Abstract
Gene expression studies indicate that body mass index (BMI) is associated with molecular pathways involved in inflammation, insulin-like growth factor activation, and other carcinogenic processes in breast tissue. The goal of this study was to determine whether BMI is associated with gene methylation in breast tissue and to identify pathways that are commonly methylated in association with high BMI. Epigenome-wide methylation profiles were determined using the Illumina HumanMethylation450 BeadChip array in the non-diseased breast tissue of 81 women undergoing breast surgery between 2009 and 2013 at the University of North Carolina Hospitals. Multivariable, robust linear regression was performed to identify methylation sites associated with BMI at a false discovery rate q value <0.05. Gene expression microarray data was used to identify which of the BMI-associated methylation sites also showed correlation with gene expression. Gene set enrichment analysis was conducted to assess which pathways were enriched among the BMI-associated methylation sites. Of the 431,568 methylation sites analyzed, 2573 were associated with BMI (q value <0.05), 57 % of which showed an inverse correlation with BMI. Pathways enriched among the 2573 probe sites included those involved in inflammation, insulin receptor signaling, and leptin signaling. We were able to map 1251 of the BMI-associated methylation sites to gene expression data, and, of these, 226 (18 %) showed substantial correlations with gene expression. Our results suggest that BMI is associated with genome-wide methylation in non-diseased breast tissue and may influence epigenetic pathways involved in inflammatory and other carcinogenic processes.
Keywords: Obesity, Methylation, Breast cancer, BMI, Epigenetics
Introduction
Breast cancer is a heterogeneous disease composed of at least five subtypes based on gene expression patterns [1, 2]. The incidence of these breast tumor subtypes varies by risk factors such as age, race, body size, and breastfeeding history [3, 4], suggesting that risk factor status may differentially influence subtype-specific carcinogenic pathways. Obesity, in particular, is a modifiable breast cancer risk factor that has well-established and complex relationships with breast cancer subtypes [5, 6]. For example, though body mass index (BMI) is associated with increased risk of breast cancer subtypes that are both positive and negative for hormone receptors [6–10], the relationship may differ by menopausal status, with hormone receptor positive tumors associated with increased risk among postmenopausal, but decreased risk among premenopausal women [10, 11].
Gene expression studies suggest several mechanisms through which BMI might influence breast carcinogenesis. In studies of non-diseased breast tissue, differences in expression by BMI were found for genes that code for insulin-like growth factor 2 (IGF-2), for the insulin-like growth factor 1 receptor (IGF-1R), and for genes involved in inflammation [12, 13]. When IGF-1R is triggered, it can activate many pathways that increase the proliferation of breast cells [14, 15] while an inflammatory response could contribute to tumor invasiveness through increased angio-genesis and tissue remodeling [16, 17]. It is unclear whether there are other mechanisms driving BMI-associated breast carcinogenesis.
The goal of this study, therefore, was to explore BMI-associated epigenetic changes in breast tissue using data from the Normal Breast Study. Specifically, we investigated whether BMI was associated with gene methylation in non-diseased breast tissue. Because gene methylation is a mechanism for controlling gene expression [18], we also assessed which of the BMI-associated methylation sites (methylation sites associated with BMI at a false discovery rate <0.05) were also correlated with gene expression. Identifying epigenetic changes that are associated with BMI and gene expression can provide insight into how BMI influences breast carcinogenesis and may direct research of molecular targets for the prevention of breast cancer.
Methods
Study population
Study subjects were participants in the Normal Breast Study. The study contacted 526 English-speaking women, at least 18 years of age, undergoing breast surgery at University of North Carolina Hospitals between 2009 and 2013. The University of North Carolina at Chapel Hill Institutional Review Board (IRB) approved this study and all participants provided informed consent according to an IRB-approved protocol.
Qualifying surgery types included total mastectomy, partial mastectomy, and excisional biopsy for women with breast tumors; prophylactic mastectomy for women at high risk of breast cancer; and reduction mammoplasty. Of the 526 patients contacted for participation, 19 declined to participate, 4 requested removal from the study after providing written consent, and breast tissue was not available for 29 patients at the time of surgery, resulting in a final study population of 474 patients. Study participants provided breast tissue at the time of surgery. Samples with sufficient quantities were snap frozen. Patient demographic and risk factor data were collected through a telephone interview conducted by the University of North Carolina's Survey Research Unit. Medical abstraction was conducted to obtain anthropometric data.
Selection criteria for the present study included having an invasive ductal carcinoma with a tissue specimen sampled ≥4 cm from tumor margins, undergoing reduction mammoplasty, or undergoing prophylactic surgery. Of the 474 Normal Breast Study participants, 324 were diagnosed with invasive breast cancer and 150 participants had tissue collected that was >4 cm from the tumor. Of the participants with tissue [4 cm from the tumor, 60 participants had a BMI ≥ 30 kg/m2, 45 had a BMI between 25 and 30 kg/m2, and 45 had a BMI < 25 kg/m2. Participants were randomly selected from each BMI group: 30 from the BMI ≥ 30 kg/m2 group, 19 from the BMI between 25 and 30 kg/m2 group, and 21 from the BMI < 25 kg/m2 group. All prophylactic surgery (n = 8) and reduction mammoplasty (n = 18) participants were included, resulting in a study population of 96. The tissue specimens for the 96 participants were all snap frozen. Two adjacent 50 mg samples were taken from each of the participants’ tissue specimens: one portion was used for the methylation assessment; the second, adjacent sample was used for histology and to determine gene expression values.
The study sample was restricted to those of white or African–American race, resulting in the exclusion of 6 individuals of other race; current smokers were excluded from the dataset because only three individuals identified as such, as were two underweight (BMI < 18.5) individuals and four individuals missing data on alcohol use. The resulting final study population was 81.
Gene methylation assessment
The Illumina HumanMethylation450 BeadChip array [19] was used to measure methylation levels in the tissue samples. The array assesses methylation at 485,577 cytosine-guanine dinucleotide (CpG) sites and has a coverage of 99 % of RefSeq genes, with an average of 17 CpG sites per gene. DNA was isolated using the DNeasy Blood and Tissue Kit from Qiagen (Valencia, CA) by following the manufacturer's spin-column protocol. Sodium bisulfite modification of the DNA was conducted using EZ DNA Methylation Gold Kit (Zymo Research, Orange, CA), after which the quantity and concentration of the DNA were assessed using a Nanodrop spectrophotometer to ensure that 500 ng of DNA was available for use for the HumanMethylation450 BeadChip platform.
Methylation levels (beta values) were calculated based on the intensity measures for unmethylated (U) or methylated (M) CpGs. Probe raw intensity values were extracted with the Illumina GenomeStudio® software (version 2011.1). To correct dye bias and normalize the methylation data, the M and U intensity values were pre-processed separately for the Infinium I and II CpG probes as follows: (1) for Infinium I probes, the red and green channel probes were separately background corrected (using the Robust Multichip Average (RMA) method [20]) and quantile normalized; (2) for Infinium II probes, dye bias between U and M intensity values were first corrected using the normalizeMethyLumiSet method in the R package “methylumi” [21], and then RMA background correction and quantile normalization were performed separately for U and M intensity values; (3) the beta value for each CpG site was recalculated as the ratio of normalized fluorescent intensities between methylated and unmethylated alleles β = M/(M + U + 100); a beta value of 0 indicates no methylation while a value of 1 indicates complete methylation. Batch effects and bisulfite conversion intensities were adjusted for and, to minimize the effect of outlier methylation values in regression modeling, any beta values that were more than three standard deviations from the mean were excluded for each CpG probe. Low performance CpG probes were also filtered by (1) excluding 13,449 CpG probes located at sites with common SNPs (minor allele frequency ≥0.05 in Europeans or Africans based on the 1000 Genomes Project data [22]); and (2) excluding 41,937 CpGs with probes mapped to multiple genomic locations [23]. After applying these exclusion criteria, 431,568 CpGs remained. The array data have been deposited in Gene Expression Omnibus under accession number GSE67919.
Gene expression assessment
Gene expression was assessed in frozen tissue samples adjacent to the tissue sampled for the methylation assay. RNA was isolated using Qiagen RNeasy kits (Valencia, CA). A Nanodrop spectrophotometer and an Agilent 2100 Bio-analyzer were used to assess RNA concentration and quality, respectively. Gene expression analysis was performed using Agilent Low Input Quick Amp labeling, 2-color (5190-2306) and Human Gene Expression 4 × 44 k v2 Microarray kits (G4845A; Santa Clara, Ca) as per the manufacturer's instruction. The data were normalized and probes with <10 dpi in any channel were excluded [13]. Probes with missing data for more than 20 % of the samples, probes that did not map to an ENTREZ ID, or individuals missing data for more than 30 % of probes were also excluded.
Statistical analysis
Robust linear regression, in which parameter estimates are less influenced by outliers, was used to assess the relationship between BMI and gene methylation [24]. BMI at the time of surgery was treated as a continuous linear variable for regression modeling. Age at first pregnancy, parity, menopausal status, alcohol use, physical activity, and fruit and vegetable intake were assessed for confounding. Only ever alcohol use (≥12 lifetime drinks) met our empirical criteria for confounding (association with BMI at p value <0.05 and association with gene methylation at false discovery rate (FDR) q value <0.05 [25]). The final adjustment set consisted of alcohol use, along with age and race, which were determined to be confounders a priori. A CpG site was considered BMI-associated if it was associated with BMI at a false discovery rate q value of <0.05.
Linear regression was also used to determine which of the BMI-associated CpG sites were correlated with gene expression. Correlation between gene methylation and expression was evaluated for 61 of the 81 study participants (20 participants were excluded after applying the gene expression quality control criteria described above). All statistical analyses were performed using the R software package (version 2.15.3). An FDR q value of <0.05 was used as the significance cut-off for regression analyses; a Pearson correlation cut-off of ≤–0.30 or ≥0.30 was used as the criteria for determining correlation with gene expression.
Gene set enrichment analysis was conducted to determine what pathways were enriched among the BMI-associated methylation sites. Using data from the National Center for Biotechnology Information (NCBI) Human Genome Build 37 Gene Annotation Database [26], each CpG was mapped to the nearest gene based on distance between the CpG and a nearby gene transcription start site (TSS). A total of 392,910 CpGs on the array are within 5 kb of the nearest TSS of 22,810 NCBI reference genes. CpGs located farther than 5 kb from the nearest TSS were excluded from the gene set enrichment analysis. To test for enrichment of gene pathways in BMI-associated CpG sites, Ingenuity Systems Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com) was used, though IPA analysis is limited by the calculation of p-values using a gene-sampling rather than subject-sampling framework [27]. We first identified all 640 canonical pathways that included at least one of the 22,810 genes represented on the array from the IPA web server, then a binomial test was performed to test whether genes significant at a FDR threshold of 0.05 were enriched in any of these pathways. Enrichment test p values were derived from 5000 rounds of permutation tests. At each permutation test, association test p values were randomly shuffled for all 392,910 CpGs and then repeated for each corresponding enrichment test. A permutation p value of <0.05 was used as a significance cut-off. We also conducted permutation-based IPA analyses on the BMI-associated sites identified in two studies [13, 28] that examined the association between BMI and gene expression in breast tissue.
Results
The 81 study participants ranged in age from 19 to 84 years, with a median age of 53. Approximately 27 % of the study participants were normal weight (18.5 kg/m2 ≥ BMI > 25 kg/m2), 32 % were overweight (25 kg/m2 ≥ BMI > 30 kg/m2), and 41 % were obese (BMI ≥ 30 kg/m2) (Table 1). An exploratory analysis was conducted to examine the effect of tissue source (reduction mammoplasty, prophylactic surgery, or invasive ductal carcinoma) on methylation value to determine whether it was appropriate to analyze these distinct tissue sources together. In the analysis, tissue source was treated as the main exposure variable and age was a covariate. No methylation sites were significantly associated with tissue source in this analysis, indicating that the three tissue sources could be analyzed together.
Table 1.
Selected population characteristics in the Normal Breast Study
| N | ||
|---|---|---|
| .Total | 81 | 100 |
| Body Mass Index (BMI) | ||
| Obese (BMI ≥ 30) | 33 | 41 |
| Overweight (25 ≤ BMI < 30) | 26 | 32 |
| Normal (18.5 ≤ BMI < 25) | 22 | 27 |
| Race | ||
| White | 55 | 68 |
| African–American | 26 | 32 |
| Age | ||
| Age < 50 | 31 | 38 |
| Age ≥ 50 | 50 | 62 |
| Menopausal status | ||
| Premenopausal | 27 | 33 |
| Postmenopausal | 50 | 62 |
| Missing | 4 | 5 |
| Ever consumed alcohol | ||
| Yes | 66 | 81 |
| No | 15 | 19 |
| Tissue Source | ||
| Prophylactic surgery | 6 | 7 |
| Reduction mammoplasty | 17 | 21 |
| Mastectomy/lumpectomy | 58 | 72 |
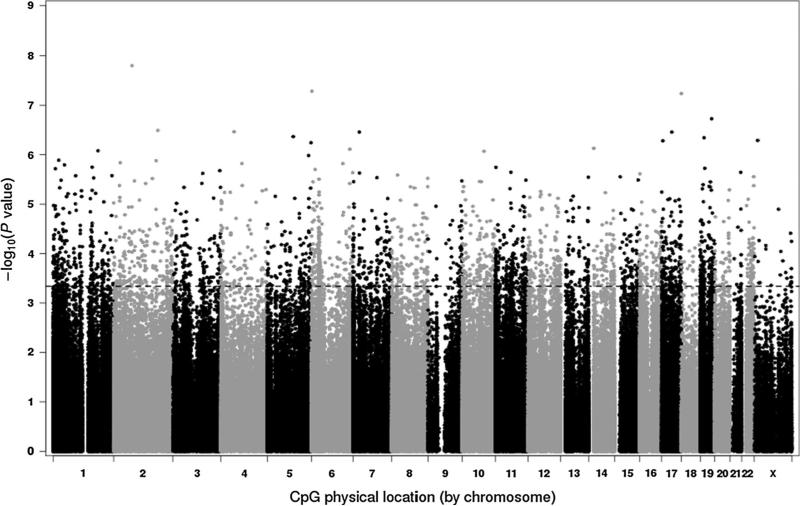
Robust linear regression analyses revealed that 2,573 CpG sites were significantly associated with BMI at FDR q value <0.05 after adjustment for age, race, and alcohol use (Supplemental Table 1). BMI-associated methylation sites were found on all chromosomes (Fig. 1), and approximately 57 % of the sites were inversely associated with BMI. Figure 2 displays scatterplots of methylation values by BMI for selected CpG sites to provide visual representations of the association between BMI and gene methylation. Of the 2573 BMI-associated probes, 10 % were located on CpG islands, which are often near promoter regions [29], 24 % were located on shores, which are up- or downstream of CpG islands [30], and 11 % were located on shelves, which are up- or downstream of shores [30]; the remaining 54 % were not located in or near CpG islands. The BMI-associated probes in this analysis were less likely to be located on CpG islands than would be expected based on the representation of CpG island probes on the 450 k platform (10 % vs 31 %) (Chi square p value <0.001).
Fig. 1.
Manhattan plot of body mass index association p values, sorted by chromosome location. Dashed line indicates false discovery rate cut-off of <0.05
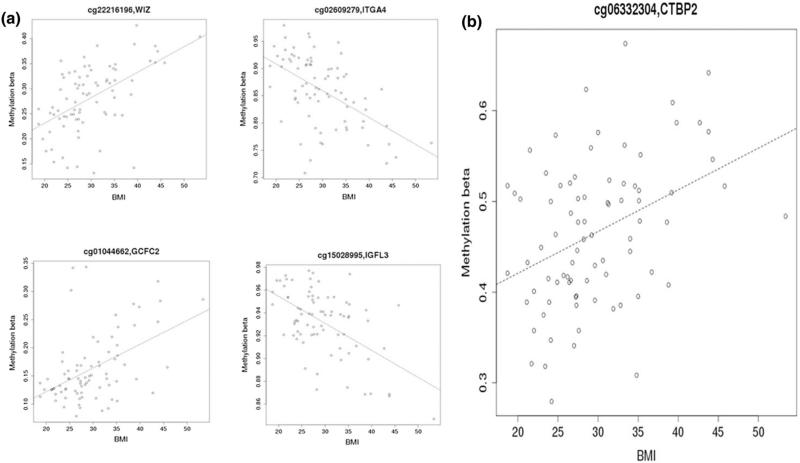
Fig. 2.
a Four scatterplots of methylation level by body mass index for selected probes. b Scatterplot of methylation level by body mass index for probe ID cg06332304 on the CTBP2 gene
Based on gene set enrichment analysis using the IPA database, many of the BMI-associated methylation sites were involved in inflammatory or immune response pathways, including genes indicated in the signaling of several interleukins, natural killer cells, B cells, T cells, and NF-kB (Supplemental Table 2). Metabolic pathways involving insulin receptor and leptin signaling and pathways that influence carcinogenesis, such as human epidermal growth factor 2 (HER-2), angiopoietin, and hepatocyte growth factor (HGF) signaling, were also significantly associated (permutation p value <0.05) with the BMI-associated CpG sites. The pathways identified from our BMI-associated methylation sites were also identified in two previously reported gene expression studies [13, 28] listed in Table 2. Many of the common pathways were related to inflammatory processes, though there were also pathways indicative of cardiovascular pathogenesis, such as the aldosterone signaling in epithelial cells pathway and the endothelin-1 signaling pathway. One gene, C-terminal binding protein 2 (CTBP2), was associated with BMI in all three studies (Supplemental Table 3); the association between BMI and methylation percentage for the CTBP2 probe identified in this study is shown in Fig. 2b.
Table 2.
Pathways Enriched in BMI-Associated Methylation Sites in the Normal Breast Studya, Ingenuity Systems Pathway Analysis
| Ingenuity Systems Pathway Analysis ID | Countb | Sizec | p valued | Gene Symbol |
|---|---|---|---|---|
| Natural Killer Cell Signaling | 18 | 68 | <0.0001 | PRKCE,PRKCB,SH3BP2,LCP2,VAV3,SYNJ2,PAK1,PTPN6,PIK3CD,VAV2, MAP2K1,PRKCZ,PRKCQ,FCGR3B,PIK3R3,INPP5D,FCER1G,GRB2 |
| Chronic Myeloid Leukemia Signaling | 21 | 88 | 0.0002 | HDAC4,CDKN1B,TGFB3,CHUK,CTBP1,CDK6,E2F5,SMAD3,CTBP2, HDAC10,HDAC1,PIK3CG, STAT5A,PIK3CD,TGFB1,MAP2K1, TFDP1,PIK3R2,CRK,PIK3R3,GRB2 |
| IL-8 Signaling | 30 | 177 | 0.0004 | PLD4,PRKCE,ARRB2,DIRAS3,ITGB2,NCF2,FLT1,PLCB2,PIK3CD, GNA12,PRKCZ,TRAF6, PIK3R2,MPO,PLD6,PRKCQ,CCND2,BRAF, PRKCB,EIF4EBP1,CHUK,CCND3,PIK3CG,GNAS,MAP2K1,LIMK2, GNG7,GNB1,FNBP1,PIK3R3 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 29 | 172 | 0.0006 | PRKCE,DIRAS3,NCF4,PPP1R10,TNFRSF11B,APOA1,RAP1A,NCF2, PIK3CD,NGFR,PRKCZ, TNFRSF1B,PIK3R2,MPO,PRKCQ,LYZ,HOXA10, PRKCB,CHUK,PPP2R5C,SERPINA1,SPI1,PIK3CG,APOC2,PTPN6,MAP2K1,IRF8,FNBP1,PIK3R3 |
| Role of NFAT in Regulation of the Immune Response | 27 | 158 | 0.0012 | CD79B,MEF2D,LCP2,XP01,CSNK1D,HLA-DMA,PLCB2,PIK3CD,GNA12, PIK3R2,PRKCQ,FCGR3B,PPP3CB,HLA-DMB,NFATC4, CHUK,CD247, PIK3CG,GNAS,MAP2K1,GNAT2,GNG7,GNB1,CD79A,PIK3R3,FCER1G,GRB2 |
| Phospholipase C Signaling | 37 | 216 | 0.0014 | PRKCE,PLD4,CD79B,MEF2D,LCP2,PLA2G4E,DIRAS3,ADCY7,RAP1A,PLCB2, PRKCZ,ADCY5, ITGB1,ARHGEF17,PLD6,PRKCQ,PPP3CB,ARHGEF10,HDAC4,PRKCB,ADCY9, NFATC4,ADCY4,CD247,ARHGEF12,HDAC10,HDAC1,GNAS,MAP2K1,GNG7, GNB1,FNBP1,CD79A,CREB5, FCER1G,GRB2,ITGA4 |
| Fc Epsilon RI Signaling | 19 | 105 | 0.0016 | PRKCE,IL13,PRKCB,LCP2,VAV3,PLA2G4E,SYNJ2,PIK3CG,PIK3CD,VAV2, MAP2K1,PRKCZ, PIK3R2,PRKCQ,GAB1,PIK3R3,INPP5D,FCER1G,GRB2 |
| Endothelin-1 Signaling | 28 | 165 | 0.0026 | ECE1,PLD4,PRKCE,PLA2G4E,PLCD4,ADCY7,PLCL2,PLA2G2C,PLCB2,PIK3CD,GNA12,CASP2, PRKCZ,ADCY5,PIK3R2,PLD6,PRKCQ,GAB1,MAPK4,BRAF,PRKCB,ADCY9, ADCY4,PIK3CG,GNAS,GNAT2,PIK3R3,GRB2 |
| IL-6 Signaling | 17 | 110 | 0.0028 | CHUK,CYP19A1,TNFRSF11B,COL1A1,SOCS1,SRF,PIK3CG,PIK3CD,NGFR, MAP2K1,IL1A, TRAF6,TNFRSF1B,PIK3R2,IL1RN,PIK3R3,GRB2 |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 18 | 90 | 0.0034 | PLD4,PRKCE,ACTA2,PRKCB,LCP2,VAV3,PAK1,PXN,PIK3CG,VAV2,PRKCZ, NCK2,PIK3R2, CRK,PRKCQ,PLD6,PIK3R3,INPP5D |
| Renal Cell Carcinoma Signaling | 13 | 68 | 0.0046 | ETS1,PAK1,RAP1A,PIK3CG,PIK3CD,TGFB1,MAP2K1,MET,PIK3R2,CRK,GAB1,PIK3R3,GRB2 |
| Leukocyte Extravasation Signaling | 28 | 181 | 0.0054 | CD44,PRKCE,ACTA2,CXCR4,NCF4,ACTN1,ITGB2,RAP1A,NCF2,PXN,PIK3CD,CLDN15, VAV2, PRKCZ,SPN,ITGB1,PIK3R2,CRK,PRKCQ,ARHGAP9,PRKCB,VAV3, CTTN,MMP17,PIK3CG, CLDN19,PIK3R3,ITGA4 |
| Aldosterone Signaling in Epithelial Cells | 23 | 147 | 0.0066 | PRKCE,DNAJB13,CRYAB,ASIC2,PLCD4,PLCL2,DNAJC11,PLCB2,PIK3CD,DNAJB6,PRKCZ, PIK3R2,SGK1,DNAJC27,PRKCQ,SLC9A1,SCNN1A,ASIC1,PRKCB,PIK3CG,MAP2K1,DNAJC15, PIK3R3 |
| Integrin Signaling | 30 | 188 | 0.0066 | TSPAN4,ACTA2,DIRAS3,ARHGAP26,PAK1,ACTN1,ITGB2,RAP1A,PXN,PARVB,ASAP1,CAPN3, PIK3CD,ITGB1,PIK3R2,CRK,NEDD9,ITGA2B,BRAF,CTTN,PIK3CG,MAP2K1, ITGB4,MYLK,NCK2,FNBP1,PIK3R3,ITGB7,ITGA4,GRB2 |
| Paxillin Signaling | 17 | 96 | 0.0114 | ACTA2,PAK1,ACTN1,ITGB2,PXN,PIK3CG,PIK3CD,ITGB4,ITGB1,NCK2,PIK3R2, CRK,PIK3R3, ITGB7,ITGA2B,GRB2,ITGA4 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 23 | 196 | 0.0130 | ALDH4A1,XPO1,ALDH3B1,CYP3A5,TNFRSF11B,HS6ST1,ALDH1A3,NGFR,IL1A,ACOX2, TRAF6,ACSL1, TNFRSF1B,HS3ST2,MGMT,ACSL6,SULT1C2,NR1H3, APOC2,CHST3,ACOX3,IL1RN,ABCB9 |
| Phospholipases | 8 | 55 | 0.0140 | PLD4,PLCB2,PLA2G4E,PLCD4,PLA2G2C,PLCL2,PLD6,CCR1 |
| Erythropoietin Signaling | 13 | 66 | 0.0144 | PRKCE,PRKCB,SOCS1,PIK3CG,STAT5A,PTPN6,PIK3CD,MAP2K1,PRKCZ,PIK3R2,PRKCQ, PIK3R3,GRB2 |
| Fcγ RIIB Signaling in B Lymphocytes | 8 | 40 | 0.0154 | PIK3CG,PIK3CD,CD79B,CD79A,PIK3R2,PIK3R3,INPP5D,GRB2 |
| FAK Signaling | 16 | 82 | 0.0164 | ACTA2,TNS1,ARHGAP26,PAK1,PXN,PIK3CG,ASAP1,PIK3CD,CAPN3,MAP2K1, ITGB1,PIK3R2, CRK,PIK3R3,GRB2,ITGA4 |
| Acute Myeloid Leukemia Signaling | 14 | 73 | 0.0176 | BRAF,EIF4EBP1,SPI1,PIK3CG,STAT5A,PIK3CD,MAP2K1,CSF1R,PIK3R2,TCF7L2,PIK3R3,CSF3R,TCF7,GRB2 |
| HGF Signaling | 17 | 96 | 0.0188 | PRKCE,ETS1,PRKCB,PAK1,RAP1A,PXN,PIK3CG,PIK3CD,MAP2K1,ELF1,PRKCZ, MET,PIK3R2, PRKCQ,GAB1,PIK3R3,GRB2 |
| Melanocyte Development and Pigmentation Signaling | 16 | 82 | 0.0196 | ADCY9,ADCY4,ADCY7,SH2B2,MITF,PIK3CG,GNAS,PTPN6,PIK3CD,MAP2K1,ADCY5,CREB5, PIK3R2,CRK,PIK3R3,GRB2 |
| Prolactin Signaling | 14 | 73 | 0.0212 | PRKCE,PRKCB,SOCS1,PIK3CG,STAT5A,PIK3CD,MAP2K1,PRKCZ,SOCS7,PIK3R2,PRKCQ, PIK3R3,TCF7,GRB2 |
| Thrombopoietin Signaling | 11 | 55 | 0.0234 | PIK3CG,STAT5A,PRKCE,PIK3CD,MAP2K1,PRKCB,PRKCZ,PIK3R2,PRKCQ,PIK3R3,GRB2 |
| Gap Junction Signaling | 24 | 149 | 0.0256 | PRKCE,ACTA2,PLCD4,CSNK1D,ADCY7,PLCL2,PRKG2,PLCB2,PIK3CD,BGLAP,PRKCZ,ADCY5, PIK3R2,PRKCQ,PPP3CB,TUBB2B,PRKCB,ADCY9,ADCY4,PIK3CG,GNAS,MAP2K1,PIK3R3, GRB2 |
| HER-2 Signaling in Breast Cancer | 15 | 75 | 0.0296 | CDKN1B,PRKCE,PRKCB,CDK6,ITGB2,PIK3CG,PIK3CD,ITGB4,PRKCZ,ITGB1,PIK3R2,PRKCQ, PIK3R3,ITGB7,GRB2 |
| Signaling by Rho Family GTPases | 33 | 219 | 0.0304 | ACTA2,DIAPH3,CDC42EP4,DIRAS3,PAK1,NCF2,CDH18,PIK3CD,GNA12,PRKCZ,ITGB1, PIK3R2,ARHGEF17,SLC9A1,CDH2,ARHGEF10,ARHGEF12,CDH8,CDH9,SEPT9,PIK3CG,GNAS, MAP2K1,LIMK2,GNAT2,PKN1,MYLK,GNG7,GNB1,FNBP1,CYFIP1,PIK3R3,ITGA4 |
| Role of NFAT in Cardiac Hypertrophy | 29 | 174 | 0.0332 | PRKCE,MEF2D,PLCD4,ADCY7,PLCL2,PLCB2,PIK3CD,TGFB1,IGF1R,PRKCZ,ADCY5,PIK3R2, PRKCQ,PPP3CB ,HDAC4,TGFB3,PRKCB,NFATC4, ADCY9, ADCY4,HDAC10,HDAC1,PIK3CG,GNAS, MAP2K1,GNG7,GNB1,PIK3R3,GRB2 |
| Angiopoietin Signaling | 10 | 63 | 0.0338 | PIK3CG,STAT5A,PIK3CD,DOK2,CHUK,PIK3R2,PAK1,CRK,PIK3R3,GRB2 |
| IL-2 Signaling | 8 | 51 | 0.0382 | PIK3CG,STAT5A,PIK3CD,MAP2K1,PIK3R2,SOCS1,PIK3R3,GRB2 |
| B Cell Receptor Signaling | 22 | 149 | 0.0402 | CD79B,PIK3CD,VAV2,PIK3R2,PRKCQ,GAB1,PPP3CB,ETS1,PRKCB,NFATC4, CHUK,VAV3, SYNJ2,CD22,PIK3CG,PTPN6,MAP2K1,CD79A,CREB5,PIK3R3,INPP5D,GRB2 |
| Insulin Receptor Signaling | 21 | 122 | 0.0408 | SCNN1A,RPTOR,ASIC1,EIF4EBP1,ASIC2,SYNJ2,PPP1R10,TRIP10,SH2B2,PIK3CG,PIK3CD, PTPRF,MAP2K1,PRKCZ,PIK3R2,SGK1,CRK,GAB1,PIK3R3,INPP5D,GRB2 |
| JAK/Stat Signaling | 10 | 67 | 0.0448 | SOCS1,PIK3CG,STAT5A,PTPN6,PIK3CD,MAP2K1,SOCS7,PIK3R2,PIK3R3,GRB2 |
Includes pathways enriched in BMI-associated probes in the present study and overlapping with pathways identified as enriched for BMI-associated expression in two published studies (References [13] and [49])
# of BMI-associated genes in the pathway
# of total gene in the pathway
Permutation p value for BMI-associated genes
Of the 2573 BMI-associated probes, 1251 mapped to loci with gene expression data on the microarray dataset. Of those 1251 sites, 226 were correlated with gene expression at a Pearson correlation coefficient equal to or greater than the absolute value of 0.30 (Supplemental Table 4). Of these 226 CpGs, 68 % showed an inverse relationship between proportion of methylation and level of gene expression. CpG sites that were on shores were more likely to be correlated with gene expression than CpG sites that were on islands, though the difference was not statistically significant (18 vs. 12 %; Chi square p value = 0.14).
Discussion
This is the first study to examine the association between BMI, genome-wide methylation, and gene expression in histologically non-diseased breast tissue. Pathways enriched among the 2573 BMI-associated sites included those involved in inflammation, insulin receptor signaling, and leptin signaling. Approximately 18 % (226/1251) of the BMI-associated methylation sites that were near gene TSS were significantly associated with gene expression. Similar to other research, we found that correlation with expression was higher for probes located on CpG shores than for probes located on CpG islands [30, 31]. The lower correlation between methylation and expression in islands relative to shores may be because methylation acts to repress gene expression through promotion of histone modification more than through interference with transcription binding on promoter regions [32].
It has been hypothesized that BMI may influence breast carcinogenesis through multiple mechanisms [12–15], and our data are consistent with these hypotheses. Our observation that BMI-associated methylation occurred preferentially in genes involved in inflammatory response (e.g., interleukin-6 and NF-kappaB signaling), energy metabolism (insulin receptor and leptin signaling), and epithelialstromal interactions (hepatocyte growth factor signaling) is in line with previous mechanistic research [15, 33–37]. Interleukin-6 (IL-6), a product of chronic inflammation, is associated with obesity and has both cancer-promoting and anti-cancer functions [33–35]. Although IL-6 has been associated with decreased risk of early-stage breast cancer, the cytokine is also correlated with cell migration, anti-apoptosis, stimulation of aromatase activity, and poor prognosis among those with metastatic disease [14, 38, 39]. The activation of NF-kB signaling is associated with angiogenesis, tumor invasiveness, and anti-apoptosis in tumor cells [36] while a pathway involving the signaling of leptin, a ligand produced by adipose tissue, could influence breast carcinogenesis through increased estrogen production via aromatase transcription [40, 41] and also through the promotion of angiogenesis and tumor invasion [5, 42]. Insulin-like growth factor receptor signaling involves activation of pathways shown to increase proliferation of breast cells [14, 15] while hepatocyte growth factor (HGF) has been shown to be associated with obesity in murine models [37], to promote cell migration in vitro [37], and to influence the invasive potential of premalignant breast cells [43]. Although these pathways identified among our BMI-associated methylation sites suggest a potential role for BMI in cancer-related methylation changes, it is unknown whether these changes induce a direct effect on epithelial cells.
Our results are consistent with studies finding that BMI is associated with gene expression changes in breast tumor tissue. Creighton et al. reported that 799 probes representing 662 genes were differentially expressed when comparing the breast tumor tissue of obese patients to that of normal and overweight patients [28]. We compared the BMI-associated genes present in our study with those listed by Creighton et al. to identify which genes were on both lists, and therefore may be good candidates for further scientific exploration. We were able to examine the overlap between 439 of the 662 genes listed by Creighton et al. and found that 42 genes were identified in both studies (Supplemental Table 3). Similarly, in our previous work examining the tissue from women undergoing reduction mammoplasty from a different population, we found 760 genes were differentially expressed when comparing the non-diseased tissue of obese and normal weight women [13]; we were able to examine the overlap between 547 of those 760 genes and found that 68 of them were identified in both studies (Supplemental Table 3). As in our present study, we previously reported enrichment for pathways involved in inflammation and immune response, including pathways that may act in macrophage infiltration [13]. Furthermore, one gene, CTBP2, which encodes for a transcriptional repressor, was identified as associated with BMI in our present methylation-based analysis, in our previous gene expression results [13], and in the gene expression results of Creighton et al. [28]. Expression of members of the CTBP family, which has been shown to promote the epithelial to mesenchymal transition and genomic instability in breast cancer cells [44], has been implicated in breast carcinogenesis [44, 45].
Our findings of a significant association between BMI and gene methylation are in contrast to the results of several previous studies that examined the relationship between BMI and methylation of specific candidate genes. These studies evaluated either non-diseased or tumor breast tissue, but did not find any significant associations [46, 47]. However, these studies were limited by small strata [47] and did not use a genome-wide approach [46, 47].
Our work was strengthened by studying epigenetic changes in benign breast tissue, which has more genomic stability than tumor tissue [48]. Methylation patterns may change after cancer initiation in tumor tissue, so studying the benign tissue allows assessment of pathways more proximal to exposure. Nonetheless, it is possible that these tissues harbor some changes related to the disease status of the patients, limiting the generalizability of our results. However, the use of tissue sampled >4 cm from an invasive tumor should minimize the potential effects of field cancerization [49–51]. Also, the inclusion of reduction mammoplasty patients with no disease and the observation in an exploratory analysis that methylation patterns were not different by tissue source in this population further mitigate this concern. This study was also strengthened by studying both methylation and expression in the same patients, allowing us to examine the extent to which differences in methylation were associated with differences in gene expression.
Our study identified numerous BMI-associated changes in normal breast tissue. Our results suggest that adiposity influences the epigenetic profile of breast tissue. BMI may affect the cellular microenvironment or breast epithelial cells through several different cancer-associated pathways. As it is unknown whether these epigenetic changes are reversible with weight loss, future research might evaluate methylation in these same pathways following weight loss and gain. Molecular markers of obesity's effects on breast tissue are important for elucidating the biologic pathways driving breast tumorigenesis and may increase our ability to prevent and treat breast cancer.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Monica D'Arcy, Xuezheng Sun, and Thomas G. Stewart for statistical support.
Funding This research was funded, in part, by the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223), the National Cancer Institute/National Institute of Environmental Health Sciences Breast Cancer and the Environment Research Program (U01 – ES019472), and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ZIA-ES049032-18, ZIA-ES049033-18). WRR was supported by the National Cancer Institute (K01-CA172717-01).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-015-3401-8) contains supplementary material, which is available to authorized users.
Conflict of interests The authors declare that they have no conflict of interests.
Ethical standards The experiments performed for this research comply with the current laws of the country in which they were performed.
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. doi:10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sommer S, Fuqua SA. Estrogen receptor and breast cancer. Semin Cancer Biol. 2001;11(5):339–352. doi: 10.1006/scbi.2001.0389. doi:10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 3.Millikan RC, Newman B, Tse CK, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109(1):123–139. doi: 10.1007/s10549-007-9632-6. doi:10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. doi:10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 5.Carmichael AR. Obesity as a risk factor for development and poor prognosis of breast cancer. BJOG. 2006;113(10):1160–1166. doi: 10.1111/j.1471-0528.2006.01021.x. doi:10.1111/j.1471-0528.2006.01021.x. [DOI] [PubMed] [Google Scholar]
- 6.Carmichael AR, Bates T. Obesity and breast cancer: a review of the literature. Breast. 2004;13(2):85–92. doi: 10.1016/j.breast.2003.03.001. doi:10.1016/j.breast.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet MM, Press MF, Haile RW, et al. Risk factors by molecular subtypes of breast cancer across a population-based study of women 56 years or younger. Breast Cancer Res Treat. 2011;130(2):587–597. doi: 10.1007/s10549-011-1616-x. doi:10.1007/s10549-011-1616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phipps AI, Buist DS, Malone KE, et al. Breast density, body mass index, and risk of tumor marker-defined subtypes of breast cancer. Ann Epidemiol. 2012 doi: 10.1016/j.annepidem.2012.02.002. doi:10.1016/j.annepidem.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritte R, Lukanova A, Berrino F, et al. Adiposity, hormone replacement therapy use and breast cancer risk by age and hormone receptor status: a large prospective cohort study. Breast Cancer Res. 2012;14(3):R76. doi: 10.1186/bcr3186. doi:10.1186/bcr3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang XR, Chang-Claude J, Goode EL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103(3):250–263. doi: 10.1093/jnci/djq526. doi:10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enger SM, Ross RK, Paganini-Hill A, et al. Body size, physical activity, and breast cancer hormone receptor status: results from two case-control studies. Cancer Epidemiol Biomark Prev. 2000;9(7):681–687. [PubMed] [Google Scholar]
- 12.Suga K, Imai K, Eguchi H, et al. Molecular significance of excess body weight in postmenopausal breast cancer patients, in relation to expression of insulin-like growth factor I receptor and insulin-like growth factor II genes. Jpn J Cancer Res. 2001;92(2):127–134. doi: 10.1111/j.1349-7006.2001.tb01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X, Casbas-Hernandez P, Bigelow C, et al. Normal breast tissue of obese women is enriched for macrophage markers and macrophage-associated gene expression. Breast Cancer Res Treat. 2012;131(3):1003–1012. doi: 10.1007/s10549-011-1789-3. doi:10.1007/s10549-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorincz AM, Sukumar S. Molecular links between obesity and breast cancer. Endocr Relat Cancer. 2006;13(2):279–292. doi: 10.1677/erc.1.00729. doi:10. 1677/erc.1.00729. [DOI] [PubMed] [Google Scholar]
- 15.Parrizas M, Saltiel AR, LeRoith D. Insulin-like growth factor 1 inhibits apoptosis using the phosphatidylinositol 3′-kinase and mitogen-activated protein kinase pathways. J Biol Chem. 1997;272(1):154–161. doi: 10.1074/jbc.272.1.154. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. doi:10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 17.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. doi:10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. doi:10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 19.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. doi:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Handle Illumina methylation data. R package.
- 22.1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. doi:10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price ME, Cotton AM, Lam LL, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6(1) doi: 10.1186/1756-8935-6-4. 4,8935-6-4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Taylor JA. Genome-wide age-related DNA methylation changes in blood and other tissues relate to histone modification, expression and cancer. Carcinogenesis. 2014;35(2):356–364. doi: 10.1093/carcin/bgt391. doi:10.1093/carcin/bgt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. doi:10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCBI News. National Center for Biotechnology Information (US); Bethesda (MD): Sep, 2009. 2009. [Google Scholar]
- 27.Wu MC, Lin X. Prior biological knowledge-based approaches for the analysis of genome-wide expression profiles using gene sets and pathways. Stat Methods Med Res. 2009;18(6):577–593. doi: 10.1177/0962280209351925. doi:10.1177/0962280209351925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creighton CJ, Sada YH, Zhang Y, et al. A gene transcription signature of obesity in breast cancer. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1595-y. doi:10.1007/s10549-011-1595-y. [DOI] [PubMed] [Google Scholar]
- 29.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103(5):1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price ME, Cotton AM, Lam LL, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenet Chromatin. 2013;6(1) doi: 10.1186/1756-8935-6-4. 4,8935-6-4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. doi:10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20(24):3139–3155. doi: 10.1038/sj.onc.1204341. doi:10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 33.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115(5):911–920. doi: 10.1016/j.jaci.2005.02.023. doi:10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 34.Antuna-Puente B, Feve B, Fellahi S, et al. Adipokines: the missing link between insulin resistance and obesity. Diabetes Metab. 2008;34(1):2–11. doi: 10.1016/j.diabet.2007.09.004. doi:10.1016/j.diabet.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Knupfer H, Preiss R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res Treat. 2007;102(2):129–135. doi: 10.1007/s10549-006-9328-3. doi:10.1007/s10549-006-9328-3. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani A, Marchesi F, Porta C, et al. Inflammation and cancer: breast cancer as a prototype. Breast. 2007;16(Suppl 2):S27–S33. doi: 10.1016/j.breast.2007.07.013. doi:10.1016/j.breast.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Sundaram S, Freemerman AJ, Johnson AR, et al. Role of HGF in obesity-associated tumorigenesis: C3(1)-TAg mice as a model for human basal-like breast cancer. Breast Cancer Res Treat. 2013;142(3):489–503. doi: 10.1007/s10549-013-2741-5. doi:10.1007/s10549-013-2741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A, Purohit A, Ghilchik MW, et al. The regulation of aromatase activity in breast fibroblasts: the role of interleukin-6 and prostaglandin E2. Endocr Relat Cancer. 1999;6(2):139–147. doi: 10.1677/erc.0.0060139. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Nichols JE, Valdez R, et al. Tumor necrosis factor-alpha stimulates aromatase gene expression in human adipose stromal cells through use of an activating protein-1 binding site upstream of promoter 1.4. Mol Endocrinol. 1996;10(11):1350–1357. doi: 10.1210/mend.10.11.8923461. [DOI] [PubMed] [Google Scholar]
- 40.Kitawaki J, Kusuki I, Koshiba H, et al. Leptin directly stimulates aromatase activity in human luteinized granulosa cells. Mol Hum Reprod. 1999;5(8):708–713. doi: 10.1093/molehr/5.8.708. [DOI] [PubMed] [Google Scholar]
- 41.Magoffin DA, Weitsman SR, Aagarwal SK, et al. Leptin regulation of aromatase activity in adipose stromal cells from regularly cycling women. Ginekol Pol. 1999;70(1):1–7. [PubMed] [Google Scholar]
- 42.Renehan AG, Frystyk J, Flyvbjerg A. Obesity and cancer risk: the role of the insulin-IGF axis. Trends Endocrinol Metab. 2006;17(8):328–336. doi: 10.1016/j.tem.2006.08.006. doi:10.1016/j.tem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Casbas-Hernandez P, D'Arcy M, Roman-Perez E, et al. Role of HGF in epithelial-stromal cell interactions during progression from benign breast disease to ductal carcinoma in situ. Breast Cancer Res. 2013;15(5):R82. doi: 10.1186/bcr3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di LJ, Byun JS, Wong MM, et al. Genome-wide profiles of CtBP link metabolism with genome stability and epithelial reprogramming in breast cancer. Nat Commun. 2013;4:1449. doi: 10.1038/ncomms2438. doi:10.1038/ncomms2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byun JS, Gardner K. C-terminal binding protein: a molecular link between metabolic imbalance and epigenetic regulation in breast cancer. Int J Cell Biol. 2013;2013:647975. doi: 10.1155/2013/647975. doi:10.1155/2013/647975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao MH, Marian C, Nie J, et al. Body mass and DNA promoter methylation in breast tumors in the Western New York Exposures and Breast Cancer Study. Am J Clin Nutr. 2011;94(3):831–838. doi: 10.3945/ajcn.110.009365. doi:10.3945/ajcn.110.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumitrescu RG, Marian C, Krishnan SS, et al. Familial and racial determinants of tumour suppressor genes promoter hyper-methylation in breast tissues from healthy women. J Cell Mol Med. 2010;14(6B):1468–1475. doi: 10.1111/j.1582-4934.2009.00924.x. doi:10.1111/j.1582-4934.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi JD, Lee JS. Interplay between epigenetics and genetics in cancer. Genomics Inform. 2013;11(4):164–173. doi: 10.5808/GI.2013.11.4.164. doi:10.5808/GI.2013.11.4.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trujillo KA, Heaphy CM, Mai M, et al. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2011;129(6):1310–1321. doi: 10.1002/ijc.25788. doi:10.1002/ijc.25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heaphy CM, Bisoffi M, Fordyce CA, et al. Telomere DNA content and allelic imbalance demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2006;119(1):108–116. doi: 10.1002/ijc.21815. doi:10.1002/ijc.21815. [DOI] [PubMed] [Google Scholar]
- 51.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118(2):229–239. doi: 10.1007/s10549-009-0504-0. doi:10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.