Highlight
Potato StUBA2 RNA-binding proteins promote hypersensitive-like cell death (dependent on the first StUBA2 RNA recognition motif) and premature leaf senescence, and increase transcripts of select pathogen-associated, senescence-associated, and autophagy-associated genes.
Key words: AKIP proteins, Arabidopsis thaliana, RNA binding proteins, senescence, Solanum tuberosum, UBA2 proteins.
Abstract
The Arabidopsis thaliana genome encodes three RNA-binding proteins (RBPs), UBP1-associated protein 2a (UBA2a), UBA2b, and UBA2c, that contain two RNA-recognition motif (RRM) domains. They play important roles in wounding response and leaf senescence, and are homologs of Vicia faba abscisic-acid-activated protein kinase-interacting protein 1 (VfAKIP1). The potato (Solanum tuberosum) genome encodes at least seven AKIP1-like RBPs. Here, two potato RBPs have been characterized, StUBA2a/b and StUBA2c, that are homologous to VfAKIP1 and Arabidopsis UBA2s. Transient expression of StUBA2s induced a hypersensitive-like cell death phenotype in tobacco leaves, and an RRM-domain deletion assay of StUBA2s revealed that the first RRM domain is crucial for the phenotype. Unlike overexpression of Arabidopsis UBA2s, constitutive expression of StUBA2a/b in Arabidopsis did not cause growth arrest and lethality at the young seedling stage, but induced early leaf senescence. This phenotype was associated with increased expression of defence- and senescence-associated genes, including pathogen-related genes (PR) and a senescence-associated gene (SAG13), and it was aggravated upon flowering and ultimately resulted in a shortened life cycle. Leaf senescence of StUBA2a/b Arabidopsis plants was enhanced under darkness and was accompanied by H2O2 accumulation and altered expression of autophagy-associated genes, which likely cause cellular damage and are proximate causes of the early leaf senescence. Expression of salicylic acid signalling and biosynthetic genes was also upregulated in StUBA2a/b plants. Consistent with the localization of UBA2s-GFPs and VfAKIP1-GFP, soluble-modified GFP-StUBA2s localized in the nucleus within nuclear speckles. StUBA2s potentially can be considered for transgenic approaches to induce potato shoot senescence, which is desirable at harvest.
Introduction
Plant development is achieved by transcriptional, post-transcriptional, and translational regulation of gene expression. Upstream regulatory regions play important roles in the initial transcription of protein-coding genes upon perception of cues, and subsequent post-transcriptional processes exert pivotal roles in modulating expression of specific transcripts through pre-mRNA splicing, alternative splicing, capping, polyadenylation, mRNA stability, and mRNA transport (Reddy et al., 2012). RNA-binding proteins (RBPs) play crucial roles in such processes.
The genome of the model plant Arabidopsis thaliana contains ~200 RBPs, some of which are involved in stress response, plant immunity, or development (Lorkovic, 2009; Woloshen et al., 2011). Many Arabidopsis RBPs are specific to plants, suggesting that these RBPs may have plant-specific functions (Lorkovic, 2009). Recent studies revealed roles of RBPs not only in various developmental processes such as floral development (Lim et al., 2004; Mockler et al., 2004; Streitner et al., 2008), but also in responses to diverse environmental stresses such as abscisic acid (ABA) (Li et al., 2002; Ng et al., 2004; Bove et al., 2008; Kim et al., 2008), wounding (Bove et al., 2008), cold stress responses (Kim et al., 2010a; Kim et al., 2010b), chromatin modification (Liu et al., 2007; Baurle and Dean, 2008), leaf senescence (Kim et al., 2008), and plant immunity (Woloshen et al., 2011).
In Vicia faba, a guard-cell-specific ABA-activated serine-threonine protein kinase (AAPK) is integral in stomatal closure in response to ABA (Li et al., 2000) and its orthologue in Arabidopsis, OST1, has been shown to be a central and limiting element in ABA signal transduction from soluble ABA receptors (Acharya et al., 2013). Expression library screening identified AAPK interacting protein 1 (VfAKIP1), and phosphorylation of VfAKIP1 by AAPK is required for the interaction of VfAKIP1 with a target mRNA, dehydrin, in V. faba (Li et al., 2002). In Arabidopsis, three UBP1-associated protein 2 (UBA2) proteins with two RNA-recognition motif (RRM) domains, UBA2a, UBA2b, and UBA2c, are homologous to VfAKIP1. UBA2a was previously identified from an interaction screen with the heterogeneous nuclear ribonucleoprotein (hnRNP) UBP1 protein, hence the designation ‘UBP1-associated protein 2’ (Lambermon et al., 2002). VfAKIP1, UBA2a, and UBA2b fused with reporter GFP change their localization from a diffuse nuclear pattern to localization in nuclear speckles upon external ABA application (Li et al., 2002; Ng et al., 2004; Riera et al., 2006; Bove et al., 2008), while UBA2c forms nuclear speckles without ABA application (Bove et al., 2008). Transient expression of each Arabidopsis UBA2 protein in Nicotiana benthamiana leaves induces a programmed-cell-death/senescing phenotype, while constitutive expression of each of these proteins causes an early lethality phenotype in Arabidopsis (Kim et al., 2008). Controlled expression of the three UBA2s under a dexamethasone-inducible system showed that elevated expression of each of the three UBA2s causes leaf senescence (Kim et al., 2008), indicating that UBA2s are positive regulators of leaf senescence. Recent work also revealed that Arabidopsis LAM-domain RBPs, LARP1b and LARP1c, are also positive regulators of leaf senescence. Overexpression of LARP1s induced leaf senescence in Arabidopsis and elevated transcript abundance of senescence- and defence-related genes, including senescence-associated genes (SAGs) and pathogen-related genes (PRs) (Zhang et al., 2012). Both LARP1c and UBA2s are involved in leaf senescence seemingly as positive regulators, but they differ in subcellular localization with LARP1c found in the cytoplasm and UBA2s in the nucleus. Although underlying mechanisms for how UBA2s induce plant leaf senescence are not well defined, it is obvious that these RBPs play crucial roles in plant leaf senescence as positive regulators.
Plant senescence occurs as the final developmental stage, leading to the death of part or all of the plant (Lim et al., 2007; Zhang and Zhou, 2012). Leaf yellowing, caused by chlorophyll loss, is one of the key indicators of the progression of leaf senescence, followed by programmed cell death. Leaf senescence is accompanied by cellular, biochemical, and molecular changes. At the molecular level, SAGs and defence-associated PR genes are upregulated with progression of senescence (Lim et al., 2007). Consistently, overexpression of LARP1c or Arabidopsis UBA2s in Arabidopsis also increases the expression of SAG and PR genes (Kim et al., 2008; Zhang et al., 2012). Leaf senescence is also triggered by plant hormones such as ethylene, jasmonic acid, ABA, and salicylic acid (SA), and can be repressed by cytokinin (Zhang and Zhou, 2012). Arabidopsis UBA2 overexpression induces ethylene accumulation (Kim et al., 2008), implying that ethylene accumulation in UBA2 overexpressors may promote or cause leaf senescence. SA accumulation also can induce not only hypersensitive-like cell death, but also the expression of SAG (Buchanan-Wollaston et al., 2005; Zhang and Zhou, 2012) and PR genes (Durrant and Dong, 2004; Zhang et al., 2010). SA is closely related to the control of levels of reactive oxygen species (ROS); ROS are upstream of SA signalling pathways but SA accumulation can induce ROS accumulation by feedback amplification (Petrov and Van Breusegem, 2012). The cell death phenotypes of UBA2s and LARP1c transgenic Arabidopsis plants might also result from elevated SA and/or ROS content, which can mediate the hypersensitive response of plants and ultimately lead to leaf senescence and cell death.
Here, the potato RBPs StUBA2a/b and StUBA2c are characterized and shown to have high homology in amino acid sequence to VfAKIP1 and Arabidopsis UBA2s. StUBA2a/b and StUBA2c were identified in silico by sequence homology to VfAKIP1 and Arabidopsis UBA2s and, like VfAKIP1 and UBA2s, contain two conserved RRMs. Transient expression of either of StUBA2a/b or StUBA2c induced a hypersensitive-like cell death phenotype in tobacco (Nicotiana tabacum) leaves, and the stable overexpression of StUBA2a/b in Arabidopsis led to early leaf senescence and ROS accumulation, and altered the expression of genes involved in SA production and autophagy. Taken together, these results suggest that StUBA2a/b and StUBA2c play a crucial role in leaf senescence.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study. Seeds were surface-sterilized and planted on half-strength Murashige and Skoog (MS) medium (Sigma) (Murashige and Skoog, 1962). After stratification for 3 days at 4°C, plates with seeds were transferred to a growth chamber (Controlled Environments Ltd.) and cultured for 2 weeks under short-day light conditions (8h of light, 20°C/16h dark, 18°C) at a light intensity of 120 µmol m−2 s−1. Then, seedlings were transferred to 16cm2 square pots filled with Miracle-Grow potting mix (Scotts) supplemented with perlite and grown under long-day light conditions (16h of light, 20°C/8h of dark, 18°C) at a light intensity of 120 µmol m−2 s−1 and 60–70% relative humidity.
Cloning of potato StUBA2a/b and StUBA2c
Amino acid sequences of RBPs VfAKIP1 (Li et al., 2000) and Arabidopsis UBA2s (Bove et al., 2008; Kim et al., 2008) were used to identify homologous EST sequences from the TIGR potato EST database (http://plantta.jcvi.org) using the Basic Local Alignment Search Tool (BLAST). Two VfAKIP1-like EST sequences were obtained from the TIGR database, and these EST sequences were used to inform cloning of VfAKIP1-like cDNAs from S. tuberosum ‘Atlantic’. Atlantic potato was obtained from the potato Germplasm Center (US Potato Genebank) and was aseptically grown in Magenta boxes for a month, followed by extraction of total RNA. cDNA synthesized from total RNA using Superscript III (Invitrogen) was used for PCR amplification of 1577bp StUBA2a/b and 1349bp StUBA2c using StUBA2a/b or StUBA2c gene-specific primer sets (Supplementary Table S1). PCR fragments of StUBA2a/b or StUBA2c were cloned into PCR-Blunt II-TOPO cloning vectors (Invitrogen) and sequenced. The amino acid sequence of StUBA2c was identical to that in the TIGR database and also that in the protein database released by the Potato Genome Sequencing Consortium (Xu et al., 2011), but StUBA2a/b showed three and seven amino acid differences compared to that in the TIGR database or potato genome protein database, likely due to single nucleotide polymorphisms among cultivars. Subsequently, SacI/XbaI digests of StUBA2a/b or StUBA2c were subcloned into SacI/SpeI sites of the modified pORE-R2 binary vector (Coutu et al., 2007) harbouring the CaMV 35S promoter to generate 35S:StUBA2a/b and 35S:StUBA2c constructs. RRM-deleted StUBA2a/b and StUBA2c were generated from pORE-R2 binary vectors harbouring 35S:StUBA2a/b or 35S:StUBA2c cassettes using gene-specific primers (Supplementary Table S1) by following the mutagenesis method described for the In-Fusion HD cloning system (Clontech Laboratories, Inc.). To generate the modified pORE-R2 with CaMV 35S promoter, a HindIII/XbaI fragment containing the CaMV 35S promoter from pGWB8 binary vector (Nakagawa et al., 2007) was cloned into the HindIII/XbaI site of the pORE-R2 vector (Coutu et al., 2007).
Constructs of guard-cell-specific pGC1:smGFP-StUBA2a/b and -StUBA2c
To examine whether StUBA2a/b and StUBA2c have roles in the ABA response of guard cells, similar to Arabidopsis UBA2a and UBA2b, and VfAKIP1 (Bove et al., 2008; Ng et al., 2004), both genes were cloned under the control of a guard-cell-specific pGC1 promoter (Yang et al., 2008). To construct the final cassette of pGC1:smGFP-StUBA2a/b or -StUBA2c, first the pGC1 promoter (−1140/+23) was PCR-amplified from genomic DNA from Arabidopsis Col-0 using gene-specific primers (Supplementary Table S1) and cloned into SacII/XhoI sites of the pORE-R2 binary vector (Coutu et al., 2007). Subsequently, smGFP, StUBA2a/b, and StUBA2c were amplified using gene-specific primers (Supplementary Table S1). The smGFP PCR fragment was cloned into the XhoI/NotI site of the pORE-R2-pGC1 vector, followed by introduction of StUBA2a/b or StUBA2c to the Not1/SpeI or Not1/KpnI sites of the pORE-R2 pGC1:smGFP vector, respectively. Images of smGFP or smGFP-StUBA2s expressed in Arabidopsis were obtained using a FV500 confocal microscope (Olympus).
Agrobacterium tumefaciens-mediated transient or stable transformation
The pORE-R2 binary vectors with 35S:StUBA2s or pGC1:smGFP-StUBA2s constructs were electroporated into Agrobacterium tumefaciens strain C58C1, which was used to generate stable transgenic Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Putative transgenic plants were screened on half-strength MS plates containing kanamycin. Kanamycin-resistant seedlings were transplanted to soil for the T2 seed set. T2 or T3 plants were used for phenotypic analysis. For the transient expression assay, Agrobacterium containing pORE-R2 binary vector harbouring target constructs, 35S:StUBA2a/b, 35S:StUBA2a/b, or 35S:GUS as a vector control, was grown overnight in lysogeny broth media supplemented with 50mg L−1 of kanamycin and 150 μM acetosyringone. Agro-infiltration was performed on 1-month-old N. tabacum leaves following the method previously described (Kim et al., 2008). The hypersensitive-like cell death phenotype was photographed 21 days after Agro-infiltration.
PCR and gene expression analysis
RNA was extracted from Arabidopsis plants or from potato using the RNasy RNA extraction kit (Qiagen). The total RNA concentration was quantified by spectrophotometric measurement, and 1 or 2 µg of total RNA was used for cDNA synthesis using either Superscript III reverse transcriptase (Invitrogen) or a cDNA EcoDry Premix-Oligo dT Kit (Clontech). PCR was carried out in 20 μL reactions containing 1 μL of cDNA and 0.1 µM of gene-specific primers (Supplementary Table S1) using Ex-Taq DNA polymerase (TaKaRa) under the following conditions: an initial denaturation step at 95°C for 1min followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, polymerization at 72°C for 0.5–2min, and a final extension at 72°C for 7min. For real-time PCR, cDNA was diluted to a concentration of 1:10, and quantitative reverse transcriptase (qRT)-PCR was performed using SYBR Premix Ex Taq (TaKaRa). Actin was used as an internal control, and the data obtained were analysed with IQ5 software (Bio-Rad). The expression analysis of autophagy-associated genes (Arabidopsis TOR, ATG8b-h, ATG9, and ATG18a) was carried out using Arabidopsis TOR or ATG gene-specific primers (Supplementary Table S1).
3,3ʹ-Diaminobenzidine staining
To determine H2O2 accumulation, leaves of 35S:StUBA2s transgenic lines and wild-type Arabidopsis were stained with 3,3ʹ-diaminobenzidine (DAB) solution according to a protocol described previously (Wohlgemuth et al., 2002). Briefly, 1-month-old leaves from the lower position were immersed overnight in staining solution containing 1mg mL−1 of DAB and de-stained by soaking in 100% ethanol for 3h.
Extraction of free SA
SA was extracted and analysed by gas chromatography-mass spectrometry (Ultra GC-Q/MS; Shimadzu Inc.) using a method described by Park et al. (2012). Three-week-old leaves (0.05g) were ground in liquid nitrogen and the powdered samples were extracted twice with 1mL of 90% methanol at 30°C for 10min. Supernatants were collected by centrifuging at 13 000rpm for 10min at 4°C and then mixed with 50 µL of 3,4,5-trimethoxycinnamic acid (100 µg mL−1) as an internal standard, followed by extraction twice with ethyl acetate. The ethyl acetate fraction was dried in a centrifugal concentrator (CVE-2000; Eyela). For derivatization of the dried extracts, 40 µL of N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide containing 1% tert-butyldimethylchlorosilane (TBDMCS) and 40 µL of pyridine were added to the dried extracts, followed by incubation at 60°C for 30min at a mixing frequency of 1200rpm using a Thermomixer Comfort (model 5355; Eppendorf AG). Each derivatized sample (1 µL) was separated on a 30 m × 0.25mm internal diameter fused-silica capillary column coated with 0.25 µm CP-SIL 8 CB low bleed (Varian Inc.). The injector temperature was 230°C, and the flow rate of helium gas through the column was 1.0mL min−1. The temperature programme was set at 150°C and maintained at 150°C for 2min, followed by a 15°C min−1 oven temperature ramp to 320°C, which was held for 10min. The column effluent was later introduced into a QP2010 Ultra mass spectrometer (Shimadzu Inc.). The transfer line and the ion-source temperatures were 250°C and 200°C, respectively. The detected mass range was 85–700 m/z. The quantity of SA was calculated based on the ratio of the major fragment ion (m/z 309) of the tert-butyldimethylsilyl (TBDMS) derivative of SA and the corresponding fragment ion (m/z 295) of the internal standard.
Results
Cloning of potato StUBA2a/b and StUBA2c
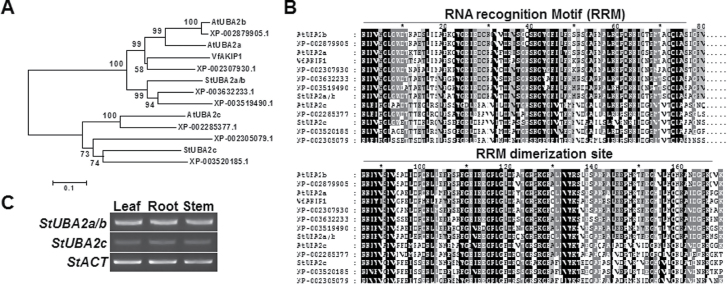
To identify VfAKIP1 homologs from S. tuberosum, amino acid sequences of VfAKIP1 and three Arabidopsis UBA2s were aligned against the TIGR potato EST database (http://plantta.jcvi.org) using BLAST. Two AKIP1-like EST sequences, TA24992_4113 and TA26496_4113, were obtained from the BLAST search. Amino acid sequences of TA24992_4113 showed higher sequence identity with VfAKIP1, and Arabidopsis UBA2a and UBA2b, while amino acid sequences of TA26496_4113 showed higher sequence identity with UBA2c. These EST sequences were used to clone 1577bp and 1349bp full-length cDNAs from potato cultivar ‘Atlantic’. The former was designated as StUBA2a/b and the latter as StUBA2c. Alignment of StUBA2s with VfAKIP1 and Arabidopsis UBA2s showed that StUBA2a/b had 42–43% amino acid sequence identity with UBA2a and UBA2b, and VfAKIP1, and that StUBA2c had 45% identity with UBA2c (Supplementary Table S2). Phylogenetic analysis using StUBA2s and various plant VfAKIP1 homologues showed distinctive separation between StUBA2a/b and StUBA2c. StUBA2a/b grouped with VfAKIP1, Arabidopsis UBA2a, and Arabidopsis UBA2b, while StUBA2c grouped with Arabidopsis UBA2c in a separate clade (Fig. 1A). The comparison of deduced amino acid sequences of the plant VfAKIP1 homologues including StUBA2s revealed that RRM domains are highly conserved in both StUBA2s (Fig. 1B; Supplementary Fig. S1). RT-PCR analysis showed that StUBA2a/b and StUBA2c expressed comparably in all tested tissues: leaf, root, and stem (Fig. 1C).
Fig. 1.
Phylogenetic analysis and comparison of potato StUBA2s and VfAKIP1 homologues in various plant species. (A). Phylogenetic tree of given VfAKIP1 homologues including two newly identified potato StUBA2s, generated using MEGA6 (Tamura et al., 2013) and tree view programs (Page, 1996). Numbers on the tree denote percent homology from 2000 bootstrap replicates. (B) Comparison of conserved RRM domains among given VfAKIP1 homologues. (C) Expression patterns of StUBA2a/b and StUBA2c in potato leaf, root, and stem.
Overexpression of StUBA2s induces early leaf senescence associated with hypersensitive-like cell death
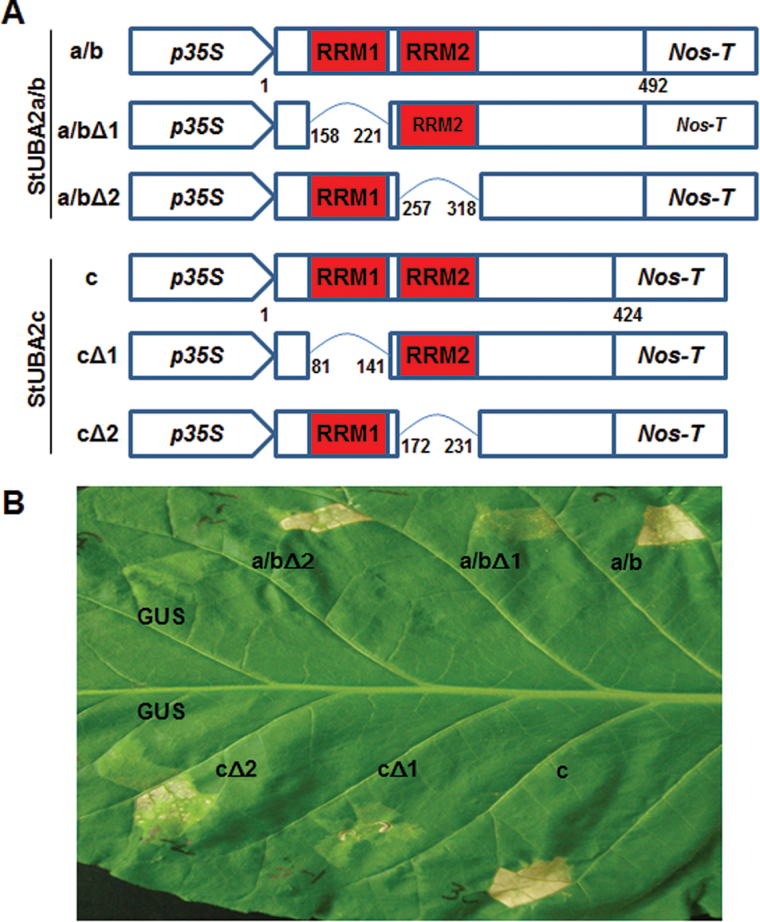
It has previously been reported that the transient expression of Arabidopsis UBA2s induces hypersensitive-like cell death in N. benthamiana leaves, while their constitutive expression under the control of the 35S promoter causes lethality at the young seedling stage (Kim et al., 2008). Because StUBA2a/b and StUBA2c had high amino acid sequence identity to VfAKIP1 and Arabidopsis UBA2s, it was of interest to test whether overexpression of StUBA2s could induce the same hypersensitive-like cell death phenotype as Arabidopsis UBA2s. First, a transient expression assay of StUBA2s was carried out. For this test, pORE-R2 binary vectors containing StUBA2s under the control of a constitutive 35S promoter (Fig. 2A) were introduced into N. tabacum leaves by Agro-infiltration. N. tabacum leaves started to show hypersensitive-like cell death symptoms within 1 week after Agro-infiltration, and symptoms were severely aggravated within 3 weeks (Fig. 2B), indicating that StUBA2s are most likely functional potato homologues of the Arabidopsis UBA2s. In parallel with full-length StUBA2a/b and StUBA2c, RRM-domain-deleted StUBA2s (Fig. 2A) were also expressed transiently to examine whether RRM domains play important roles in the induction of hypersensitive-like cell death. As a result, it was found that the first RRM domain in both StUBA2s is crucial for the induction of hypersensitive-like cell death (Fig. 2B).
Fig. 2.
Transient expression of potato StUBA2a/b and StUBA2c induces hypersensitive-like cell death in N. tabacum leaves. (A) Schematics of StUBA2a/b, StUBA2c, and RRM deletion constructs in a modified pORE-R2 vector carrying the 35S promoter. (B) Hypersensitive-like cell death caused by StUBA2a/b and StUBA2c. All constructs were transiently expressed in tobacco leaves by Agro-infiltration. Hypersensitive-like cell death was induced by the transient expression of 35S:StUBA2a/b (a/b), 35StUBA2a/bΔ2 (a/bΔ2), 35S:StUBA2c (c), or 35S:StUBA2cΔ2 (cΔ2) but not by 35StUBA2a/bΔ1 (a/bΔ1) or 35S:StUBA2cΔ1 (cΔ1). A 35S:GUS (GUS) construct was used as a control. Pictures were taken 20 days after Agro-infiltration (this figure is available in colour at JXB online).
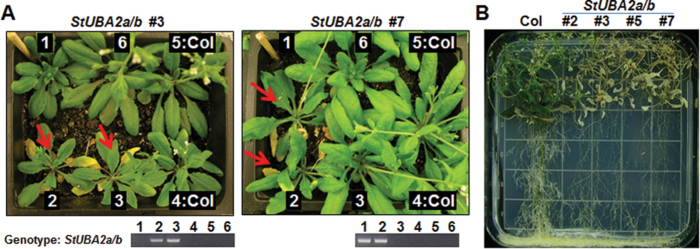
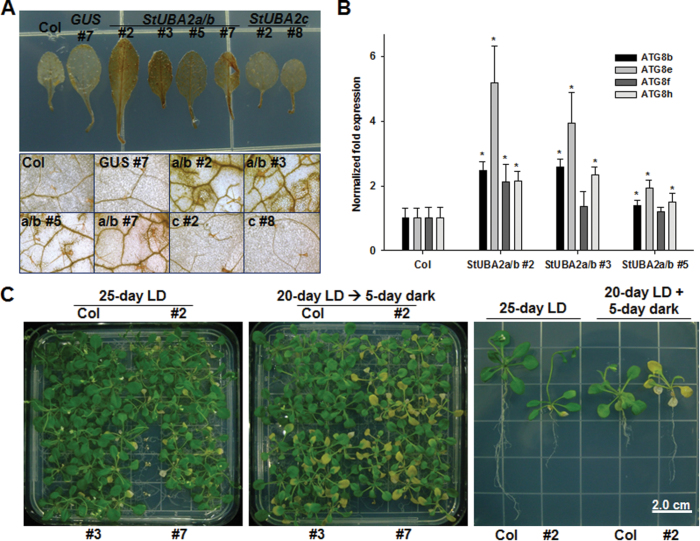
To further investigate the roles of StUBA2 proteins, stable 35S:StUBA2a/b and 35S:StUBA2c transgenic Arabidopsis plants were generated using the same binary vector as used for the transient expression assay. Unlike constitutive expression of Arabidopsis UBA2s, which terminated growth of transgenic Arabidopsis plants at the young seedling stage (Kim et al., 2008), the overexpression of StUBA2a/b did not cause a lethal phenotype during early development, which made it possible to perform phenotypic analysis at later stages. Segregating T2 StUBA2a/b transgenic plants were planted in soil along with wild-type (Col) plants as control. Some of the T2 StUBA2a/b overexpressor lines started to show a yellowing phenotype in old leaves 6 weeks after transplanting (Fig. 3A). Genotyping of segregating 35S:StUBA2a/b #3 and #7 T2 plants using StUBA2a/b specific primers showed that the presence of the transgene coincided with the leaf-yellowing phenotype (Fig. 3A). The senescing phenotype was re-examined in T3 35S:StUBA2a/b transgenic plants, in which it was found that the phenotype was aggravated with increasing plant age (Fig. 3B; Supplementary Fig. S2). The T2 transgenic plants shown in Fig. 3A were used for a survey of transcript levels of transgene, and defence- and senescence-associated genes as shown in Fig. 4A. The extent of the hypersensitive-like cell death in 35S:StUBA2a/b transgenic lines coincided with the levels of transgene expression, which may explain why no such phenotype was detected in plants with low expression level of the StUBA2c transgene as summarized in ‘Phenotype’ (Fig. 4A). Except for T2 plants used in Fig. 3A and Fig. 4A, T3 transgenic lines were used for all phenotypic analyses.
Fig. 3.
Early cell death/senescence of Arabidopsis plants correlates with presence of the 35S:StUBA2a/b transgene. (A) Phenotype and genotype were examined for 1-month-old 35S:StUBA2a/b T2 transgenic lines, #3 and #7. Six plants were planted in each pot, four plants randomly picked from each 2-week-old T2 segregating transgenic line and two untransformed Arabidopsis Col-0 plants, and numbered from 1 to 6 as shown. Early senescing phenotype was observed in PCR-confirmed transgenic plants as indicated by red arrows. Genomic DNA from individual plants was used for PCR-genotyping using StUBA2a/b gene-specific primers to detect the presence of transgene StUBA2a/b. (B) The senescing phenotype was re-examined in T3 35S:StUBA2a/b transgenic lines, and these lines exhibited early death of 3-month-old 35S:StUBA2a/b plants on an MS plate (this figure is available in colour at JXB online).
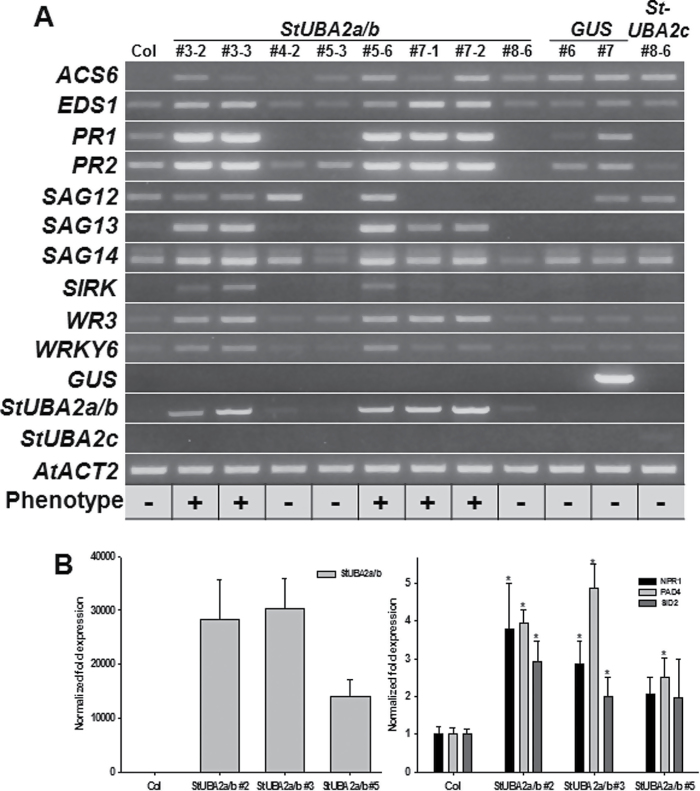
Fig. 4.
Overexpression of StUBA2a/b altered expression patterns of senescence- and defence-associated genes. (A) Transcript level of StUBA2a/b correlated with early senescing phenotype (‘Phenotype’), accompanied by expression changes of various genes. Total RNA from wild-type and T2 segregating 35S:StUBA2a/b transgenic plants including those shown in Fig. 3A was isolated and used for RT-PCR analyses. (B) Elevated transcript levels of genes involved in SA signalling or biosynthesis in T3 35S:StUBA2a/b plants. qRT-PCR was carried out using StUBA2a/b gene-specific primers or primers specific to NPR1, PAD3, and SID2 involved in SA signalling or biosynthesis (Supplementary Table S1). Transcript levels were normalized by transcript levels of Arabidopsis ACT2. Among StUBA2a/b transgenic lines, #2, #3, and #7 lines were heterozygous, and #5 was homozygous. Five-week-old rosette leaves were used for RNA extraction for qRT-PCR. T3 transgenic plants with kanamycin resistance were used for experiments unless otherwise stated except for those in Fig. 3A, Fig. 4A, and Supplementary Fig. S2B, where T2 plants were used. Asterisk indicates a significant difference at P < 0.05.
In addition to UBA2s and StUBA2s, constitutive expression of another set of Arabidopsis RBPs, LARP1b and LARP1c, was recently reported to induce precocious leaf senescence in Arabidopsis (Zhang et al., 2012). These gain-of-function mutants showed reduced chlorophyll content and hypersensitive-like cell death starting from leaves at a lower position, which is very similar to the phenotype observed in StUBA2a/b transgenic plants. Transcript level of the native LARP1c increased along with the progression of leaf senescence, and overexpression of LARP1c induced the expression of SAG12 and 13, suggesting that LARP1c and SAGs may be involved in plant senescence concomitantly (Zhang et al., 2012). Therefore, an examination of whether overexpression of StUBA2a/b or StUBA2c induced the upregulation of LARP1b and LARP1c (Supplementary Fig. S3) was performed, but this was not the case. Also, leaf senescence induced by StUBA2a/b did not show any correlation with SAG12 expression (Fig. 4A), implying that the expression of StUBA2a/b and Arabidopsis UBA2s induce leaf senescence through overlapping but non-identical mechanism(s) from LARP1c.
Upregulation of stress- and defence-associated genes in StUBA2a/b Arabidopsis
Plants expressing StUBA2a/b show phenotypes similar to both age-dependent leaf senescence and the hypersensitive response. To better understand the pathways induced by StUBA2a/b expression, the transcript levels of genes associated with pathogen response, leaf senescence, SA, and autophagy were analysed (Figs. 4 and 5B; Supplementary Table S3). Leaf senescence is accompanied by changes in the expression of genes such as SAGs and defence-related genes. Consistently, leaf senescence caused by the overexpression of Arabidopsis UBA2s also induces the altered expression of various stress-responsive genes (Kim et al., 2008), providing the plausible hypothesis that constitutive expression of StUBA2a/b also could alter the expression of such genes. StUBA2a/b transgenic lines #3, #5, and #7 exhibited altered expression of enhanced disease susceptibility 1 (EDS1), PR1 and PR2, SAG13, SAG14, and WOUND-RESPONSIVE 3 (WR3) among tested genes (Fig. 4A). Upregulation of these genes is consistent with previous findings concerning Arabidopsis UBA2 genes, indicating that StUBA2a/b is most likely a UBA2a or UBA2b functional homologue in potato. Transcript levels of the genes 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 6 (ACS6) and WRKY6 were not consistent in two separate RT-PCR analyses, suggesting that the expression of these genes is not only influenced by the StUBA2a/b transgene, but also by other factors such as plant age.
Fig. 5.
Early senescence of 35S:StUBA2a/b transgenic plants is associated with H2O2 accumulation and the expression of autophagy-associated genes. (A) H2O2 accumulation in 35S:StUBA2a/b transgenic plants grown on a half-strength MS plate for one month under permissive long-day light conditions (16h light, 22°C/12h dark, 20°C) at light intensity of 100 µmol m−2s−1. (B) Altered expression of autophagy-associated genes in 35S:StUBA2a/b transgenic plants. qRT-PCR was carried out using ATG8b, ATG8e, ATG8f, and ATG8h gene-specific primers (Supplementary Table S1). Asterisk indicates a significant difference at P < 0.05. (C) Senescence of 35S:StUBA2a/b transgenic plants was enhanced under darkness. Seeds of transgenic lines and wild type (Col) were sown on half-strength MS plates and grown for 25 days under the same permissive long-day light conditions as described above. For darkness, the plate was wrapped with aluminium foil on the 20th day and otherwise maintained in the same conditions for 5 more days. Because StUBA2a/b #2, #3, and #7 seeds from T2 heterozygous parents were sown in kanamycin-free MS media in this experiment, some progeny (WT) from StUBA2a/b #2, #3, and #7 lines did not show the senescing phenotype.
Recently, it was reported that Arabidopsis RNA-binding protein-defence related 1 (Arabidopsis RBP-DR1), with three RRM domains, is involved in pathogen defence through SA signalling (Qi et al., 2010). To examine whether StUBA2a/b also affects SA signalling, transcript levels of genes involved in SA biosynthesis and signalling (Lu, 2009; Ng et al., 2011) were examined. NPR1, PAD4, and SID2 were upregulated in StUBA2a/b transgenic lines and their transcript levels showed positive correlation with StUBA2a/b expression levels (Fig. 4B; Supplementary Fig. S4), indicating that StUBA2a/b expression may be involved in SA signalling and biosynthesis. To examine whether upregulation of SA signalling genes was due to or resulted in increased SA content, SA was measured by gas chromatography-mass spectrometry using a GCMS-PQ2010 Ultra (Shimadzu). SA content in StUBA2a transgenic lines was found to be significantly higher than that in wild-type plants (Supplementary Fig. S5).
Correlation of hypersensitive-like cell death in StUBA2a/b plants with H2O2 accumulation and autophagy
Excessive ROS accumulation can damage plant cells and can cause necrosis either directly or indirectly via programmed cell death (Van Breusegem and Dat, 2006). To test whether hypersensitive-like cell death caused by StUBA2 expression is associated with ROS accumulation, 5-week-old rosette leaves of wild-type, GUS (vector control), StUBA2a/b, and StUBA2c transgenic lines were stained with DAB. H2O2 accumulation was detected in StUBA2a/b transgenic plants (Fig. 5A). Consistent with the hypersensitive-like cell death phenotype, StUBA2c transgenic plants expressing low levels of the transgene did not exhibit H2O2 accumulation. Because higher levels of H2O2 can cause severe oxidative damage in Arabidopsis, followed by induction of autophagy (Perez-Perez et al., 2012), whether StUBA2a/b expression alters transcript levels of genes involved in autophagy was examined. RT-PCR and qRT-PCR were carried out to examine the expression of seven Arabidopsis autophagy-related genes: TOR, ATG8 (b, e, f, and h), ATG9, and ATG18a. Four genes, Arabidopsis ATG8b, e, h, and Arabidopsis ATG9 were upregulated in StUBA2a/b plants (Fig. 5B; Supplementary Fig. S6), indicating that StUBA2a/b expression plausibly induces autophagy through ROS accumulation. However, Arabidopsis TOR, a key negative regulator for the induction of autophagy, was not influenced by StUBA2a/b expression compared to wild type (Supplementary Fig. S6), suggesting that those genes with altered expression resulting from StUBA2a/b transgene expression may be regulated by other mechanisms independent from Arabidopsis TOR. Because the leaf senescence of Arabidopsis autophagy mutants is enhanced under darkness (Liu and Bassham, 2012), the response of StUBA2a/b transgenic plants to darkness was determined. Five days of darkness enhanced the yellowing symptom of StUBA2a/b plants (Fig. 5C), but no changes were detected in autophagosome formation compared to wild type (data not shown), as assayed by monodansylcadaverine staining of roots treated with 5-day darkness (Contento et al., 2005; Liu and Bassham, 2010).
Subcellular localization of StUBA2s
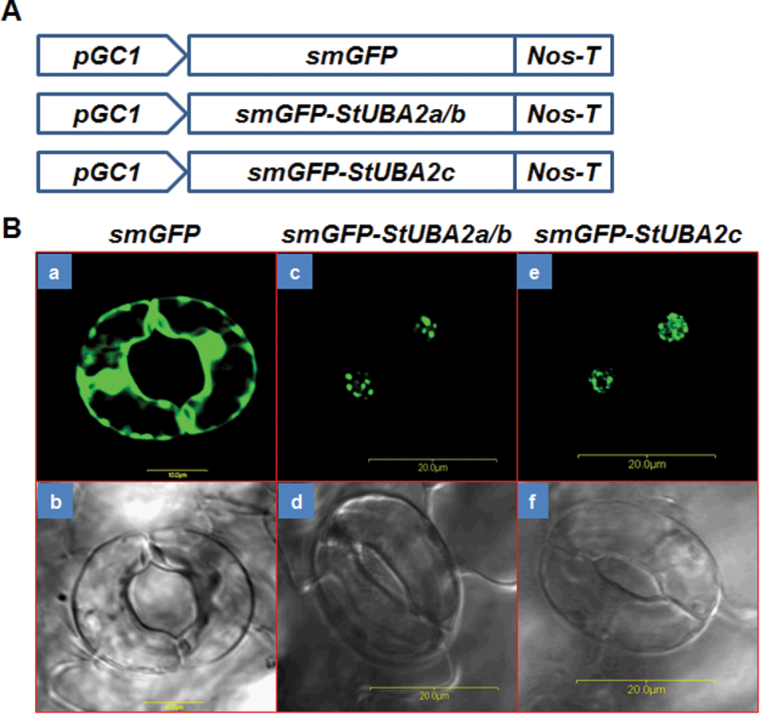
In V. faba, VfAKIP1 is involved in ABA signalling in guard cells (Li et al., 2002) and ABA induces rapid subnuclear relocalization of VfAKIP1, UBA2a, and UBA2b proteins into nuclear speckles (Li et al., 2002; Ng et al., 2004). To examine StUBA2 localization, smGFP-StUBA2s under the control of the pGC1 promoter (Fig. 6A), a guard-cell-specific promoter (Yang et al., 2008), were transformed into Arabidopsis. Nuclear speckles were observed in guard cells of both pGC1:smGFP-StUBA2a/b and pGC1:smGFP-StUBA2c transgenic plants (Fig. 6B). Because it was reported that exogenous ABA application can rearrange or enhance VfAKIP1-GFP subnuclear localization into nuclear speckles, leaves of wild-type and transgenic plants were submerged in ABA solution as described previously (Li et al., 2002) and examined under a confocal microscope for smGFP-StUBA2s-marked nuclear speckles. However, no changes were observed in smGFP-StUBA2s localization after ABA treatment (Supplementary Fig. S7), similar to UBA2c-GFP fusion proteins (Bove et al., 2008). These results suggest that StUBA2a/b and StUBA2c can reside in nuclear speckles, but that this localization is not affected by ABA, although it is difficult to exclude the possibility that this level of endogenous ABA may already be sufficient for StUBA2 proteins to form stable foci.
Fig. 6.
Localization of smGFP-StUBA2s in guard cells. (A) Diagram of constructs used for guard-cell-specific StUBA2a/b and StUBA2c expression in modified pORE-R3 vector. pGC1, guard-cell-specific promoter (Yang et al., 2008); smGFP, green fluorescence protein cloned from pORE-R3. (B) Confocal images were taken from leaves of 2-week-old seedlings. (a) smGFP image; (b) bright field image of (a); (c) smGFP-StUBA2a/b image; (d) bright field image of (c); (e) smGFP-StUBA2c image; (f) bright field image of (e) (this figure is available in colour at JXB online).
Discussion
It has been reported that VfAKIP1 and Arabidopsis UBA2s are hnRNP-like RBPs involved in wounding, senescence (Bove et al., 2008; Kim et al., 2008), and ABA signalling in guard cells (Bove et al., 2008; Li et al., 2002; Ng et al., 2004; Riera et al., 2006). Despite described roles of VfAKIP1 and Arabidopsis UBA2s in stress and senescence, no other VfAKIP1 homologs have been identified and characterized from other plant species. In this study, the physiological roles of two VfAKIP1-like proteins from S. tuberosum ‘Atlantic’ were investigated, with a particular interest in leaf senescence. Controllable shoot senescence could confer an agronomic benefit. Potato growers routinely remove potato shoots prior to harvest to facilitate harvesting, improve setting of skin colour, and minimize disease infection from the shoots (Chen et al., 2009). Therefore, timely potato shoot decay upon harvest could have a beneficial economic impact on potato farming, which may be achievable through altered expression of gene(s) related to plant senescence such as StUBA2s or Arabidopsis UBA2s.
Leaf senescence of 35S:StUBA2a/b plants was enhanced upon flowering
In agreement with the hypersensitive-like cell death and leaf senescence caused by overexpression of Arabidopsis UBA2s (Kim et al., 2008), transient expression or stable expression of StUBA2a/b or StUBA2c under the constitutive 35S promoter induced hypersensitive-like cell death in tobacco (Fig. 2B). StUBA2a/b transgenic Arabidopsis plants showed slight growth reduction, but did not exhibit lethality or severe growth arrest at a young seedling stage as was observed in 35S:UBA2s transgenic Arabidopsis plants (Kim et al., 2008). Rather, the leaf senescence of StUBA2a/b transgenic plants started to appear at a much later developmental stage than in 35S:UBA2s plants, close to the time of flower initiation, and was aggravated upon flowering, suggesting that the mode of StUBA2a/b action may be closely related to the change of phase associated with flowering. The phenotype also apparently required a certain threshold in transcript/protein level to exert its role, as evidenced by RT-PCR results in which the transcript level of transgene StUBA2a/b correlated with the degree of the senescing phenotype (Fig. 4A). Also, older leaves of StUBA2a/b plants exhibited higher H2O2 accumulation than young leaves (Supplementary Fig. S8), implying that certain levels of ROS accumulation may be required for the initiation of hypersensitive-like cell death preceding leaf senescence in StUBA2a/b transgenic plants.
The SAG12 gene is well known as a senescence-associated marker and its expression is tightly correlated with age-dependent leaf senescence/cell death (Kim et al., 2009). Age-dependent leaf senescence of wild-type Arabidopsis plant ecotype ‘Col-0’ starts about 28 days after germination and requires the activation of ORESARA1 (ORE1) transcription factor (Kim et al., 2009), of which transcript level is itself regulated by ETHYLENE INSENSITIVE 2 (EIN2). ORE1 and SAG12 expression show tight correlation with progression of aged-leaf senescence (Kim et al., 2009). Aged-leaf senescence and SAG12 expression were delayed in the ore1 mutant and further delayed in an ore1 and ein2 double mutant (Kim et al., 2009), indicating that ORE1 and SAG12 coincidently express in accordance with leaf senescence. Opposite to delayed age-dependent leaf senescence in ore1 or ein2 mutants, leaf senescence was accelerated in the senescence-associated ubiquitin ligase1 (saul1) mutant, in which ORE1 and SAG12 started to accumulate much earlier than in wild type (Vogelmann et al., 2012). Since the senescing phenotype of StUBA2a/b and Arabidopsis UBA2 overexpressing plants did not correlate with SAG12 expression, these results strongly suggest that leaf senescence induced by this family of hnRNPs may not be through the age-dependent leaf senescence pathway as also reported by Kim et al. (2008). Intriguingly, the transcript level of ORE1 but not EIN2 was upregulated in StUBA2a/b plants (Supplementary Fig. S9), implying that leaf senescence caused by StUBA2a/b and UBA2s likely is related to the upregulation of ORE1, but independent from ORE1-EIN2-SAG12-associated age-dependent leaf senescence mechanism(s). Therefore it would be interesting to examine whether cell death in Arabidopsis UBA2s and StUBA2a/b overexpression plants can be sustained in an ore1 mutant background, in which leaf senescence is delayed.
Do ROS production and autophagy correlate with hypersensitive-like cell death in StUBA2a/b transgenic plants?
StUBA2a/b expression in Arabidopsis caused H2O2 accumulation in leaves (Fig. 5A), and homolysis of H2O2 to 2OH− can damage plant cells (Becana et al., 1998), suggesting that the hypersensitive-like cell death phenotype of StUBA2a/b plants could be attributed to elevated H2O2. As one ROS, H2O2, is known to be involved in various signalling processes in plants and is generated from various sources, including as a by-product of reactions in chloroplasts, mitochondria, and peroxisomes (Tripathy and Oelmuller, 2012). Because ROS accumulation is harmful to cells, death or signalling depends on how cells regulate ROS homeostasis. Ironically, however, cell death is also an essential part of the life cycle by which multicellular organisms can recycle nutrients to maintain proper growth and development (Van Breusegem and Dat, 2006). ROS and autophagy have long been known to be associated with cell death, but recent studies reveal that they are also involved in signalling and acclimation under adverse stress conditions (Perez-Perez et al., 2012). H2O2 accumulation activates autophagy, which is involved in the recycling of reusable molecules and damaged intracellular components or toxic molecules. StUBA2a/b expression in Arabidopsis altered the expression of autophagy-associated genes, ATG8 (ATG8b, e, and h) and ATG9 (Fig. 5B; Supplementary Fig. S6), implying that H2O2 resulting from StUBA2a/b expression indeed changed autophagy signalling by increasing the expression of ATG genes. It was reported that nitrogen remobilization in several ATG mutants including atg5 was significantly reduced (Guiboileau et al., 2012), indicating that autophagy plays important roles in nitrogen recycling and remobilization in plants. Nutrition deficiency also can induce ROS production in plants, which can activate autophagy. As shown in Fig 3A, early leaf senescence at the lower position in StUBA2a/b plants is similar to symptoms observed in atg mutants (Yoshimoto et al., 2009; Guiboileau et al., 2012), and three ATG8 genes were upregulated in StUBA2a/b transgenic plants under normal conditions. Together, these results raise questions about whether StUBA2s are involved in autophagy-mediated regulation of nitrogen uptake or remobilization, which will be interesting topics for future studies.
StUBA2a/b expression induced the expression of genes involved in senescence, defence, and SA signalling
Constitutive expression of StUBA2a/b in Arabidopsis increased the expression of various genes involved in defence and senescence, such as PRs and several SAGs, as shown in Fig. 4A, which is consistent with the results observed in transgenic Arabidopsis with inducible overexpression of Arabidopsis UBA2s (Kim et al., 2008), as well as in transgenic tobacco expressing LARP1c (Zhang et al., 2012). In addition, StUBA2a/b also elevated the transcript levels of SID2, NPR1, and PAD4 involved in SA biosynthesis or signalling in Arabidopsis (Fig. 4B; Supplementary Fig. S4), suggesting that StUBA2a/b also affects SA signalling. SID2 is upstream in the SA biosynthesis pathway where it catalyses conversion of chorismate to isochorismate (Chen et al., 2009), and NPR1 and PAD4 can induce SA accumulation by feedback amplification (Lu, 2009; Ng et al., 2011). It is well known that SA accumulation can induce hypersensitive-like cell death. Therefore, SA content may be elevated in StUBA2a/b plants, which would promote cell death, as was the case here (Supplementary Fig. S5).
Nuclear speckles formed by smGFP-StUBA2s are not reorganized by exogenous ABA
Nuclear speckles are interchromatin granule clusters enriched in pre-mRNA splicing factors (Spector and Lamond, 2011). Nuclear speckles are believed to play crucial roles in gene expression as sites of pre-mRNA processing such as mRNA splicing (Reddy et al., 2012). VfAKIP1 is relocalized to nuclear speckles by ABA treatment and also plays important roles in ABA-mediated stomatal regulation (Li et al., 2002; Ng et al., 2004). To examine StUBA2 localization and ABA-response in guard cells, Arabidopsis transgenic plants were generated that constitutively expressed smGFP-StUBA2s in guard cells. Because it was shown that VfAKIP1 and Arabidopsis UBA2a and UBA2b fused to GFP relocalize to nuclear speckles following a few minutes of ABA treatment, localization of smGFP-StUBA2s with and without exogenous ABA application was compared. smGFP-StUBA2s fusion proteins were visualized as nuclear speckles (Fig. 6B). However, reorganization of nuclear speckles upon exogenous ABA application was not observed in pGC1:smGFP-StUBA2s plants (Supplementary Fig. S7). Also, the accelerated leaf senescence phenotype was not observed in pGC1:smGFP-StUBA2s plants.
In summary, potato RBP StUBA2a/b is a positive regulator of a leaf senescence, which likely is accelerated by hypersensitive-like cell death caused by H2O2 accumulation and the activation of SA signalling. This hypersensitive-like cell death/senescence phenotype occurs earlier than age-dependent leaf senescence. Furthermore, StUBA2a/b also induces genes involved in autophagy signalling that are related to nitrogen mobilization, which is yet to be examined. Thus, leaf senescence induced by potato RBP StUBA2a/b appears to arise from activation of components of several distinct cell-death pathways. Because timely leaf senescence upon potato harvest could confer a tremendous economic impact on potato farming, StUBA2s potentially can be considered for transgenic approaches that can induce potato shoot senescence at harvest.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. List of primers used in this study
Table S2. Identity and similarity among StUBA2s and other homologous proteins.
Table S3. Characteristics of genes used for expression assays.
Fig. S1. Alignment of full-length StUBA2s with homologous proteins in several plant species. (A) StUBA2a/b and its homologous proteins. (B) StUBA2c and its homologous proteins.
Fig. S2. Early leaf senescence of 35S:StUBA2a/b plants grown aseptically in Magenta boxes, or in soil. (A). Senescing phenotype observed in 6-week-old T3 35S:StUBA2a/b plants in MS media. # numbers refer to independent transgenic lines. Each T3 transgenic line was germinated on an MS plate containing kanamycin and then kanamycin-resistant plants were transferred to antibiotic-free MS media. (B). Senescing phenotype observed in 2-month-old T2 transgenic plants grown in soil in a growth chamber. These plants are the same as those shown in Fig. 3A, but at a later developmental stage. Numbers indicate individual T2 plants from either 35S:StUBA2a/b #3 or 35S:StUBA2a/b #7.
Fig. S3. Transcript levels of LARP1 RBP family are not altered by the overexpression of StUBA2a/b.
Fig. S4. Elevated transcript levels of genes involved in SA signalling or biosynthesis in T3 35S:StUBA2a/b plants. RT-PCR was carried out using gene specific primers (Supplementary Table S1). Among StUBA2a/b transgenic lines, #2, #3, and #7 lines were heterozygous, and #5 was homozygous. StUBA2c #2 and #8 and GUS #7 lines were homozygous. GUS #7 line was used as vector control.
Fig. S5. SA content in 3-week-old 35S:StUBA2a/b and wild-type plants grown in MS media under 120 µmol−1 m−2 s−1 light with 16h/8h light/dark conditions. Representative ion chromatogram of SA extracted from (A) Col and (B) StUBA2a/b #3 as TBDMS derivatives separated on a 30 m × 0.25mm internal diameter fused-silica capillary column coated with 0.25 µm CP-SIL 8 CB low bleed. The upper trace was recorded in SIM mode (m/z 309, quantification ion of SA). Internal standard: 3,4,5-trimethoxycinnamic acid. (C) SA contents in the specified genotypes. Asterisk indicates a significant difference at P < 0.05.
Fig. S6. Altered expression of autophagy-associated genes in 35S:StUBA2a/b transgenic plants. RT-PCR was carried out using autophagy-associated gene-specific primers (Supplementary Table S1).
Fig. S7. smGFP-StUBA2s localization was not changed by exogenous ABA application.
Fig. S8. H2O2 accumulation in young or old leaves of 35S:StUBA2a/b transgenic plants.
Fig. S9. Age-dependent leaf senescence marker gene ORE1 is upregulated by the overexpression of StUBA2a/b. # numbers refer to independent T3 lines. (A) RT-PCR analysis of ORE1, SID2, AAO3, and EIN2 genes. (B) qRT-PCR analysis of AAO3, SID2, and ORE1 genes. For both panels, transcript level of Actin2 was used as the reference. Asterisk indicates a significant difference at P < 0.05.
Acknowledgements
This research was supported by NSF grant MCB-0345251 to SMA and a grant to DYK from Research Program for Agricultural Science and Technology Development (PJ011233), Rural Development Administration, Republic of Korea.
References
- Acharya BR, Jeon BW, Zhang W, Assmann SM. 2013. Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytologist 200, 1049–1063. [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C. 2008. Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS One 3, e2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becana M, Moran JF, Iturbe-Ormaetxe I. 1998. Iron-dependent oxygen free radical generation in plants subjected to environmental stress: toxicity and antioxidant protection. Plant and Soil 201, 137–147. [Google Scholar]
- Bove J, Kim CY, Gibson CA, Assmann SM. 2008. Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis . Plant Molecular Biology 67, 71–88. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ. 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis . Plant Journal 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. 2009. Biosynthesis of salicylic acid in plants. Plant Signaling & Behavior 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC. 2005. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant Journal 42, 598–608. [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD. 2007. pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Research 16, 771–781. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. 2004. Systemic acquired resistance. Annual Review of Phytopathology 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataille MP, Avice JC, Masclaux-Daubresse C. 2012. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis . New Phytologist 194, 732–740. [DOI] [PubMed] [Google Scholar]
- Kim CY, Bove J, Assmann SM. 2008. Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytologist 180, 57–70. [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis . Science 323, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim WY, Kwak KJ, Oh SH, Han YS, Kang H. 2010a. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. Journal of Experimental Botany 61, 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Kim JY, Jung HJ, Oh SH, Han YS, Kang H. 2010b. Comparative analysis of Arabidopsis zinc finger-containing glycine-rich RNA-binding proteins during cold adaptation. Plant Physiology and Biochemistry 48, 866–872. [DOI] [PubMed] [Google Scholar]
- Lambermon MH, Fu Y, Wieczorek Kirk DA, Dupasquier M, Filipowicz W, Lorkovic ZJ. 2002. UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Molecular and Cellular Biology 22, 4346–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CK, Gygi SP, Shimazaki K, Assmann SM. 2002. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418, 793–797. [DOI] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM. 2000. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287, 300–303. [DOI] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Hong CB, Kim HJ, Park CM. 2004. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillen P, Baurle I, Swiezewski S, Dean C. 2007. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Molecular Cell 28, 398–407. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2010. TOR is a negative regulator of autophagy in Arabidopsis thaliana . PLoS One 5, e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2012. Autophagy: pathways for self-eating in plant cells. Annual Review of Plant Biology 63, 215–237. [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ. 2009. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends in Plant Science 14, 229–236. [DOI] [PubMed] [Google Scholar]
- Lu H. 2009. Dissection of salicylic acid-mediated defense signaling networks. Plant Signaling & Behavior 4, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, Chory J, Lin C. 2004. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proceedings of the National Academy of Sciences USA 101, 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Ng CK, Kinoshita T, Pandey S, Shimazaki K, Assmann SM. 2004. Abscisic acid induces rapid subnuclear reorganization in guard cells. Plant Physiology 134, 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, Lu H. 2011. Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis . Genetics 189, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. 1996. TreeView: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Park SY, Ha SH, Lim SH, Jung JY, Lee SM, Yeo Y, Kim JK. 2012. Determination of phenolic acids in Korean rice (Oryza sativa L.) cultivars using gas chromatography-time-of-flight mass spectrometry. Food Science and Biotechnology 21, 1141–1148. [Google Scholar]
- Perez-Perez ME, Lemaire SD, Crespo JL. 2012. Reactive oxygen species and autophagy in plants and algae. Plant Physiology 160, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov VD, Van Breusegem F. 2012. Hydrogen peroxide – a central hub for information flow in plant cells. AoB Plants 2012, pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Joe A, Sato M, Nguyen le V, Glazebrook J, Alfano JR, Cohen JD, Katagiri F. 2010. A putative RNA-binding protein positively regulates salicylic acid-mediated immunity in Arabidopsis . Molecular Plant-Microbe Interactions 23, 1573–1583. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS, Gohring J, Barta A. 2012. Localization and dynamics of nuclear speckles in plants. Plant Physiology 158, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M, Redko Y, Leung J. 2006. Arabidopsis RNA binding protein UBA2a relocalizes into nuclear speckles in response to abscisic acid. FEBS Letters 580, 4160–4165. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harbor Perspectives in Biology 3, a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schoning JC, Alfano JR, Staiger D. 2008. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana . Plant Journal 56, 239–250. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy BC, Oelmuller R. 2012. Reactive oxygen species generation and signaling in plants. Plant Signaling & Behavior 7, 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. 2006. Reactive oxygen species in plant cell death. Plant Physiology 141, 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann K, Drechsel G, Bergler J, Subert C, Philippar K, Soll J, Engelmann JC, Engelsdorf T, Voll LM, Hoth S. 2012. Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiology 159, 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth H, Mittelstrass K, Kschieschan S, Bender J, Weigel HJ, Overmyer K, Kangasjarvi J, Sandermann H, Langebartels C. 2002. Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell and Environment 25, 717–726. [Google Scholar]
- Woloshen V, Huang S, Li X. 2011. RNA-binding proteins in plant immunity. Journal of Pathogens 2011, 278697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, et al. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. 2008. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis . Plant Cell 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jia J, Yang M, Yan C, Han Y. 2012. Overexpression of a LAM domain containing RNA-binding protein LARP1c induces precocious leaf senescence in Arabidopsis . Molecules and Cells 34, 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou C. 2012. Signal transduction in leaf senescence. Plant Molecular Biology 82, 539–545. [DOI] [PubMed] [Google Scholar]
- Zhang J, Du X, Wang Q, Chen X, Lv D, Xu K, Qu S, Zhang Z. 2010. Expression of pathogenesis related genes in response to salicylic acid, methyl jasmonate and 1-aminocyclopropane-1-carboxylic acid in Malus hupehensis (Pamp.) Rehd. BMC Research Notes 3, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.