. thaliana PIN1, and Z. mays PIN1a and SoPIN1 proteins is given in Supplementary Table S3. Motifs S1, S2, HC2-4, HC2-1-1, and S11 are found in the PIN1 clade proteins but not in the SoPIN1 clade, whereas S7 and S20 are found only in the SoPIN1 clade but not the PIN1 clade. The distribution of these motifs is shown in Supplementary Fig. S4. The motifs denoted HC are found within highly conserved regions of the hydrophilic loop of the proteins analysed. Only A. thaliana PIN1 and AtqSoPIN1 have the HC1-4 motif. Differences within these HC motifs are candidates to determine the particular function and localization of the distinct classes of PIN proteins.
Fig. 4.
Dendrogram showing the relationship between PIN1 amino acid sequences from monocotyledonous and dicotyledonous species and A. tequilana. The agave sequences are boxed. PIN1 and ‘Sister of PIN1’ (SoPIN1) clades containing dicotyledonous or monocotyledonous species are indicated.
Expression of agave PIN1-related genes during bulbil formation
To determine the patterns of expression of AtqPIN1 and AtqSoPIN1 during bulbil formation, RT-qPCR was carried out throughout the different stages of bulbil development. As shown in Fig. 5A and B, AtqPIN1 and AtqSoPIN1 showed opposing patterns of expression from S0 to S4 stages. AtqPIN1 showed the highest level of expression in non-induced tissue (S0 stage) followed by a decrease 7 d after induction (S1 stage). Expression continued to decrease in S2 and S3 stages, reaching the lowest level in the S4 stage (28 d after induction). An increase was observed at the S5 (35 d after induction) stage when auxin gradients at bulbil SAMs are established (Fig. 5A). This result is in agreement with results from the immunolocalization analysis where PIN protein is observed when vegetative meristems are formed (Fig. 2e, f).
Fig. 5.
RT-qPCR expression profiles of agave PIN1 genes in pedicel tissue at different stages of bulbil formation in the presence or absence of NPA or IAA. (A and C) AtqPIN1. (B and D) AtqSoPIN1. S0–S5, stages of bulbil formation; C-, application of lanolin-only control; N-, application of NPA; I-, application of IAA.
In contrast, AtqSoPIN1 showed a lower level of expression in non-induced pedicel tissue (S0) in comparison with leaf tissue and increased gradually in stages S1, S2, and S3, to reach a maximum in S4 before decreasing slightly in S5 (Fig. 5B). These contrasting patterns of expression of the A. tequilana PIN1 genes suggest that they could be differentially regulated by auxin and play different functional roles.
Analysis of A. tequilana PIN1 gene expression in pedicel tissue to which exogenous IAA, NPA, or lanolin alone had been applied to the cut surfaces where flower buds had been removed confirmed the differential patterns of expression of AtqPIN1 and AtqSoPIN1 during bulbil induction (Fig. 5C, D). Interestingly, the application of exogenous auxin, NPA, or lanolin alone did not affect the patterns of expression of AtqPIN1 in stages S1–S2. AtqPIN1 was expected to increase in the S5 stage in control and NPA-treated samples; however, the lack of increase observed may be due to the development of both vegetative and floral structures in the plants analysed in Fig. 5C and D, whereas very few or no floral structures were observed in the samples that were used for analysis in Fig. 5A and B. In contrast, application of exogenous auxin strongly affected the pattern of AtqSoPIN1 expression in pedicel tissue. An increase in expression was observed in untreated (Fig. 5B), lanolin-only, and NPA-treated samples (Fig. 5D, C-S0 to C-S5 and N-S0 to N-S5); however, no change in expression was detected with the addition of auxin, suggesting that auxin inhibits expression of AtqSoPIN1.
Analysis of the components of auxin metabolism in pedicel tissue during bulbil formation
To determine the role of auxin synthesis and turnover during bulbil development, the levels of free auxin (IAA), a conjugated form of auxin (IAAsp), and the auxin precursors TRP (tryptophan) and TRA (tryptamine) were determined by UHPLC-Q-TOF MS (calibration parameters are shown in Supplementary Table S4 at JXB online). Tissue analysed included pedicel tissue in the different stages of bulbil development, and leaf and root tissue as controls. No free IAA was detected in pedicel tissue at any of the stages of bulbil development (Fig. 6). Free IAA was only detected in root tissue. The auxin precursors TRP and TRA were detected in roots and all pedicel tissues tested. TRP but not TRA was also detected in leaf tissue, and varying concentrations of both molecules were observed during bulbil development, with the highest concentrations observed in non-induced pedicel tissue (S0). On bulbil induction, the levels of both precursors decreased ~5- and 2.6-fold for TRP and TRA, respectively. TRP levels increased from stage S1 until S4, and remained at the S4 level in the S5 stage. In contrast, levels of TRA continued to decrease until S3 to an ~5-fold lower level than in non-induced samples at S3, and then increased ~3.7-fold until stage S5. The auxin conjugate IAAsp was also detected in all tissues, with the highest concentration found in root tissue. A low level of IAAsp was observed in non-induced pedicel tissue and slowly increased through S1–S5 to reach an almost 2-fold higher concentration in S5.
Fig. 6.
Quantification of free and conjugated forms of IAA and IAA precursors (TRP and TRA) by UHPLC-Q-TOF MS in different stages of bulbil formation in A. tequilana. S0–S5: different stages of bulbil formation; DW, dry weight. Leaf and root samples are included as controls.
Heterologous expression of agave PIN genes in Arabidopsis thaliana
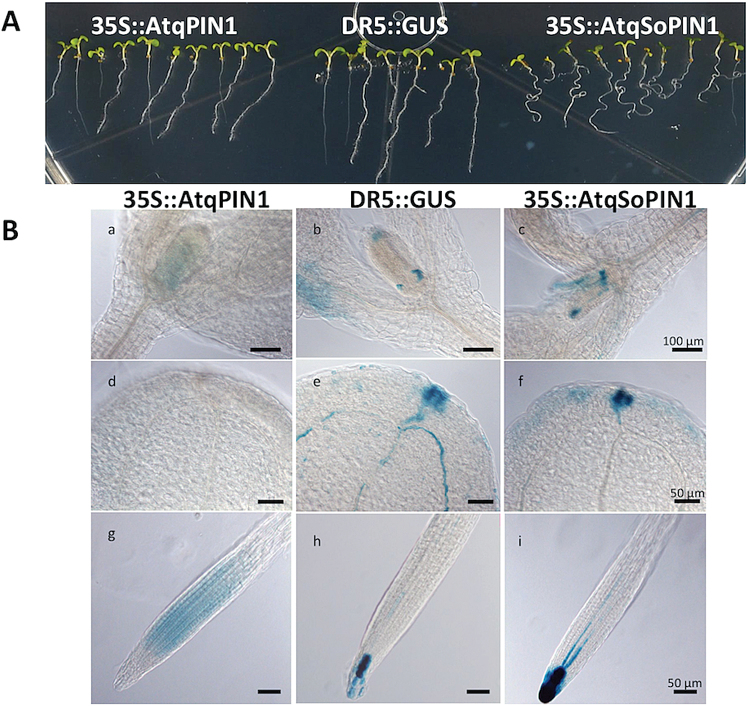
To examine the function of the AtqPIN genes, DR5::GUS (β-glucuronidase) Arabidopsis lines that express AtqPIN1 and AtqSoPIN1 under control of the 35S promoter were produced. Ectopic expression of AtqSoPIN1 showed an altered gravitropic response in comparison with the DR5::GUS control line (Fig. 7A), whereas expression of AtqPIN1 in A. thaliana had no effect on gravitropism but showed slightly shorter roots in comparison with the DR5::GUS line. The pattern of auxin response shown by GUS staining was similar in AtqSoPIN1 and control DR5::GUS lines in the aerial parts of the plant (Fig. 7B b, c, e, f), but in AtqPIN1-expressing lines points of auxin accumulation were more diffuse or absent (Fig. 7B a, d, g). In root tissue, significant differences in patterns of auxin localization were observed. AtqPIN1 lines showed a diffuse pattern of auxin distribution in the region above the quiescent centre and meristematic region, whereas in AtqSoPIN1-expressing plants auxin is strongly concentrated in the root tip both at and below the quiescent centre in comparison with the DR5::GUS control which shows auxin localization in cells very close to the quiescent centre (Fig. 7B g–i). These data suggest that, although closely related, AtqPIN1 proteins are functionally distinct at least in the heterologous system of A. thaliana.
Fig. 7.
Heterologous expression of A. tequilana PIN1 and SoPIN1 genes in A. thaliana. (A) Gravitropic responses of DR5::GUS A. thaliana seedlings expressing AtqPIN1 or AtqSoPIN1 under control of the 35S promoter and DR5::GUS control. (B) Auxin localization based on GUS-stained tissues of DR5::GUS A. thaliana plants expressing AtqPIN1 or AtqSoPIN1 under control of the 35S promoter and DR5::GUS control. (This figure is available in colour at JXB online.)
Discussion
Many species of agave such as A. tequilana produce bulbils when sexual reproduction is unsuccessful, and this may occur throughout the inflorescence or on particular umbels. Other species such as A. desmettiana appear to flower normally but are naturally infertile and only produce bulbils rather than seeds. Under field conditions, bulbil induction in both species results in development of predominantly indeterminate, vegetative meristems from which viable independent plantlets are formed. However, plants induced to form bulbils under greenhouse conditions produced predominantly non-viable propagules with a mixture of floral and vegetative organs. The floral organs produced under greenhouse conditions are invariably misshapen and non-viable in reproductive terms. These results confirm the report of Abraham-Juarez et al. (2010), which described the formation of new meristems in the bracteolar regions of induced pedicel tissue; however, they also demonstrate that the identity of the meristems formed (determinate or indeterminate) depends on environmental factors. Since temperature, substrate, and manipulation were essentially the same for both garden- and greenhouse-grown plants, changes in light quality or intensity are most probably responsible for the differences observed in bulbil morphology. Interestingly, a recent report described the importance of red light for the efficient production of bulbils in Pinellia ternata (Chen et al., 2013).
Contrasting expression patterns of the KNOX and MADS box genes support the change in meristem identity under different conditions. Light has been shown to have an effect in STM KNOX1 gene expression, as reported by Soucek et al. (2007), where activation of isopentenyl transferase repressed STM especially at low light. These findings suggest that high levels of cytokinin repress KNOX1 genes in plants grown in low light. However, the formation of distorted and non-viable floral organs suggests that during the development of the secondary structures, meristem identity was not clearly defined, leading to a mixture of vegetative and floral organs, although ultimately a determinate meristem was produced.
Exogenous application of auxin to the cut surfaces in the absence of the flower bud suppresses bulbil formation, whereas removal of the auxin source allows new vegetative structures to form. Immunolocalization experiments showed localization of PIN proteins with the flux in normal pedicel tissue with intact flowers and in induced pedicel tissue in the latter stages of bulbil induction. Since the PIN1a antibody was raised to a large region of the maize PIN1a protein, with partial homology between AtqSoPIN1 and AtqPIN1 (Supplementary Fig. S3 at JXB online), it cannot be determined precisely which proteins were detected in A. tequilana tissue. However, based on the model of O’Connor et al. (2014), the observation that the PIN1 proteins are oriented with the flux suggests that the observed protein is probably AtqPIN1.
The search for PIN1 ESTs in the A. tequilana transcriptome database identified an AtqPIN1 gene related to maize ZmPIN1a, b, c genes and an AtqSoPIN1 gene related to members of a recently described new PIN clade SoPIN1 (O’Connor et al., 2014) which is absent in the Brassicacea. Both AtqPIN1 and AtqSoPIN1 transcripts were expressed throughout the different stages of bulbil formation but showed contrasting patterns of expression. Whereas AtqPIN1 transcripts decreased in response to flower bud excision both during bulbil induction in untreated pedicels and after auxin treatment, AtqSoPIN1 expression increased with bulbil induction, and this response was suppressed with auxin treatment. These results suggest that the apical tissues are necessary to maintain AtqPIN1 expression; however, application of exogenous auxin does not maintain AtqPIN1 expression although bulbil formation is inhibited, but does suppress AtqSoPIN1 expression. A more detailed study of PsPIN1 expression in decapitated and auxin-treated stem stumps of pea (Balla et al., 2011) revealed that after decapitation and auxin treatment the PsPIN1 expression increased 5-fold during the first 12hours but after 5 days dropped below the level observed in intact plants although axillary bud formation remained inhibited. Unchanged expression of AtqSoPIN1 after auxin treatment is especially interesting since it is in accordance with the observation of O’Connor et al. (2014) where BdSoPIN1 is highly expressed in the epidermis and localized at the points of lemma initiation. This process is similar to the formation of auxin maxima during the initial stages of bulbil development, therefore no change in AtpSoPIN1 would be expected if bulbil formation is inhibited.
Although removal of floral buds leads to an initial decrease in AtqPIN1 transcript levels, the pattern of PIN protein immunolocalization in vascular tissue remains unchanged when exogenous auxin is applied. In untreated pedicel samples, at the S5 stage, AtqPIN1 gene expression increased in correlation with the polarized pattern of immunolocalized PIN protein associated with the formation of new SAMs. However, unexpectedly, no increase in AtqPIN1 expression was observed in lanolin-only or NPA-treated samples at the S5 stage, although polarized PIN protein was observed at the S5 stage in the typical conformation, indicating a downward flux of auxin from meristem regions.
AtqSoPIN1 expression remains unchanged when exogenous auxin is applied because no auxin maxima are formed but increases in expression when basipetal auxin transport is interrupted and new meristems are formed. The highest levels of expression in the S5 stage of bulbil development for both lanolin-only and NPA-treated plants support this conclusion. AtqPIN1 expression decreases in untreated samples, and only in the S5 stage is a downward flux of auxin established, reflected by both AtqPIN1 expression and AtqPIN1 localization. Since the stages of bulbil formation are determined by morphology and not time after induction, it is unlikely that the lack of increased expression of AtqPIN1 and AtqPIN1 protein in lanolin-only and NPA-treated samples is due to delayed development where the true S5 stage was reached later in this particular series of experiments. The interpretation of this is therefore that the lack of AtqPIN1 expression and AtqPIN1 protein is probably due to the presence of predominantly determinate floral meristems in these samples.
Suppression of bulbil formation is maintained in pedicel tissue receiving exogenous auxin, although AtqPIN1 expression decreases, suggesting that synthesis of new AtqPIN1 protein is not necessary for this process. In contrast, increasing levels of AtqSoPIN1 expression suggest that newly synthesized SoPIN1 protein could be involved in the auxin flux needed to concentrate this hormone close to the bracteoles where new meristems initiate and develop to form bulbils.
At the amino acid sequence level, AtqPIN1 and AtqSoPIN1 are distinguished by the presence or absence of specific motifs that in general correlate with their classification in distinct clades. One exception, however, is motif HC1-4 within a highly conserved region of the hydrophilic loop that is only strongly supported in AtqSoPIN1 and the A. thaliana PIN1 protein. Perhaps unexpectedly, ectopic expression of AtqSoPIN1 in A. thaliana produced morphological effects such as altered gravitropism and patterns of auxin localization similar to ectopic expression of the endogenous A. thaliana PIN1 gene as reported by Mravec et al. (2008), suggesting that AtqSoPIN1 may be functionally more closely related to A. thaliana PIN1. In contrast, ectopic expression of the orthologous AtqPIN1 protein that does not carry the HC1-4 motif in A. thaliana showed a very different pattern of auxin localization in both aerial and root tissue, and no altered gravitropism, suggesting that this motif may contribute to the functional differences between the agave PIN proteins observed in A. thaliana. Because of the absence of SoPIN1 in Brassicacea, the PIN1 protein in these species must be able to act in pumping auxin both against the gradient, thus forming auxin maxima, and also with the flux, draining excess auxin from this maxima. The similar auxin distribution pattern of ectopic expression of AtqSoPIN1 and AtPIN1 in A. thaliana with strong auxin maxima could suggest that the HC1-4 motif absent in AtqPIN1 plays an important role in the ability for auxin maxima formation.
PIN1 and SoPIN1 clades may have overlapping functions depending on the specific motifs present in each protein in different species, and perhaps this is how Brassicacea family plants were able to deal with the loss of SoPIN1. It will be interesting in future studies to analyse in detail the distribution of PIN1 and SoPIN1 proteins and the expression patterns of AtqPIN1/SoPIN1 in A. tequilana roots.
The lack of motif 11 in the SoPIN1 proteins is also interesting since phosphorylation of conserved serine and threonine residues in this motif determines the correct polarity of the A. thaliana PIN1 protein (Zhang et al., 2010). Lack of motif 11 could play a role in the different polarities (up the gradient and with the flux) described for PIN1 and SoPIN1 proteins in the model proposed by O’Connor et al. (2014).
No free auxin could be detected in the pedicel samples analysed, probably due to the limits of detection of the method used. However, variations in levels of auxin precursors and conjugated auxin correlate with the observed changes in AtqPIN protein and AtqPIN1/SoPIN1 transcript levels and help to shed light on the changes in auxin metabolism during the process of bulbil formation. Before induction at time S0, levels of auxin precursors are highest whereas the auxin conjugate IAAsp is found at a relatively low concentration. The sharp drop in precursor levels following flower bud removal could reflect the response of plant tissue to synthesize and concentrate auxin locally, leading to the development of new meristems and vascular tissue, therefore consuming the precursor molecules, leading to a drop in concentration. As bulbil development progresses, auxin levels would increase to the S5 stage where the normal auxin localization patterns associated with apical meristem development are observed. The gradual rise in the level of the auxin conjugate could indicate that as auxin levels rise, more auxin is converted to the conjugate form either for storage or eventually to be destined for degradation. In regard to auxin levels, the initiation of bulbil formation may be compared with breaking of seed dormancy or the stimulation of axillary meristems since these processes also rely on a localized drop in auxin levels (Sorce et al., 2009; He et al., 2012; Liu et al., 2013). The fluctuation in auxin levels may allow the activation by other plant hormones such as cytokinin or ABA of cells close to the bracteoles with the potential to form new meristems.
Based on results in this and previous reports, a model for induction of bulbils in A. tequilana is proposed (Fig. 8). The model proposes that the basipetal auxin gradient from the flower bud suppresses formation of bulbils at the bracteoles. Under natural light conditions, removal of flower buds leads to a drop in auxin flux in pedicel vascular tissue, stimulating the development of new meristems and vegetative bulbils at the bracteoles. Under reduced light conditions, a mixture of determinate floral meristems producing non-viable floral structures and indeterminate meristems producing vegetative bulbils at the bracteoles develops. Putative roles for each agave PIN protein in bulbil formation can also be proposed as follows. Polarized AtqPIN1 is present in vascular tissue leading to a downward auxin flux from the flower bud. Removal of the flower bud interrupts this auxin flux and AtqPIN1 expression decreases. In contrast, AtqSoPIN1 expression increases, allowing auxin to accumulate close to the bracteoles where new meristems will form. In the final stage of bulbil induction, AtqSoPIN1 allows accumulation of auxin at the SAM of developing bulbil meristems, reinstating AtqPIN1 expression and thus the downward auxin flux and formation of new vascular tissue.
Fig. 8.
Model for the role of PIN1-mediated auxin flux in induction of secondary structures at the bracteoles in A. tequilana following removal of floral buds. (A) Auxin suppresses secondary structure formation. (a) A normal auxin gradient from the flower bud suppresses formation of secondary structures at the bracteoles. (b) Under natural light conditions, removal of flower buds leads to a drop in auxin flux in pedicel vascular tissue, stimulating the development of new meristems and vegetative bulbils at the bracteoles. (c) Under reduced light conditions, removal of flower buds leads to a drop in auxin flux in pedicel vascular tissue, stimulating the development of a mixture of determinate floral meristems producing non-viable floral structures and indeterminate meristems producing vegetative bulbils at the bracteoles. (B) Putative role of agave PIN1 proteins in bulbil formation. (a) Polarized AtqPIN1 is present in vascular tissue, leading to a downward auxin flux from the flower bud. (b) Removal of the flower bud interrupts the auxin flux. AtqPIN1 expression decreases and AtqSoPIN1 expression increases, allowing auxin to accumulate close to the bracteoles where new meristems will form (dots). (c) AtqSoPIN1 allows accumulation of auxin at the SAM of developing bulbil meristems and AtqPIN1 allows downward auxin flux and formation of vascular tissue. (This figure is available in colour at JXB online.)
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Stages of vegetative bulbil formation S0–S5 and corresponding histological images.
Figure S2. qRT-qPCR expression profiles of agave KNOX and MADS genes during the different stages of bulbil formation in pedicel tissue which produces floral structures at bracteoles.
Figure S3. Alignment of amino acid sequences of PIN1 proteins from A. thaliana, corn, rice sorghum, and agave.
Figure S4. Distribution within the variable hydrophilic domain of conserved amino acid motifs found in PIN1 and SoPIN1 proteins.
Table S1. Primers used for RT-qPCR.
Table S2. Levels of identity between complete amino acid sequences and hydrophilic domains of PIN1 and SoPIN1 proteins from maize and agave.
Table S3. Percentage identity of conserved amino acid motifs found in PIN1 and SoPIN1 proteins.
Table S4. Calibration parameters and sensitivity of the UHPLC-Q-TOF MS method.
Acknowledgements
We are grateful to Luis Herrera Estrella and the two anonymous reviewers for critical reading of the manuscript, Katia Gil Vega for technical assistance, and to Dr Javier Arroyo of Tequila Real de Pénjamo for providing A. tequilana plants and his continual enthusiastic support for our research. This work was carried out with financial support from CONACyT grant no. 132160 from which MJAJ received a post-doctoral fellowship, and JNSV and RHC received undergraduate scholarships.
References
- Abraham-Juarez MJ, Martinez-Hernandez A, Leyva-Gonzalez MA, Herrera-Estrella L, Simpson J. 2010. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. Journal of Experimental Botany 61, 4055–4067. [DOI] [PubMed] [Google Scholar]
- Arizaga S, Ezcurra E. 1995. Insurance against reproductive failure in a semelparous plant—bulbil formation in Agave macroacantha flowering stalks. Oecologia 101, 329–334. [DOI] [PubMed] [Google Scholar]
- Arizaga S, Ezcurra E. 2002. Propagation mechanisms in Agave macroacantha (Agavaceae), a tropical arid-land succulent rosette. American Journal of Botany 89, 632–641. [DOI] [PubMed] [Google Scholar]
- Arizaga S, Ezcurra E, Peters E, de Arellano FR, Vega E. 2000. a . Pollination ecology of Agave macroacantha (Agavaceae) in a Mexican tropical desert. I. Floral biology and pollination mechanisms. American Journal of Botany 87, 1004–1010. [PubMed] [Google Scholar]
- Arizaga S, Ezcurra E, Peters E, de Arellano FR, Vega E. 2000. b Pollination ecology of Agave macroacantha (Agavaceae) in a Mexican tropical desert. II. The role of pollinators. American Journal of Botany 87, 1011–1017. [PubMed] [Google Scholar]
- Balla J, Kalousek P, Reinohl V, Friml J, Prochazka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. The Plant Journal 65, 571–577. [DOI] [PubMed] [Google Scholar]
- Bennett T, Brockington SF, Rothfels C, et al. 2014. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Molecular Biology and Evolution 31, 2042–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn-Courseau I. 2010. Auxin: a major regulator of organogenesis. Comptes Rendus Biologies 333, 290–296. [DOI] [PubMed] [Google Scholar]
- Borland AM, Griffiths H, Hartwell J, Smith JA. 2009. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. Journal of Experimental Botany 60, 2879–2896. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu GS, Guo QS, Wang CL, Liu ZY. 2013. [Effect of light quality on growth and quality of Pinellia ternata]. Zhongguo Zhong Yao Za Zhi 38, 4257–4262. [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. 2004. Real-time RT–PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal 38, 366–379. [DOI] [PubMed] [Google Scholar]
- Delgado Sandoval SC, Abraham Juarez MJ, Simpson J. 2012. Agave tequilana MADS genes show novel expression patterns in meristems, developing bulbils and floral organs. Sexual Plant Reproduction 25, 11–26. [DOI] [PubMed] [Google Scholar]
- Diaz-Martinez M, Nava-Cedillo A, Guzman-Lopez JA, Escobar-Guzman R, Simpson J. 2012. Polymorphism and methylation patterns in Agave tequilana Weber var. ‘Azul’ plants propagated asexually by three different methods. Plant Science 185–186, 321–330. [DOI] [PubMed] [Google Scholar]
- Ebi M, Kasai N, Masuda K. 2000. Small inflorescence bulbils are best for micropropagation and virus elimination in garlic. Hortscience 35, 735–737. [Google Scholar]
- Felsenstein J. 1986. Jackknife, bootstrap and other resampling methods in regression-analysis—discussion. Annals of Statistics 14, 1304–1305. [Google Scholar]
- Garces HM, Koenig D, Townsley BT, Kim M, Sinha NR. 2014. Truncation of LEAFY COTYLEDON1 protein is required for asexual reproduction in Kalanchoe daigremontiana. Plant Physiology 165, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry HS. 1982. Agaves of continental North America. Tucson: University of Arizona Press. [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. 2012. Multiple AUX/IAA–ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philosophical Ttransactions of the Royal Society B: Biological Sciences 367, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G, Aleman S, Infante D. 2003. Asexual genetic variability in Agave fourcroydes II: selection among individuals in a clonally propagated population. Plant Science 165, 595–601. [Google Scholar]
- He D, Mathiason K, Fennell A. 2012. Auxin and cytokinin related gene expression during active shoot growth and latent bud paradormancy in Vitis riparia grapevine. Journal of Plant Physiology 169, 643–648. [DOI] [PubMed] [Google Scholar]
- Kim CJ, Cho YG, Park YG, Nam SW, Kim SY, Lee SH, Yoo NJ, Lee JY, Park WS. 2005. Pin1 overexpression in colorectal cancer and its correlation with aberrant beta-catenin expression. World Journal of Gastroenterology 11, 5006–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Leyser O. 2005. The fall and rise of apical dominance. Current Opinion in Genetics and Development 15, 468–471. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH. 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 110, 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MG, Mancilla-Margalli NA, Mendoza-Diaz G. 2003. Molecular structures of fructans from Agave tequilana Weber var. azul. Journal of Agricultural and Food Chemistry 51, 7835–7840. [DOI] [PubMed] [Google Scholar]
- Mai YX, Wang L, Yang HQ. 2011. A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis. Journal of Integrated Plant Biology 53, 480–492. [DOI] [PubMed] [Google Scholar]
- Mancilla-Margalli NA, Lopez MG. 2006. Water-soluble carbohydrates and fructan structure patterns from Agave and Dasylirion species. Journal of Agricultural and Food Chemistry 54, 7832–7839. [DOI] [PubMed] [Google Scholar]
- Mathew D, Ahmed Z, Singh N. 2005. Formulation of flowering index, morphological relationships, and yield prediction system in true garlic aerial seed bulbil production. Hortscience 40, 2036–2039. [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. 2008. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135, 3345–3354. [DOI] [PubMed] [Google Scholar]
- Muller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Annals of Botany 107, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel PS. 1976. Water relations and photosynthesis of a desert CAM plant, Agave deserti. Plant Physiology 58, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak O, Henykova E, Sairanen I, Kowalczyk M, Pospisil T, Ljung K. 2012. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. The Plant Journal 72, 523–536. [DOI] [PubMed] [Google Scholar]
- O’Connor DL, Runions A, Sluis A, Bragg J, Vogel JP, Prusinkiewicz P, Hake S. 2014. A division in PIN-mediated auxin patterning during organ initiation in grasses. PLoS Computational Biology 10, e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA. 2013. From perception to attenuation: auxin signalling and responses. Current Opinion in Plant Biology 16, 561–568. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, et al. 2012. Different auxin response machineries control distinct cell fates in the early plant embryo. Developmental Cell 22, 211–222. [DOI] [PubMed] [Google Scholar]
- Sorce C, Lombardi L, Giorgetti L, Parisi B, Ranalli P, Lorenzi R. 2009. Indoleacetic acid concentration and metabolism changes during bud development in tubers of two potato (Solanum tuberosum) cultivars. Journal of Plant Physiology 166, 1023–1033. [DOI] [PubMed] [Google Scholar]
- Soucek P, Klima P, Rekova A, Brzobohaty B. 2007. Involvement of hormones and KNOXI genes in early Arabidopsis seedling development. Journal of Experimental Botany 58, 3797–3810. [DOI] [PubMed] [Google Scholar]
- Valenzuela A. 2003. El agave tequilero: su cultivo e industria de México. México: Mundiprensa Editores. [Google Scholar]
- Valenzuela A. 2011. A new agenda for blue agave landraces: food, energy and tequila. Global Change Biology Bioenergy 3, 15–24. [Google Scholar]
- Walck JL, Cofer MS, Hidayati SN. 2010. Understanding the germination of bulbils from an ecological perspective: a case study on Chinese yam (Dioscorea polystachya). Annals of Botany 106, 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CN, Cronk QC. 2003. Meristem fate and bulbil formation in Titanotrichum (Gesneriaceae). American Journal of Botany 90, 1696–1707. [DOI] [PubMed] [Google Scholar]
- Wang CN, Moller M, Cronk QC. 2004. Altered expression of GFLO, the Gesneriaceae homologue of FLORICAULA/LEAFY, is associated with the transition to bulbil formation in Titanotrichum oldhamii. Development Genes and Evolution 214, 122–127. [DOI] [PubMed] [Google Scholar]
- Wang P, Cheng T, Wu S, Zhao F, Wang G, Yang L, Lu M, Chen J, Shi J. 2014. Phylogeny and molecular evolution analysis of PIN-FORMED 1 in angiosperm. PLoS One 9, e89289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencik A, Rolcik J, Friml J. 2010. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proceedings of the National Academy of Sciences, USA 107, 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.