Abstract
In this paper, we propose a novel predictor for the conversion from mild cognitive impairment (MCI) to Alzheimer’s disease (AD). This predictor is based on the shape diffeomorphometry patterns of subcortical and ventricular structures (left and right amygdala, hippocampus, thalamus, caudate, putamen, globus pallidus, and lateral ventricle) of 607 baseline scans from the Alzheimer’s Disease Neuroimaging Initiative database, including a total of 210 healthy control subjects, 222 MCI subjects, and 175 AD subjects. The optimal predictor is obtained via a feature selection procedure applied to all of the 14 sets of shape features via linear discriminant analysis, resulting in a combination of the shape diffeomorphometry patterns of the left hippocampus, the left lateral ventricle, the right thalamus, the right caudate, and the bilateral putamen. Via 10-fold cross-validation, we substantiate our method by successfully differentiating 77.04% (104/135) of the MCI subjects who converted to AD within 36 months and 71.26% (62/87) of the non-converters. To be specific, for the MCI-converters, we are capable of correctly predicting 82.35% (14/17) of subjects converting in 6 months, 77.5% (31/40) of subjects converting in 12 months, 74.07% (20/27) of subjects converting in 18 months, 78.13% (25/32) of subjects converting in 24 months, and 73.68% (14/19) of subject converting in 36 months. Statistically significant correlation maps were observed between the shape diffeomorphometry features of each of the 14 structures, especially the bilateral amygdala, hippocampus, lateral ventricle, and two neuropsychological test scores—the Alzheimer’s Disease Assessment Scale-Cognitive Behavior Section and the Mini-Mental State Examination.
Keywords: Alzheimer’s disease, lateral ventricles, linear discriminant analysis, mild cognitive impairment, prediction, principal component analysis, shape diffeomorphometry, subcortical structures
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that is predominantly diagnosed in people over 65 years of age [1]. It is the most common form of dementia, which is characterized by trouble with thinking and language as well as a loss of long-term memory. The cause and the mechanism of progression for most AD cases are still unknown. AD worsens during its progression until death and currently there are no treatments that can cure or reverse this progression. Definite diagnosis of AD can only be made based on histopathologic evidence obtained from a biopsy or autopsy [2]. Mild cognitive impairment (MCI) is a syndrome that is regarded as a risk state for dementia [3] and is associated with an increased risk of progression to probable AD [4]. More than half of the individuals with MCI deteriorate to dementia within 5 years [3] at a rate of about 10% to 15% per year. Considerable heterogeneity exists among MCI patients: some convert to AD with varying progression rates, and others remain stable for a long period of time or even revert to normal cognitive status. In this article, the former is named MCI converters (MCI-C) and the latter is named MCI non-converters (MCI-NC). Specifically, in this study, the MCI-C subjects converted to the AD status within 36 months from their baseline while the MCI-NC did not.
The ability to identify an MCI patient’s risk of developing AD is important for clinical decision making and timing therapy. Structural neuroimaging measures have been shown to be sensitive to the degeneration that occurs in MCI and AD [5], which may provide robust biomarkers for predicting the conversion from MCI to AD. Methods of detecting MCI that represents prodromal AD would aid clinical practice by allowing attention to be focused on those with the highest risk of conversion. However, accurate prediction of the MCI-to-AD conversion is very challenging [6], especially when utilizing only the baseline information. This difficulty is due to the “lag” between brain atrophy and cognitive decline. The MCI-to-AD prediction can be regarded as a classification problem between the MCI-C and the MCI-NC. During the last decade, there have been many methods developed, using structural imaging, to differentiate between MCI-C and MCI-NC [6–20]. A majority of the studies focusing on the prediction of MCI-to-AD conversion were based on the information from a single scan, the baseline dataset. Recently, longitudinal structural features have been proposed as better biomarkers for the prediction of MCI conversion. For a more comprehensive literature survey of previous work on predicting the MCI-to-AD conversion using structural imaging biomarkers, we refer the reader to [15].
There are typically three different types of structural MCI-to-AD predictors. The most widely used one is the cortical thickness [6,7,13,15,17–20] because this measurement is very sensitive to small structural changes in the cortex. The second measurement is the volume of specific structure of interest, such as the hippocampus [6,14,19] and the amygdala [14]. The third type of predictors is the biomarkers obtained from whole brain based analyses. To be specific, methods such as the voxel-based morphometry analysis have been suggested to locate the most discriminative regions that are then taken as the biomarkers for the MCI-to-AD prediction [6,8,10–12,15].
In recent years, the single-scan based shape diffeomorphometry patterns of subcortical and ventricular structures have been implicated to provide important anatomical information in characterizing the difference between healthy controls (HC) and MCI as well as AD [21–28]. In diffeomorphometry, one utilizes the diffeomorphisms stemming from a fixed coordinate system (the template space) to study the shape morphometrics [29] of the target groups rather than the shapes themselves. In our previous study [28], we have demonstrated the diffeomorphometric abnormalities detected in MCI and AD populations, when compared with HC, of the fourteen subcortical and ventricular structures: the left and right amygdala, hippocampus, thalamus, caudate, putamen, globus pallidus, and lateral ventricle. In a continuation of the previous work, we herein propose a method for the prediction of MCI-to-AD conversion using the diffeomorphometry-based shape features of those same fourteen structures in the baseline data consisting of 210 HC subjects, 175 AD subjects, and 222 MCI subjects, among which 87 MCI subjects did not deteriorate to AD within 36 months from baseline.
The shape features are obtained in the framework of large deformation diffeomorphic metric mapping (LDDMM) [30], in which a diffeomorphism is computed as the end point of an energy-minimizing path (a geodesic) through the group of diffeomorphisms. Given a fixed template, the anatomical variability in the targets is encoded by the geodesics from the template to each target. The fundamental “conservation of momentum” property of these geodesics [31] allows for representing the entire flow of a geodesic with the initial momentum configuration. This means that, once a template is fixed, the space of initial momenta becomes an appropriate linear vector space [32–34] for studying shape. Anatomical patterns that are specific to MCI and AD can therefore be studied by applying linear statistical techniques, such as principal component analysis (PCA), to the initial momentum vectors, which was already demonstrated in [26,28].
In this experiment, the primary training process, including feature extraction and dimensionality reduction, was performed on the baseline HC and AD subjects while the optimization over possible structure combinations utilized the addition of the baseline MCI subjects. The testing process was carried out on the baseline MCI subjects. Such a design comes from the observation that the differences, in terms of the shape diffeomorphometry patterns, between MCI-C and MCI-NC are similar to those detected between AD and HC, which has been shown in [28]. It has also been reported that other patterns of change, within the brain, of MCI-C are similar to those of AD while those of MCI-NC are similar to HC [35]. Predicting the conversion from MCI to AD using classifiers built from HC-versus-AD information has also been successfully demonstrated in [35–38].
In this article, we first describe the procedure for generating the possible HC-versus-AD linear discriminant analysis (LDA) classifiers to be used in the prediction of MCI-to-AD conversion, which are built from the diffeomorphometry-based shape features of subsets of the fourteen subcortical and ventricular structures from 210 HC subjects and 175 AD subjects, all of which come from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) baseline dataset. We then demonstrate the optimization procedure over possible combinations of structure-specific shape features by utilizing data from 222 MCI subjects in a 10-fold cross-validation fashion, thus yielding 10 optimal LDA MCI-to-AD predictors. After that, we cross-validate the predictors on the 222 MCI subjects, including 135 MCI-C subjects and 87 MCI-NC subjects, the conversion of which is determined by a follow-up of 36 months. At the end of this paper, we evaluate the correlation between the vertex-based shape diffeomorphometry patterns of the fourteen structures and two neuropsychological test scores for the Alzheimer dementia – the Alzheimer’s Disease Assessment Scale-Cognitive Behavior Section (ADAS-cog) [39] and the Mini-Mental State Examination (MMSE) [40].
MATERIALS AND METHODS
Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu/). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal aging individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see http://www.adni-info.org/. In this study, we included data from 210 HC subjects, 222 subjects with MCI, and 175 subjects with AD. Within the MCI group, 135 subjects converted to AD (MCI-C) within a follow-up of 36 months. The conversion time within the MCI-C group is heterogeneous since an MCI patient may convert at any time over the course of 6 months to 3 years. In the case of this study, 17 subjects converted in 6 months, 40 in 12 months, 27 in 18 months, 34 in 24 months, and the remaining 17 in 36 months. For the group MCI-NC, we only included those MCI subjects that were followed for at least 3 years, yielding a total of 87 subjects in MCI-NC. Clinical and demographic data for the three groups (HC, MCI, and AD) are presented in Table 1. In brief, subjects are 55–92 years old, and are not depressed. The control subjects have MMSE scores of 25–30 and a Clinical Dementia Rating (CDR) score of 0. The subjects with MCI have MMSE scores of 23–30, a CDR of 0.5, preserved ability to perform daily living activities, and an absence of dementia. The subjects with AD have MMSE scores of 20–28, a CDR of 0.5 or 1.0, and meet the criteria for probable AD. The three groups did not differ significantly in terms of age (F = 2.53, p = 0.081). All groups differed on MMSE and CDR-sum of boxes (CDR-SB) as expected based on diagnostic criteria (all p < 0.001). Table 2 lists the demographics of the two sub-groups of MCI: MCI-C and MCI-NC. According to Student’s t-tests, the two MCI sub-groups did not differ significantly in age (p = 0.734), but had statistically significant difference in terms of both MMSE (p = 5.262e−5) and CDR-SB (p = 2.364e−4).
Table 1.
Demographic data for the three groups: Healthy controls (HC), mild cognitive impairment (MCI), and Alzheimer's disease (AD). The statistics for age, Mini-Mental State Examination (MMSE), and Clinical Dementia Rating-sum of boxes (CDR-SB) are displayed as mean ± SD
| Parameter | HC Group (n = 210) |
MCI Group (n = 222) |
AD Group (n = 175) |
|---|---|---|---|
| Subject age (y) | 76.25 ± 5.01 | 74.73 ± 7.55 | 75.28 ± 7.49 |
| No. of male subjects | 109 | 150 | 94 |
| MMSE score | 29.12 ± 1.02 | 27.57 ± 1.76 | 23.43 ± 2.01 |
| CDR-SB score | 0.03 ± 0.12 | 1.3 ± 0.60 | 4.23 ± 1.64 |
HC, healthy controls; MCI, mild cognitive impairment; AD, Alzheimer's disease; MMSE, Mini-Mental State Examination; CDR-SB, Clinical Dementia Rating-sum of boxes.
Table 2.
Demographic data for the two subgroups of MCI: MCI-C and MCI- NC. The statistics for age, MMSE, and CDR-SB are displayed as mean ± SD
| Parameter | MCI-C (135) | MCI-NC (87) |
|---|---|---|
| Subject age (y) | 74.39 ± 7.04 | 74.73 ± 7.57 |
| No. of male subjects | 81 | 69 |
| MMSE score | 26.68 ± 1.72 | 27.57 ± 1.76 |
| CDR-SB score | 1.84 ± 0.96 | 1.30 ± 0.60 |
MCI, mild cognitive impairment; MCI-C, MCI converters; MCI-NC, MCI non-converters; MMSE, Mini-Mental State Examination; CDR-SB, Clinical Dementia Rating-sum of boxes.
Image protocol and volumetric segmentation
The raw MR scans, in the format of DICOM, were downloaded from the public ADNI website (http://adni.loni.usc.edu/data-samples/mri/). Locally, the raw MR data were automatically corrected for spatial distortion due to gradient nonlinearity [41] and B1 field inhomogeneity [42]. Each subject was scanned twice with a 3D MPRAGE protocol at 1.5 Tesla. The two T1-weighted images were rigid-body aligned to each other, averaged to improve the signal-to-noise ratio, and then resampled to isotropic 1-mm voxels. Description of the rigid-body alignment algorithm is detailed in [43], wherein it served as a first step of a longitudinal registration pipeline. Averaging the two scans of the same subject allowed an increase in the signal-to-noise ratio. Volumetric segmentations of the fourteen subcortical and ventricular structures were obtained using FreeSurfer [44]. Based on the transformation of the full brain mask into atlas space, the total cranial vault value was estimated from the atlas scaling factor [45] to control individual differences in head size. The quality of the automated volumetric segmentations has been reviewed by technicians who have been trained by an expert neuroanatomist with more than 10 years of experience. Images that have been degraded due to motion artifacts, technical problems (a change in scanner model or change in the radiofrequency coil during the time-series), or significant clinical abnormalities (e.g., hemispheric infarction) were excluded from our analysis.
Baseline shape processing
The processing of the baseline shapes of all the fourteen structures for each individual subject was detailed in [28], for which we will briefly summarize the steps here. For each structure, the 2-D surface that contours the boundary of the 3-D volume segmentation was approximated by a diffeomorphically deformed atlas surface to ensure correct topology and smoothness. The deformation was created by aligning the atlas segmentations (manually labeled) and the subject segmentations (from FreeSurfer) using a multi-channel LDDMM-image mapping. More details and validation of this surface-generation methodology can be found in [28,46].
To obtain the baseline shape diffeomorphometry features associated with the surface of each structure for each individual subject, we created a common template surface for each structure using the algorithm proposed and validated in [47]. Briefly, for each single structure, every subject surface was first rigidly aligned (rotation and translation) to a common spatial position. The rigid registration algorithm computes an optimal transformation between the vertex sets of two surfaces by minimizing a score which combines registration and soft assignment. After rigid registration, each observed subject surface is modeled as a random deformation of a hidden template plus additive Gaussian noise. Given this model, the template is estimated from the subject surfaces using an approximation of the expectation-maximization algorithm, subject to some topology constraints. It is enacted by ensuring that the hidden template surface is a diffeomorphic deformation of a reference shape, called the hyper-template.
The LDDMM-surface mapping algorithm [48] was then employed to align the common template surface to each subject surface, creating the deformation associated with each structure surface of each subject. The readers are referred to [28] for more details about the mathematical foundations and the experimental steps of this procedure.
Generation of the baseline shape diffeomorphometry features
For each single structure, we generate its baseline shape diffeomorphometry features separately as follows. From each LDDMM-surface mapping, we obtain initial momentum vectors α0 that are defined at all vertices of the template surface. The initial momentum vectors characterize the shape variations in the subject surface relative to the template surface. Similar to the “conservation of momentum” concept in physics, given the vertices on the template surface and the initial momentum vectors α0, the evolution of the entire diffeomorphic flow is uniquely determined [31]. The initial momentum vectors α0 encode the geodesic connecting the template surface to the subject surface. Therefore, we selected the initial momentum vectors α0 to be our features for prediction. These vectors form an extremely high dimensional space and thus, for each structure, we performed PCA on the initial momentum vectors of all the HC and the AD subjects to construct an orthonormal basis. The initial momentum vectors of all the MCI subjects were projected onto that PCA basis. It is worth noting that, in our context, the inner product used in the PCA is derived from the Riemannian metric that leads to the geodesic equation in LDDMM. Details of PCA on the initial momentum space can be found in [32–34]. To reduce the dimension of the feature space, we retained only the first M principal components (PCs) that account for 95% of the total variance. We then selected only those PCs that show significant group difference between HC and AD. To make this selection, for each PC, we performed a Student’s t-test between the PC coefficients for HC and those for AD, and selected the PCs with a p-value less than 0.05. We also retained the corresponding PC coefficients for all the MCI test subjects. This established our training and testing features for cross-validation.
Selecting the optimal baseline classifier and validation
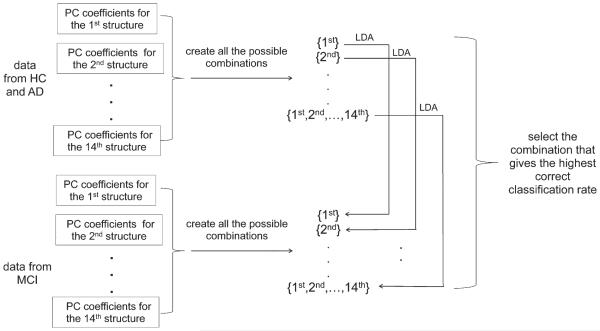
After performing PCA on the initial momentum space and an ensuing selection of PC coefficients, we had feature matrices for fourteen structures. In each feature matrix, every row contains the PC coefficients for a different subject while each column contains the coefficients for all of the projections onto a specific PC basis vector. As we have shown in [28], it is plausible that the combination of features from a subset of structures is the most discriminating. To find the optimal combination, we tested all of the possible classifiers we could build from the PCs of the fourteen structures. We used linear discriminant analysis (LDA) to construct our classifiers. Considering each possible combination, we built a total of 16383 (214–1) different classifiers and compared their classification performance with each other. The procedure of sifting the optimal LDA classifier for the MCI-to-AD prediction is demonstrated in Fig. 1.
Fig. 1.
Flowchart demonstrating the procedure of selecting the optimal linear discriminant analysis (LDA) classifier in classifying MCI-C from MCI-NC. PC, principal component.
To estimate the accuracy of our predictor in classifying MCI-C versus MCI-NC, we adopted 10-fold cross-validation; about 22 MCI subjects (8 from MCI-NC and 14 from MCI-C) were excluded before we selected the optimal LDA classifier. To be specific, we excluded those 22 MCI subjects at the very beginning and then sifted the optimal LDA classifier based on the feature information from the other 200 MCI subjects (called “data from MCI” in Fig. 1) and the 210 HC and 175 AD subjects (the “data from HC and AD” in Fig. 1), as described in the previous section. We then used that sifted optimal LDA classifier to determine the subgrouping of the MCI subjects excluded at the beginning. The 10-fold cross-validation procedure is demonstrated in Fig. 2. It is important to notice that we would obtain a unique optimal classifier for each of the 10-fold tests. Since the test subjects were removed from both the initial PCA and the process of selecting the optimal classifier, we avoid any bias or overestimation of the accuracy of the predictor in forecasting the conversion from MCI to AD.
Fig. 2.
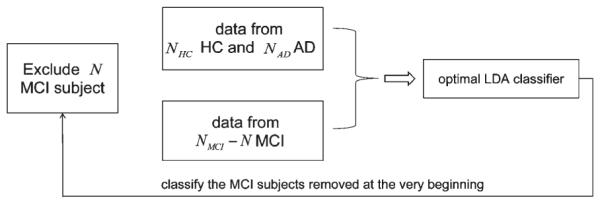
Flowchart demonstrating the procedure of performing 10-fold cross-validation without being biased by the subjects removed at the beginning. This process is repeated for the N MCI subjects in each fold. NHC indicates the number of HC subjects, NMCI indicates the number of MCI subjects, and NAD indicates the number of AD subjects.
Comparison with the hippocampal volume based biomarker
The hippocampal volume has been suggested to play a critical role in predicting the MCI-to-AD conversion [6,14,19]. We therefore treat it as a benchmark for evaluating our shape diffeomorphometry based MCI-to-AD predictor. We compared the prediction accuracy from our shape diffeomorphometry informatics with that from the hippocampal volumes. Following the work presented in [19] and [6], we combined the volumes of the left and right hippocampus together. Furthermore, we did not use the HC and AD hippocampal volumes as the training features. Instead, we performed 10-fold cross-validation with respect to the MCI hippocampal volumes. To be consistent with our shape-based procedure and the two aforementioned works, we again used LDA as the classification technique. It is noticeable that in [19], the hippocampal segmentations were not obtained from FreeSurfer but from the authors’ own approach.
Correlating the shape diffeomorphometrics and neuropsychological test scores
Imaging based biomarkers would not be useful in clinical trials if they could not be significantly associated with cognitive deterioration. In this study, we evaluated the correlation between the shape diffeomorphometrics of each structure and two neuropsychological test scores (ADAS-cog and MMSE). ADAS-cog measures a number of cognitive domains, including components of memory, language, and praxis. This scale is scored from 0 to 70 with higher values indicating greater cognitive impairment. MMSE provides a continuous scale to assess primary cognitive functions that affect the dementia of the Alzheimer type, including orientation, registration, attention, recall, language, and constructional praxis [40]. The MMSE score ranges from 0 to 30. In contrast to ADAS-cog, lower MMSE scores indicate more severe cognitive impairment.
For the correlation analysis, instead of the initial momentum vectors α0, we used a scalar field—the log-determinant of the Jacobian matrix of the diffeomorphism from the LDDMM-surface mapping. The log-determinant of the Jacobian is defined at each vertex of the template surface, quantifying the factor by which the diffeomorphism expands or shrinks the surface area at each vertex [28]. We calculated the Pearson product-moment correlation coefficients (PCCs) between the scalar field at each vertex and the two cognitive test scores, co-varying for the age, gender, and the estimated intracranial volume. Statistical significance of the correlation at each vertex is measured by a p-value obtained from non-parametric permutation tests, in which a total of 40,000 permutations were performed. Since multiple correlation tests were performed simultaneously at all vertices of the template surface, for each structure, we corrected for multiple comparison by adjusting the p-values in a way that controls the familywise error rate (FWER) at a level of 0.05 based on the “maximum statistic” method described in [49].
RESULTS
The PCA procedure resulted in 48 PCs for the left hippocampus, 49 PCs for the right hippocampus, 21 PCs for the left amygdala, 24 PCs for the right amygdala, 44 PCs for the left lateral ventricle, 39 PCs for the right lateral ventricle, 61 PCs for the left thalamus, 60 PCs for the right thalamus, 45 PCs for the left caudate, 45 PCs for the right caudate, 58 PCs for the left putamen, 59 PCs for the right putamen, 30 PCs for the left globus pallidus, and 29 PCs for the right globus pallidus. After a further reduction via the Student’s t-tests between HC and AD, we retained 9 PCs for the left hippocampus, 11 PCs for the right hippocampus, 5 PCs for the left amygdala, 6 PCs for the right amygdala, 16 PCs for the left lateral ventricle, 15 PCs for the right lateral ventricle, 8 PCs for the left thalamus, 16 PCs for the right thalamus, 5 PCs for the left caudate, 7 PCs for the right caudate, 9 PCs for the left putamen, 15 PCs for the right putamen, 6 PCs for the left globus pallidus, and 7 PCs for the right globus pallidus.
From the 10-fold cross-validation, we obtained 10 optimal LDA classifiers. For 7 out of 10, the optimal LDA classifiers came from the same combination of structures: the left hippocampus, the left lateral ventricle, the right thalamus, the right caudate, and the bilateral putamen. For 2 out of 10, the optimal combination came from four structures: the left hippocampus, the left lateral ventricle, the right thalamus, and the right caudate. For the remaining classifier, the optimal combination came from six structures: the bilateral hippocampus, the left amygdala, the right thalamus, the right caudate, and the left globus pallidus. Applying the optimal LDA selection procedure to the complete MCI sample (all 222 MCI subject), we obtained a combination of the left hippocampus, the left lateral ventricle, the right thalamus, the right caudate, and the bilateral putamen. Based on this experimental result, to predict the conversion for future MCI scans using the shape diffeomorphometry patterns built from the HC and AD population, we consider the combination of those six structures to be the optimal LDA predictor.
According to the 10-fold cross-validation results, we are capable of achieving a total classification accuracy of 74.77%, a sensitivity of 77.04% (classifying MCI-C) and a specificity of 71.26% (classifying MCI-NC). The area under the receiving operating characteristic curve (AUC) is found to be 73.81%. The accuracy of our approach in predicting the outcome for the MCI-C subjects varies depending on the subject’s conversion time. To be specific, we correctly predicted 82.35% (14/17) of the MCI subjects converting in 6 months, 77.5% (31/40) of the MCI subjects converting in 12 months, 74.07% (20/27) of the MCI subjects converting in 18 months, 78.13% (25/32) of the MCI subjects converting in 24 months, and 73.68% (14/19) of the MCI subjects converting in 36 months. Generally, the closer the MCI subject is about to convert, the more accurately the classifier can predict that conversion.
Using the hippocampal volume to predict the MCI-to-AD conversion for our MCI dataset, we successfully predicted the outcome for 57 out of 87 MCI-NC subjects and 83 out of 135 MCI-C subjects, yielding a specificity of 65.52% and a sensitivity of 61.48%. For the MCI-C subjects, the hippocampal volume predicted 58.82% (10/17) of the MCI subjects converting in 6 months, 62.5% (25/40) of the MCI subjects converting in 12 months, 62.96% (17/27) of the MCI subjects converting in 18 months, 68.75% (22/32) of the MCI subjects converting in 24 months, and 47.37% (9/19) of the MCI subjects converting in 36 months. A comparison of the prediction accuracy given by using our shape diffeomorphometry patterns and the hippocampal volumes for the dataset in this study as well as the prediction results reported in [19] and [6] are listed in Table 3. Compared with the hippocampal volume biomarker, our shape diffeomorphometry biomarker boosted the overall prediction accuracy by 12%. Comparing the prediction accuracy from the hippocampal volume biomarker applied to three different subsets of ADNI dataset (the first three rows in Table 3), we observed that all three prediction results are in the same range with only slight differences. These differences may have been caused by variations in the MCI dataset used in each study, especially the MCI-C subjects; [6] only examined MCI-Cs with conversion periods of up to 18 months which are generally considered as relatively easy cases for prediction. Also the hippocampal volumes examined in [19] were not obtained from FreeSurfer but from the authors’ own segmentation approach. This difference in the hippocampal definition may have also contributed to the slight differences in the prediction accuracy of the three works.
Table 3.
Comparisons of the shape diffeomorphometry and the hippocampal volume as biomarkers for the prediction of MCI to AD
| Study | Features | Conversion period | N (MCI-NC, MCI-C) | Acc | Sen | Spe |
|---|---|---|---|---|---|---|
| Cuingnet et al. [6] | Hippocampal volume | 0-18 months | 134, 76 | 0.67 | 0.62 | 0.69 |
| Wolz et al. [19] | Hippocampal volume | 0-48 months | 238, 167 | 0.65 | 0.63 | 0.67 |
| Proposed | Hippocampal volume | 0-36 months | 87, 135 | 0.63 | 0.66 | 0.61 |
| Shape diffeomorphometry | 0-36 months | 87, 135 | 0.75 | 0.77 | 0.71 |
MCI, mild cognitive impairment; MCI-C, MCI converters; MCI-NC, MCI non-oonverters; Acc, accuracy, Sen, sensitivity, Spe, specificity.
For each of the 14 subcortical and ventricular structures, the vertex-based correlation maps between the two neuropsychological test scores (ADAS-cog and MMSE) and the shape diffeomorphometry patterns, as quantified by the log-determinant of the Jacobian matrix, are demonstrated in Fig. 3. In that figure, PCCs are assigned non-zero values only for vertices where the shape diffeomorphometry correlated statistically significantly with the corresponding score after performing multiple comparison correction by controlling the FWER at a level of 0.05.
Fig. 3.
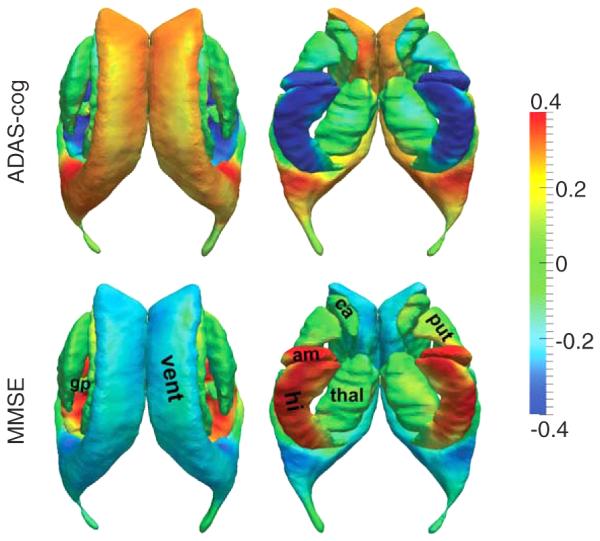
Statistically significant correlation maps between the vertex-based shape diffeomorphometrics of the 14 subcortical and ventricular structures and the values of ADAS-cog (top panel) and MMSE (bottom panel). Colors represent the Pearson product-moment correlation coefficients. For each correlation map, two views are presented. vent, lateral ventricle; am, amygdala; hi, hippocampus; ca, caudate; thal, thalamus; put, putamen; gp, globus pallidus.
As shown in Fig. 3, for a majority of vertices on each structure surface, especially the surfaces of the bilateral hippocampus, amygdala, and lateral ventricle, significant correlations were observed between the shape diffeomorphometrics and the values of ADAS-cog and MMSE. The shape diffeomorphometrics of the bilateral hippocampus and amygdala were mostly negatively correlated with the ADAS-cog value and positively correlated with the MMSE value. The opposite trend was observed for the lateral ventricle; the shape diffeomorphometrics of the bilateral ventricles were positively correlated with the ADAS-cog value and negatively correlated with the MMSE value. This observation implies that the atrophy of a majority of regions on the hippocampus and the amygdala, as well as the ventricular expansion, is indicative of the cognitive deterioration that occurs in the progression toward AD. Inhomogeneous correlation maps were observed across the surface of the basal ganglia structures and the thalamus; some vertices were positively correlated with the ADAS-cog score while others were negatively correlated. This observation may explain why, globally, the basal ganglia and the thalamus are not significantly different in the MCI and AD population when compared to normally aging subjects in general. We also notice that the spatial maps of the ADAS-cog and MMSE correlations are highly consistent with each other, revealing vertex-wise reverse correlation trends on the template surfaces.
DISCUSSION
In this paper, we proposed a new structural imaging based biomarker for the prediction of conversion from MCI to AD. The features were extracted from the shape diffeomorphometry patterns of fourteen subcortical and ventricular structures (the left and right amygdala, hippocampus, thalamus, caudate, putamen, globus pallidus, and lateral ventricle) of baseline HC and AD subjects from the ADNI database. Shape diffeomorphometry patterns offer new biomarkers in addition to the more conventional structural biomarkers: cortical volumes [6,14,18–20,50] and cortical thickness [6,7,17–20,50]. So far, there have been very few studies exploring the possibility of predicting MCI-to-AD conversion via the shape diffeomorphometry patterns in those fourteen subcortical and ventricular structures. That said, the shapes of subcortical and ventricle structures have been shown to be affected by the dementia of Alzheimer type [9,21–26,28,51,52], which may suggest discriminant features for the prediction of MCI-to-AD. The application of diffeomorphometry patterns to the classification of AD from HC has already been successfully demonstrated in several other studies [11,26,27] and one can see the possibility for this given the fact that, for a fixed template, the evolution of the target is completely determined by the deformation. In the same sense, the diffeomorphisms stemming from a single template to different targets can be used to characterize the morphometrics in those targets. Statistical inference based on the metric, the geodesic length of the flows of diffeomorphisms connecting human biological shapes, enables machine learning of the statistical representation of shape [53]. Indeed, we have found that the shape diffeomorphometrics of the 14 structures, especially the amygdala-hippocampus memory circuit and the adjacent lateral ventricles, are statistically significantly correlated with two neuropsychological test scores—ADAS-cog and MMSE (Fig. 3). This clearly suggests that the shape diffeomorphometrics can be indicative of the cognitive impairment in the pathology of AD.
In recent years, compared to single-structure approaches, multi-structure based diffeomorphometry has become widely employed in application to shape abnormality detection in various disorders [28,54,55]. In this study, we developed and validated a shape diffeomorphometry based biomarker from multiple subcortical and ventricular structures to predict the conversion from MCI to AD. To the best of our knowledge, this is the most detailed application of machine learning to diffeomorphometry markers from multiple structures and the first one applied to the MCI-to-AD prediction. Instead of using the shape patterns from all of the fourteen structures, we developed an automated feature selection procedure, as demonstrated in Fig. 1, to select the optimal subset for predicting the MCI-to-AD conversion. In our experiment, the optimal LDA predictor arises from a combination of the shape diffeomorphometry patterns of a subset containing six of the original structures: the left hippocampus, the left lateral ventricle, the right thalamus, the right caudate, and the left and right putamen. In reaching this conclusion, all the shape diffeomorphometry patterns were obtained from the HC and AD baseline datasets while the MCI baseline dataset was used in the optimization procedure to determine which combination yields the highest prediction accuracy. According to a 10-fold cross-validation, our pipeline achieved a classification accuracy of 74.77%, a sensitivity of 77.04%, a specificity of 71.26% and an AUC of 73.81%, which are superior or comparable to results reported in other classification methods (Tables 4 and 5).
Table 4.
A comparison of the success rates of various methods in predicting the conversion of MCI-C patients grouped by conversion times. The total number of MCI-C subjects used in each study is given in parenthesis
| m06 | ml2 | ml8 | m24 | m36 | m48 | |
|---|---|---|---|---|---|---|
| Eskildsen et al. [15] | ||||||
| cortical thickness | 78.7% (122) | 75.2% (128) | - | 69.4% (61) | 69% (29) | - |
| cortical thickness+age | 77.1% (122) | 76.6% (128) | - | 70.5% (61) | 72.4% (29) | - |
| Zhang et al. [12] | 50% (2) | 100% (9) | 87.5 (8) | 80% (10) | 50% (6) | 66.67% (3) |
| Proposed method | 82.4% (17) | 77.5% (40) | 74.07% (27) | 78.1% (32) | 73.7% (19) | - |
MCI-C, mild cognitive impairment converters.
Table 5.
A comparison of the proposed method with previous work on predicting conversion from MCI to AD. Some results are directly obtained from the ones summarized in [15]
| Study | Structures | Features | Conversion period | N (MCI-NC, MCI-C) | Acc | Sen | Spe | AUC |
|---|---|---|---|---|---|---|---|---|
| Zhang et al. [12] | Whole brain | VBM+PET | 0-48 months | 50, 38 | 0.78 | 0.79 | 0.78 | 0.77 |
| Wee et al. [20] | Cortical | Thickness+Volume | 0-36 months | 111, 89 | 0.75 | 0.63 | 0.84 | 0.84 |
| Cho et al. [7] | Cortex | Thickness | 0-18 months | 131, 72 | 0.71 | 0.63 | 0.76 | NA |
| Chupin et al. [14] | Hippocampus and amygdala |
Volume | 0-18 months | 134, 76 | 0.64 | 0.6 | 0.65 | NA |
| Cuingnet et al. [6] | Hippocampus | Atlas based | 0-18 months | 134, 76 | 0.67 | 0.62 | 0.69 | NA |
| - | Whole brain | VBM (grey matter) | - | 0.71 | 0.57 | 0.78 | NA | |
| - | Cortex | Cortical thickness | - | - | 0.7 | 0.32 | 0.91 | NA |
| Davatzikos et al. [8] | Whole brain | VBM | 0-36 months | 170, 69 | 0.56 | 0.95 | 0.38 | 0.73 |
| Koikkalainen et al. [16] | Whole brain | TBM, combination of classifiers |
0-36 months | 215, 154 | 0.72 | 0.77 | 0.71 | NA |
| Misra et al. [10] | Whole brain | VBM, ROIs | 0-36 months | 76,27 | 0.82 | - | - | 0.77 |
| Querbes et al. [17] | Cortex | Cortical thickness | 0-24 months | 50,72 | 0.73 | 0.75 | 0.69 | NA |
| Westman et al. [18] | Cortical and subcortical |
Thickness and volume | 0-12 months | 256, 62 | 0.58 | 0.74 | 0.56 | NA |
| Wolz etal. [19] | Hippocampus | Atlas based | 0-48 months | 238, 167 | 0.65 | 0.63 | 0.67 | NA |
| - | Whole brain | TBM | - | 0.64 | 0.65 | 0.62 | NA | |
| - | Whole brain | Manifold learning | - | 0.65 | 0.64 | 0.66 | NA | |
| - | Cortex | Cortical thickness | - | - | 0.56 | 0.63 | 0.45 | NA |
| - | Combination | Combination | - | - | 0.68 | 0.67 | 0.69 | NA |
| Liu et al. [50] | Hippocampus, amygdala, caudate |
Volume | 0-12 months | 79,21 | 0.69 | 0.76 | 0.68 | NA |
| - | Inferior temporal and lateral orbitofrontal |
Thickness | - | - | 0.59 | 0.62 | 0.59 | NA |
| Singh et al. [11] | Whole brain | Volume deformation Pattern |
0-48 months | 73,54 | 0.66 | 0.65 | 0.67 | 0.72 |
| Leung et al. [9] | Hippocampus | Shape atrophy rates | 0-12 months | 128, 86 | NA | NA | NA | 0.67 |
| Proposed method | Subcortical | Shape diffeomorphometry |
0-36 months | 87, 135 | 0.75 | 0.77 | 0.71 | 0.74 |
| structure and laterary ventricle |
Acc, accuracy; Sen, sensitivity; Spe, specificity; AUC, area under the receiver operating characteristic curve; MCI-C, MCI converters; MCI-NC, MCI non-converters; VBM, voxel-based morphometry; PET, positron emission tomography; NA, not available; TBM, tensor-based morphometry; ROI, region of interest.
The performance of the LDA classifier, in the prediction of MCI-to-AD conversion, is affected by the conversion times of the MCI-C subjects (82.35% for MCI subjects converting in 6 months, 77.5% for MCI subjects converting in 12 months, 74.07% for MCI subjects converting in 18 months, 78.13% for MCI subjects converting in 24 months, and 73.68% for MCI subjects converting in 36 months). As expected, the accuracy for the prediction of MCI-C subjects that converted within 6 months after the baseline is the highest while the accuracy for those who had not converted until 36 months later from baseline is the lowest. This suggests that it is relatively easier to predict the conversion of MCI subjects who will convert sooner since their patterns have more similarities with those of AD subjects.
In a majority of existing studies on the prediction of MCI-to-AD conversion, we find a lack of details on the variability of prediction accuracy with respect to the conversion times of the MCI-C subjects. This is largely due to the fact that many studies focused entirely on the MCI-C subjects with a follow-up time of no longer than 18 months or had a relatively small number of subjects with longer conversion times. In our study, we presented the prediction results for MCI-C subjects with a variety of conversion times, and compared them with two recently published works that proceeded similarly [12,15]. The comparison is shown in Table 4. Compared with the results reported in [15], our approach is capable of achieving a higher prediction accuracy at each conversion time. However, more subjects were examined in [15]. The small number of MCI-C subjects investigated in [12] at each conversion time (2 MCI subjects that converted within 6 months, 9 MCI subjects that converted with 12 months, 8 MCI subjects that converted within 18 months, 10 MCI subjects that converted within 24 months, 6 MCI subjects that converted within 36 months, and 3 MCI subjects that converted within 48 months) makes it difficult to compare with our proposed method, considering that at least twice as many MCI-C subjects were included in our study at each conversion time. These small numbers of testing subjects are explained by the use of longitudinal and multimodality data in the classification in [12], which largely restricted the number of available subjects. A comprehensive comparison between the proposed methods and recent work on MCI-to-AD predictors is summarized in Table 5.
According to the comparison results illustrated in Table 3 and results from previous publications [6,19], the structural definitions can make a difference to the prediction accuracy; in this case, the hippocampal volumes produced by different segmentation methods vary from each other in terms of the MCI-to-AD prediction accuracy. This observation motivates us in future to adopt more advanced segmentation methods for producing more precise subcortical and ventricular structures [56], which may potentially enhance the prediction power of our shape diffeomorphometry biomarker. Another improvement may be found in looking to the class of structures we analyze. Currently, many studies are focusing on extracting features from whole brain regions [6,8,10,11,15,16,19]. It is thus natural to consider extending the proposed framework to incorporate the shape diffeomorphometry patterns from regions other than the fourteen structures analyzed herein.
In this study, the shape PCs were computed for each structure separately and then combined directly. This may potentially ignore important information since different structures may be correlated. For example, the expansion of the lateral ventricle is directly related with the compression of the hippocampus and the amygdala, which should be taken into consideration when combining the features from those three structures. Future work should focus on taking the correlation between the shape features from different regions into consideration when designing the feature matrix, similar to the work done in [20]. In addition, instead of directly grouping the shape features from different structures and then performing a single LDA, the technique of multi-kernel learning, such as those demonstrated in [57,58], may improve the results. It is likely that the clearest avenue for further exploration in this work lies in our utilization of only the baseline information. In the future, an important extension will be to incorporate longitudinal information [10,12,13,57] into the prediction procedure.
In conclusion, we have developed and validated a novel structural imaging based biomarker, for the prediction of MCI-to-AD conversion, using patterns of shape diffeomorphometry in the framework of LDDMM. These patterns were extracted from a subset of subcortical structures and the lateral ventricles, based on a training dataset of 210 HC and 175 AD subjects. The optimal combination of structure-specific shape diffeomorphometry patterns was determined using 222 MCI subjects. The procedure was validated on a total of 135 progressive MCI subjects and 87 MCI subjects who remained stable after a follow-up of 3 years. The prediction accuracy on each subgroup of MCI-C, grouped according to the conversion time, was demonstrated to be superior or comparable to previously reported results.
ACKNOWLEDGMENTS
This work is partially supported by NIH R01 EB000975, NIH P41 RR15241, and R01 EB008171.
Anders M. Dale is a founder and holds equity in CorTechs Labs, Inc, and also serves on its Scientific Advisory Board. Michael I. Miller owns an equal share in Anatomyworks LLC. The terms of this arrangement have been reviewed and approved by the Johns Hopkins University as well as the University of California, San Diego in accordance with their conflict of interest policies.
Authors’ disclosures available online (http://www.jalz.com/disclosures/view.php?id=2551).
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI Acknowledgement_List.pdf
REFERENCES
- [1].Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88:1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group∗ under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–939. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- [3].Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K, Belleville S, Brodaty H, Bennett D, Chertkow H, Cummings JL, de Leon M, Feldman H, Ganguli M, Hampel H, Scheltens P, Tierney MC, Whitehouse P, Winblad B, International Psychogeriatric Association. Expert Conference on mild cognitive impairment Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- [4].Morris JC, Storandt M, Miller JP, McKeel DW, Price JL, Rubin EH, Berg L. Mild cognitive impairment represents early-stage Alzheimer disease. Arch Neurol. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- [5].Ramani A, Jensen JH, Helpern JA. Quantitative MR imaging in Alzheimer disease. Radiology. 2006;241:26–44. doi: 10.1148/radiol.2411050628. [DOI] [PubMed] [Google Scholar]
- [6].Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- [7].Cho Y, Seong J, Jeong Y, Shin SY. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32:2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leung KK, Shen KK, Barnes J, Ridgway GR, Clarkson MJ, Fripp J, Salvado O, Meriaudeau F, Fox NC, Bourgeat P, Ourselin S. Increasing power to predict mild cognitive impairment conversion to Alzheimer’s disease using hippocampal atrophy rate and statistical shape models. Med Image Comput Comput Assist Interv. 2010;13:125–132. doi: 10.1007/978-3-642-15745-5_16. [DOI] [PubMed] [Google Scholar]
- [10].Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. Neuroimage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Singh N, Wang AY, Sankaranarayanan P, Fletcher PT, Joshi S, Alzheimer’s Disease Neuroimaging, Initiative Genetic, structural and functional imaging biomarkers for early detection of conversion from MCI to AD. Med Image Comput Comput Assist Interv. 2012;15:132–140. doi: 10.1007/978-3-642-33415-3_17. [DOI] [PubMed] [Google Scholar]
- [12].Zhang D, Shen D, Alzheimer’s Disease Neuroimaging, Initiative Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McEvoy LK, Holland D, Hagler DJ, Jr, Fennema-Notestine C, Brewer JB, Dale AM, Alzheimer’s Disease Neuroimaging Initiative Mild cognitive impairment: Baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology. 2011;259:834–843. doi: 10.1148/radiol.11101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chupin M, Gerardin E, Cuingnet R, Boutet C, Lemieux L, Lehericy S, Benali H, Garnero L, Colliot O, Alzheimer’s Disease Neuroimaging, Initiative Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Eskildsen SF, Coupé P, García-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Koikkalainen J, Lotjonen J, Thurfjell L, Rueckert D, Waldemar G, Soininen H, Alzheimer’s Disease Neuroimaging Initiative Multi-template tensor-based morphometry: Application to analysis of Alzheimer’s disease. Neuroimage. 2011;56:1134–1144. doi: 10.1016/j.neuroimage.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P, Alzheimer’s Disease Neuroimaging Initiative Early diagnosis of Alzheimer’s disease using cortical thickness: Impact of cognitive reserve. Brain. 2009;132:2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Westman E, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, Weiner MW, Lovestone S, Spenger C, Wahlund LO, AddNeuroMed consortium, Alzheimer’s Disease Neuroimaging Initiative AddNeuroMed and ADNI: Similar patterns of Alzheimer’s atrophy and automated MRI classification accuracy in Europe and North America. Neuroimage. 2011;58:818–828. doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- [19].Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lotjonen J, Alzheimer’s Disease Neuroimaging Initiative Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wee C, Yap P, Shen D, Alzheimer’s Disease Neuroimaging, Initiative Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer’s disease: Hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- [22].Qiu A, Fennema-Notestine C, Dale AM, Miller MI, Alzheimer’s Disease Neuroimaging, Initiative Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Y, Song Y, Rajagopalan P, An T, Liu K, Chou Y, Gutman B, Toga AW, Thompson PM. Surface-based TBM boosts power to detect disease effects on the brain: An N=804 ADNI study. Neuroimage. 2011;56:1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20:667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- [25].Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. Neuroimage. 2006;30:52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imaging. 2007;26:462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Feng J, Tang X, Tang M, Priebe C, Miller M. Metric space structures for computational anatomy. In: Wu G, Zhang D, Shen D, Yan P, Suzuki K, Wang F, editors. Machine Learning in Medical Imaging, Lecture Notes in Computer Science. Vol. 8184. Springer International Publishing Switzerland; 2013. pp. 123–130. [Google Scholar]
- [28].Tang X, Holland D, Dale AM, Younes L, Miller MI, for the Alzheimer’s Disease Neuroimaging, Initiative Shape abnormalities of subcortical and ventricular structures in mild cognitive impairment and Alzheimer’s disease: Detecting, quantifying, and predicting. Hum Brain Mapp. 2014;35:3701–3725. doi: 10.1002/hbm.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Miller MI, Younes L, Trouvé A. Diffeomorphometry and geodesic positioning systems for human anatomy. Technology (Singap World Sci) 2014;2:36. doi: 10.1142/S2339547814500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Miller MI, Trouve A, Younes L. On the metrics and euler-lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- [31].Miller MI, Trouve A, Younes L. Geodesic shooting for computational anatomy. J Math Imaging Vis. 2006;24:209–228. doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vaillant M, Miller MI, Younes L, Trouve A. Statistics on diffeomorphisms via tangent space representations. Neuroimage. 2004;23(Suppl 1):S161–S169. doi: 10.1016/j.neuroimage.2004.07.023. [DOI] [PubMed] [Google Scholar]
- [33].Qiu A, Younes L, Miller MI. Principal component based diffeomorphic surface mapping. IEEE TransMedImaging. 2012;31:302–311. doi: 10.1109/TMI.2011.2168567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tward DJ, Ma J, Miller MI, Younes L. Robust diffeomorphic mapping via geodesically controlled active shapes. Int J Biomed Imaging. 20132013:205494. doi: 10.1155/2013/205494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy inMCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu C, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Wahlund L, Spenger C, Toga AW, Lovestone S, Simmons A. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage. 2011;56:212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin JS, Alzheimer’s Disease Neuroimaging, Initiative Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer Disease: Quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mohs RC, Rosen WG, Davis KL. The Alzheimer’s disease assessment scale: An instrument for assessing treatment efficacy. Psychopharmacol Bull. 1983;19:448–450. [PubMed] [Google Scholar]
- [40].Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- [41].Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- [42].Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- [43].Holland D, Dale AM. Alzheimer’s Disease Neuroimaging, Initiative (2011) Nonlinear registration of longitudinal images and measurement of change in regions of interest. Med Image Anal. 15:489–497. doi: 10.1016/j.media.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van derKouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- [45].Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: Reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- [46].Qiu A, Miller MI. Multi-structure network shape analysis via normal surface momentum maps. Neuroimage. 2008;42:1430. doi: 10.1016/j.neuroimage.2008.04.257. [DOI] [PubMed] [Google Scholar]
- [47].Ma J, Miller MI, Younes L. A bayesian generative model for surface template estimation. Int J Biomed Imaging. 2010:974957. doi: 10.1155/2010/974957. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vaillant M, Glaunés J. Surface matching via currents. Inf Process Med Imaging. 2005;19:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- [49].Groppe DM, Urbach TP, Kutas M. Mass univariate analysis of event-related brain potentials/fields I: A critical tutorial review. Psychophysiology. 2011;48:1711–1725. doi: 10.1111/j.1469-8986.2011.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Y, Paajanen T, Zhang Y, Westman E, Wahlund L, Simmons A, Tunnard C, Sobow T, Mecocci P, Tsolaki M, Vellas B, Muehlboeck S, Evans A, Spenger C, Lovestone S, Soininen H. Combination analysis of neuropsychological tests and structural MRI measures in differentiating AD, MCI and control groups—The AddNeuroMed study. Neurobiol Aging. 2011;32:1198–1206. doi: 10.1016/j.neurobiolaging.2009.07.008. [DOI] [PubMed] [Google Scholar]
- [51].Ferrarini L, Palm WM, Olofsen H, van Buchem MA, Reiber JH, Admiraal-Behloul F. Shape differences of the brain ventricles in Alzheimer’s disease. Neuroimage. 2006;32:1060–1069. doi: 10.1016/j.neuroimage.2006.05.048. [DOI] [PubMed] [Google Scholar]
- [52].Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Postell E, Lee DS, Wang M, Mori S, O’Brien R, Albert M. The diffeomorphometry of temporal lobe structures in preclinical Alzheimer’s disease. Neuroimage Clin. 2013;3:352–360. doi: 10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller MI, Younes L, Trouvé A. Diffeomorphometry and geodesic positioning systems for human anatomy. Technology (Singap World Sci) 2014;2:36. doi: 10.1142/S2339547814500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Younes L, Ratnanather JT, Brown T, Aylward E, Nopoulos P, Johnson H, Magnotta VA, Paulsen JS, Margolis RL, Albin RL, Miller MI, Ross CA, PREDICT-HD Investigators and. Coordinators of the Huntington Study Group Regionally selective atrophy of subcortical structures in prodromal HDas revealed by statistical shape analysis. Hum Brain Mapp. 2014;35:792–809. doi: 10.1002/hbm.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Postell E, Lee DS, Wang MC, Mori S, O’Brien R, Albert M, BIOCARD Research Team The diffeomorphometry of temporal lobe structures in preclinical Alzheimer’s disease. Neuroimage Clin. 2013;3:352–360. doi: 10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tang X, Oishi K, Faria AV, Hillis AE, Albert MS, Mori S, Miller MI. Bayesian parameter estimation and segmentation in the multi-atlas random orbit model. PLoS One. 2013;8:e65591. doi: 10.1371/journal.pone.0065591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: An analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]