Abstract
Loss of the cytoprotective protein Peroxiredoxin 6 (Prdx6) in cells that are aging or under oxidative stress is known to be linked to pathobiology of many age-related diseases. However, the mechanism by which Prdx6 activity goes awry is largely unknown. Using Prdx6-deficient (Prdx6−/−) cells as a model for aging or redox active cells, human/mouse LECs facing oxidative stress, and aging lenses, we found a significant increase in the levels of Sumo1 conjugates. These cells displayed increased levels of Sumo1 and reduced expression of Prdx6. Specifically we observed that Prdx6 is a target for aberrant Sumoylation signaling, and that Sumo1 modification reduces its cellular abundance. LECs overexpressing Sumo1 showed reduced expression and activity of Prdx6 and its transactivator Sp1, mRNA and protein with increased levels of ROS; those cells were vulnerable to oxidative stress-induced cell death. A significant reduction in Prdx6, Sp1 protein and mRNA expression was observed in redox-active Prdx6−/− cells and in aging lenses/LECs. The reduction was correlated with increased expression of Sumo1 and enrichment of the inactive form (dimeric) of Sumo1 specific protease Senp1. Experiments with Sumo1-fused Prdx6 and Prdx6 promoter-linked to CAT reporter gene constructs pointed that Sumo1 dysregulated Prdx6 activity by reducing its abundance and attenuating its transcription, in contrast delivery of Senp1 or Prdx6 reversed the process. Data underpins that ROS-evoked aberrant Sumoylation signaling affects Prdx6 activity by reducing Prdx6 abundance as well as transcription. Our finding may provide a foundation for a strategy to repair deleterious oxidative signaling generated by reduced activity of Prdx6.
Keywords: Prdx6; Sumo1; Senp1; Oxidative stress; ROS; Cell survival; Transcription, Sp1
Introduction
The biological activity of protein is altered by its posttranslational modification at physiological levels to perform their genetically allotted function. This occurs by conjugation of small chemical groups or polypeptides like phosphate or Small Ubiquitin-like Modifier (Sumo) to their predefined amino acid residues or motif(s) in protein [1, 2]. Recently, Sumoylation has emerged as a complex dynamical posttranslational process with an increasingly large variety of targets and larger influence on its substrate activity. Four isoforms, Sumo 1, Sumo2, Sumo3 and Sumo4 [3], are Ubiquitin-like proteins, and are bound to target proteins bearing Sumo binding motif by an enzymatic pathway similar to the ubiquitylation pathway [1, 4-6]. Sumoylation was first identified as a posttranslational modification that alters the activity of Ran GTPase activating protein (RanGAP) [7, 8]. Since discovery of Sumo modification, many proteins have been identified as its targets. Sumo modification has been shown to significantly alter protein activity, which can modulate protein stability[3, 4, 9], affect protein-protein interaction and modify protein localization and trafficking [10-14]. Moreover, most of the transcription proteins are targets for Sumo modification, and in general, Sumoylation of these proteins leads to repression of their transcriptional activity [15-21]. However, activation of transcriptional activity has also been observed [22-26].
The Sumoylation process is reversible, and deconjugation involves Sumo-specific proteases, predominately at a core consensus motif in proteins (ψ-K-X-[D/E], where ψ is hydrophobic amino acids (I,V, L), K is a target lysine, X can be any amino acid and D/E asparate or glutamate) [27, 28]. Sumos are synthesized as inactive forms that must be matured by Sumo-specific peptidases (Senps). Activation of Sumoylation occurs by an ATP-dependent heterodimerization of Sumo1 activating enzyme 1 (SAE1) and SAE2, followed by activated Sumo protein transfer onto ubiquitin conjugating enzyme 9 (UBC9), which in conjunction with Sumo E3 (ligating enzymes) facilitates Sumo conjugation to the substrate. However, the Sumoylation reaction is mechanistically similar to ubiquitination.
Protein Sumoylation can be reversed by a family of sentrin/Sumo-specific proteases (Senps) [29-31]. Sumo proteases or specific isopeptidases, members of the Senp family, ensure the reversibility of this modification [15, 32]. Seven Senps are known at present, all with preferences for particular Sumo paralogs [33] and all showing distinct patterns of cellular localization [34]. However, many Sumo activities take place in the nuclear compartment, such as controlling DNA replication/stability and gene transcription. Recently, several extranuclear proteins have been shown to be targets for Sumo, and Sumo modification leads to gain or loss of their functions [35, 36].
Mounting evidence indicates that higher levels of oxidative stress lead to aberrant Sumoylation that ultimately alters nuclear or extra nuclear protein functions. Recently, extranuclear roles of Sumoylation have been documented [4-6]. Sumoylation regulates signal transduction by modifying growth factor mediated signaling, protein trafficking, and protein-protein interaction [32, 37]. In above scenario Sumoylation also controls cell fate by enhancing protein stability or reducing protein degradation [9]. Sumoylation has been shown to play a role in antioxidant protein modification, which affects abundance and function of the antioxidants [38, 39], leading to redox status of cells.
The multifunctional protective protein Peroxiredoxin 6 (Prdx6) is a member of a newly derived family of antioxidants which provide cytoprotection by detoxifying ROS in situations of internal/external environmental stress [40-48]. Six members of the Peroxiredoxin family have been identified, ranging in molecular size from 20-30 kDa and residing in the cells/tissues of eukaryotes and prokaryotes as well as plants [40, 42-44, 49]. Their expression and localization patterns are varied, as are their reaction intermediates, but the peroxiredoxins are uniquely involved in cytoprotection against stressors, and participate in signaling pathways mainly by optimizing ROS expression. All Prdxs consist of conserved cysteine (Cys) residues at the amino terminal, corresponding to Cys47 and known as an active redox Cys. Based on Cys residues, the Prdx family is classified into three groups; 2-Cys, atypical 2-Cys and 1-Cys [40-44, 50-53]. Prdx1, 2, 3 and 4 contain 2 conserved Cys; Prdx5 is atypical 2-Cys, while Prdx6 has only 1-Cys residue. Prdx6 is thus unique in the Peroxiredoxin family and is bifunctional, having both GSH peroxidase and Ca2+-independent phospholipase A2 (aiPLA2) activities [42, 45, 51, 52]. This is the only member of Prdx family that utilizes GSH as an electron donor to catalyze the reduction of peroxides [42, 51, 54]. Prdx6 has the ability to reduce phospholipid hydroperoxides, and is thereby capable of maintaining phospholipid turnover and repairing membrane damage caused by oxidative stress [46-48, 51, 52, 54]. We have shown that a deficiency of Prdx6 leads to failure of cellular homeostasis due to enhanced oxidative load and aberrant signaling [40-44]. Similarly, decline in expression and activity of Prdx6 causes increased oxidative stress-evoked abnormal signaling and cellular injuries, which in turn initiate a pathogenic state [40, 42, 43, 51, 55, 56]. Furthermore, oxidative stress has been identified as a major cause of age-related diseases, including neurodegenerative disorders and cataractogenesis [28, 40-44, 46-48, 51, 57, 58]. Oxidative stress-induced etiology and progression of diseases may result either from diminished expression and activity of natural antioxidants due to aging or from increased generation of reactive oxygen species (ROS) [40, 44]. Based upon recent emerging evidence showing that adverse Sumoylation signaling initiates pathogenic process and current work, we think that ROS-driven oxidative stress-induced aberrant Sumoylation signaling causes dysregulation of Prdx6, at least in lens epithelial cells (LECs).
Posttranslational modifications regulate protein function through various mechanisms. Regulation can occur at the level of individual targets, through interplay between stress-induced Sumoylation and target proteins via modulation of the conjugation/deconjugation machinery [59, 60]. Furthermore, Sumoylation can be regulated by ROS at the level of Sumo1 expression conjugation or deconjugation [61], possibly leading to aberrant expression of Sumo. Since Sumo itself is a limiting factor for conjugation [62], its overexpression or underexpression influences the status of target substrate Sumoylation globally. As the level of ROS can affect its function (pathologic or survival signaling), tight regulation of ROS is critical. To this, the physiological expression and function of an antioxidant defense protein such as Prdx6 should be essential.
In the present work, we analyzed the functional effect of the dynamical process of Sumoylation/deSumoylation on regulation of Prdx6 and the cellular response to oxidative stress. We also examined whether ROS-evoked oxidative stress-induced overstimulated Sumoylation signaling modulates Prdx6 activity in relation to its protective activity and cellular responses. Our work revealed that indeed, ROS-evoked aberrant Sumoylation signaling in eye lens/LECs, jeopardized Prdx6 protective activity in two ways; by increasing Prdx6 Sumoylation and thereby reducing its abundance, and by attenuating its transcription. The findings offer a new dimension for devising means to protect cells from ROS-evoked aberrant Sumoylation signaling.
Results
Aging human lenses and Prdx6-deficient mouse LECs (redox active cells), a model for aging, show significantly increased Sumo1 conjugates with enhanced ROS level
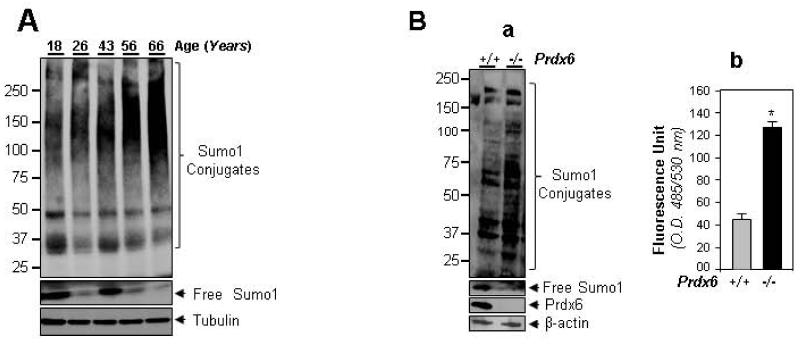
During advancing of age, a decline in antioxidant defenses causes increased oxidative stress [42-44, 63, 64]. One feature of such stress is overstimulation of posttranslational modification of proteins. Recently aberrant Sumoylation protein has been shown to adversely affect protein biological activity by altering proteome regulation [15, 65-67]. Therefore, we first analyzed whether aging human (h) lenses and Prdx6-deficient (Prdx6−/−) LECs contained increased Sumo1 protein conjugates. We obtained eye lenses of variable ages from the Lions Eye Bank, Nebraska Medical Center and NDRI, PA., and mouse (m) lens epithelial cells derived from Prdx6−/− and Prdx6+/+ mice. Total Sumo1 conjugates were visualized by immunoblotting using anti-Sumo1 antibody (Fig. 1A and B, a). We observed significantly increased Sumo conjugation with advancing of age. Apparently the level of free Sumo is also decreased, demonstrating that most of the protein Sumoylation is increased with aging. We surmised that increased Sumo conjugates should be related to oxidative stress-evoked aberrant Sumoylation signaling. Because faithfully measuring the levels of ROS was cumbersome, we utilized Prdx6-deficient cells to establish causal correlation between aging or oxidative stress and aberrant Sumoylation [42, 43]. We quantified the levels of ROS wild-type in Prdx6+/+ and Prdx6−/− LECs, and immunoblotted the extracted proteins with Sumo1 antibody. As expected, we found that an increase of Sumo1 conjugates (and a decrease of free Sumo1) (Fig. 1B, a) was related to increased ROS levels (Prdx6−/− vs Prdx6+/+, Fig. 1B, b). The study also indicated that deficiency of antioxidant and thereby enhanced endogenous production of ROS regulated Sumoylation signaling, and increased levels of ROS dysregulated Sumoylation signaling.
Fig. 1.
(A). Human lenses revealed age-dependent increases of Sumo1 conjugates. Total proteins were isolated from freshly obtained eye lenses and equal amounts of protein were loaded onto 4-20% SDS PAGE gel. Sumo1 conjugates and free Sumo1 were visualized by anti-Sumo1 antibody. (B). Prdx6-deficient mLECs bore increased Sumo1 conjugates and elevated expression of ROS. Total cell lysates were prepared from Prdx6+/+ and Prdx6−/− cells and immunoblotted with Sumo1 antibody (a), and quantification of ROS levels (b) Prdx6−/− with H2-DCF-DA showed involvement of oxidative stress (b). Histogram values are mean ± SD from three independent experiments. O.D., optical density.* Statistically significant difference (p<0.001 vs control).
Human LECs and Prdx6-deficient mLECs displayed increased Sumoylation of most of the proteins with enhanced expression of ROS and cell death in response to oxidative stress
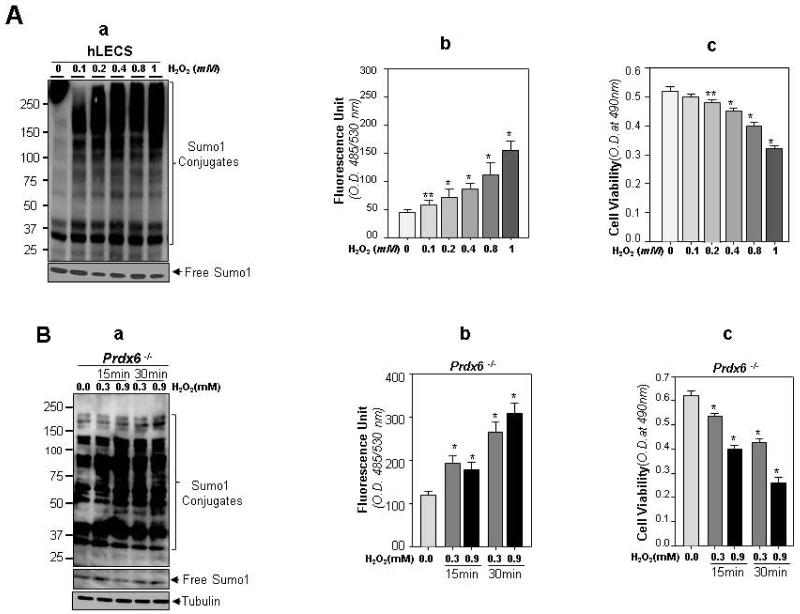
In light of the aberrant modulation of Sumo conjugation by stresses, Saitoh and Hinchey (2000) [65] suggested that stressors increase global Sumoylation. We posited that selective Sumoylation can be associated affinity of Sumo1 conjugation motif in protein as well as Sumo abundance in cell-types. Thus we assessed specifically protein Sumoylation pattern in hLECs and redox-active Prdx6−/− LECs (a model for aging), in response to oxidative stress. Cultured hLECs exposed to different concentrations of H2O2 for 30 min were analyzed by immunoblotting with Sumo1 antibody. Data indicated differential Sumo1 conjugation of proteome in response to oxidative stress. A significant increase in Sumo1 conjugates was observed in cells treated with 0.2 to 1 mM H2O2, and the increase was correlated with higher expression of ROS as quantified by H2DCF dye (Fig. 2A, b) and decreased cell viability (Fig. 2A, c). This suggests that ROS modulated Sumoylation signaling. ROS modulation of Sumo conjugation to its substrate was also observed previously [15].
Fig. 2.
(A) Oxidative stress induced Sumo1 conjugation in hLECs, and these cells displayed higher levels of ROS and reduced viability. Cultured hLECs were treated with different concentrations of H2O2 for 30 min. Complete medium (DMEM supplemented with 15% FBS) was replaced with DMEM containing 0.2% BSA prior to the H2O2 treatment. (A, a). Total cell lysates were prepared and immunoblotted with anti-Sumo1 antibody to measure free Sumo1 and Sumo1 conjugates. (A, b). Cells were cultured and subjected to oxidative stress. ROS levels were monitored. (A, c). MTS assay was conducted to monitor cell viability against oxidative stress. Data represent means ± SD of three independent experiments. * p<0.001; **p <0.05 vs control, statistically significant.
(B) Redox active Prdx6−/− LECs exposed to oxidative stress showed further increases in Sumo1 conjugates, correlated with increased ROS and reduced viability. Total cell lysates were prepared from Prdx6−/− cells untreated or treated with different concentrations of H2O2 for different time intervals and were immunoblotted with anti-Sumo1 antibody (B, a). ROS production (B, b) and cell viability (B, c) were measured in H2O2 treated Prdx6−/− cells by using H2-DCF-HA dye and MTS dye assays. Data represent the mean ± SD of three independent experiments. *, p<0.001 vs control
We next tested whether Prdx6−/− cells showed enhanced Sumo1 conjugates and were more susceptible to ROS-induced cell death caused by acute oxidative stress. Prdx6−/− cells exposed to variable concentrations of H2O2 for 15 and 30 minute were examined for Sumoylation pattern by immunoblotting. Sumo1 conjugation of most of proteins was significantly increased with 15 min of exposure at concentrations of 0.3 and 0.9 mM H2O2 (acute stress) (Fig. 2B, a). Importantly, an increase in Sumo1 conjugates was directly related to enhanced oxidative load (Fig. 2B, b) and reduced cell survival (Fig. 2B, c). These data imply the involvement of oxidative stress-induced aberrant Sumoylation signaling in reduced viability of Prdx6−/− cells or cells during oxidative stress. Additionally, immunoblot analyses (Figs. 1, A and B and 2 A,a and B,a) revealed an abundance of Sumo1 conjugates ranging from 25kDa to >250 kDa protein SDS-PAGE. On SDS-Page, Prdx6 is detected at ~24 to 28kDa [40-44] and Sumo1 ~ 15kDa [25]. If Prdx6 is mono Sumoylated at least, at a site, this should be positioned at ~40 kDa protein bands on membrane. Therefore, our next study examined whether Prdx6 is Sumoylated and whether its aberrant Sumoylation in response to oxidative stress modulates its biological activity, possibly causing loss of Prdx6 expression and activity.
Prdx6 is modified by Sumo1 both in vitro and in vivo
Strikingly, from the previous experiments with wild-type Prdx6+/+ and Prdx6−/− LECs, it was evident that Prdx6 played a key role in maintaining Sumoylation signaling at physiological condition, while its deficiency caused increased Sumo conjugates leading to ROS-evoked LEC death (Fig. 2B, c). Recently Sumoylation of extranuclear proteins including antioxidants has been reported. Sumoylation of these proteins dramatically alters their biological functions [35, 36]. To determine whether Prdx6 is Sumoylated in LECs and to test whether Prdx6 is a substrate for covalent Sumo1 conjugation, we first constituted Sumoylation reactions in vitro using recombinant Prdx6 fused TAT-HA tag as described earlier [50, 68, 69], according to the manufacturer’s protocol (Active Motif Cat No. 40120). Briefly, different concentrations of recombinant Prdx6 protein (TAT-HA-Prdx6) with Sumo1 WT protein or Sumo1 mutant protein were incubated at 30°C for 3h with activating enzyme E1 and conjugation enzyme E2 (UBC9). Reaction product immunoblotted with anti-Sumo1 and anti-Prdx6 polyclonal antibodies revealed a slower migrating band in the presence of Sumo1 WT protein, (Fig. 3; upper panel, lanes 1 and 2) and anti-Prdx6 antibody (Fig. 3; lower panel, lanes 1 and 2). Conversely, these protein bands were absent from control reactions performed in the presence of a Sumo1 mutant that could not form covalent conjugation (Fig. 3; right panel; upper and lower panel, lanes 3 and 4). Also, data revealed that Prdx6 is not Sumoylated in absence of ATP (data not shown), suggesting that Sumo1 and Prdx6 interaction was ATP-mediated. Furthermor, we also found that Prdx6 was Sumoylated by Sumo1 as shown by in vitro Sumoylation assay and that UBC9 as essential as an interacting partner for the Sumo1/Prdx6-Sumoylation process. Considering the broad range of substrates including emerging extranuclear proteins in which the Sumoylation pathway is involved, it would not be surprising if Prdx6 is a target for Sumo modification. Furthermore, it has recently been reported that Sumoylation can occur by both covalent or noncovalent interaction of Sumo to Sumo-interacting motif(s) (SIMs) present in the substrate [70, 71], emphasizing that Sumoylation/deSumoylation is more complex process, and requires further exploration to understand its complexity in maintaining cellular physiology by regulating gene function.
Fig. 3.
In vitro Sumoylation assay revealed that Prdx6 was modified by Sumo1. The in vitro Sumoylation assay was performed according to the manufacturer’s protocol. Briefly, a combination of E1 enzyme, E2 (Ubc9) enzyme, Sumo1 wild-type (WT) protein and/or Sumo1 mutant protein and different concentrations of recombinant Prdx6 protein (TAT-HA-Prdx6) were mixed with 20μl reaction mixture containing Sumoylation buffer. Following incubation at 30°C for 3h, reaction product was incubated at 90°C with 2X SDS-gel loading buffer and was immunoblotted using anti-Sumo1 and anti-Prdx6 polyclonal antibodies. Concentration-dependent Sumoylation of recombinant wild-type Prdx6 protein was seen, as shown in figure, lanes 1 and 2 (* denotes the Sumoylation band). Mutant Sumo1 failed to conjugate to Prdx6, suggesting Prdx6 was Sumoylated in vitro.
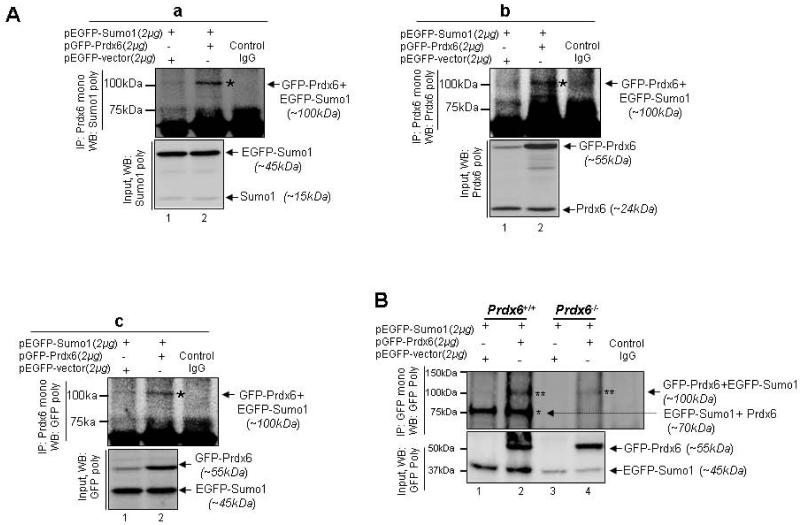
We next aimed to examine if Prdx6 is Sumoylated in vivo, we overexpressed hLECs and mLECs with Prdx6 and Sumo1, since most of Sumo target proteins are modified at very low steady state levels (a dynamic process). We first cotransfected hLECs with either pEGFP-Sumo1 plus pEGFP-Vector or pEGFP-Sumo1 plus pGFP-Prdx6 and processed the cells for immunoprecipitation (IP) using anti-Prdx6 monoclonal antibody. Probing immunoprecipitates with anti-Sumo1, anti-Prdx6 and anti-GFP polyclonal antibodies revealed a discrete slower migrating band in monoclonal Prdx6-IPs from extracts coexpressing GFP-Prdx6 and EGFP-Sumo1 (Fig. 4A, a; upper panel, lane 2; EGFP-Sumo1+GFP-Prdx6, ~100kDa) recognized with Sumo1 polyclonal, Prdx6 polyclonal (Fig. 4A, b; upper panel lane 2), and GFP polyclonal (Fig. 4A, c; upper panel lane 2) antibodies. 10% inputs were visualized as shown in Fig. 4A (lower panel). Importantly, this protein band did not appear in Prdx6-IPs performed with extracts transfected with pEGFP-Sumo1 plus pEGFP empty vector (lane 1) or IPs done with control IgG (Fig. 4A; lane 3).
Fig. 4.
Extrinsic or intrinsic Prdx6 was Sumoylated in LECs in vivo. (A) hLECs (8X105) were cotransfected with pEGFP-Sumo1 (2μg) along with pGFP-Prdx6 (2μg) or pEGFP-vector (2μg). After 48h, total cell lysates were prepared and subjected to immunoprecipitation (IP) using monoclonal anti-Prdx6 or control IgG. 10% Input and IP samples were resolved onto 4-20% SDS-PAGE and immunoblotted with anti-Sumo1 (A, a) and anti-Prdx6 (A, b) and anti-GFP (A, c) polyclonal antibodies. Sumoylated band was visualized with all three antibodies at ~100kDa (pEGFP-Sumo1 plus pGFP-Prdx6; lane 2, * denotes Prdx6 Sumoylated band). In input, Prdx6 and GFP-Prdx6 were seen with anti-Prdx6 antibody, Sumo1 and EGFP-Sumo1 with anti-Sumo1 antibody and EGFP-Sumo1 and GFP Prdx6 with anti-GFP polyclonal serum.
(B) Sumoylation assays using Prdx6+/+ and Prdx6−/− validated that Prdx6 was Sumoylated in vivo. Prdx6-deficient and Prdx6 wild-type LECs were overexpressed with pEGFP-Sumo1 (2μg) along with pGFP-Prdx6 (2μg) or pEGFP-vector (2μg) for 48h. Total cell lysates were subjected to immunoprecipitation (IP) using monoclonal anti-GFP or control IgG. 10% Input and IP samples were immunoblotted with polyclonal anti-GFP serum. Endogenous Prdx6 Sumoylated protein band could be detected with polyclonal anti-GFP serum at ~70kDa (Prdx6 plus pEGFP-Sumo1, lanes 1 and 2) in Prdx6+/+ cells only, while exogenous Prdx6 Sumoylated bands were visualized at ~100kDa (pGFP-Prdx6 plus pEGFP-Sumo1, lanes 2 and 4) in both Prdx6+/+ and Prdx6−/− cells. Absence of the endogenous Prdx6 Sumoylation band in Prdx6−/− LECs demonstrated that indeed Prdx6 was Sumoylated.
Next we also run parallel experiments in WT Prdx6+/+ and Prdx6−/− LECs if the Prdx6 is Sumoylated in these cells as observed in human LECs. We transfected Prdx6+/+ and Prdx6−/− cells with pEGFP-Sumo1 plus pEGFP-vector or pEGFP-Sumo1 plus pEGFP-Prdx6 followed by immunoprecipitation using anti-GFP monoclonal antibody as described in materials and methods. Immunoblotting of immunoprecipitates from Prdx6+/+ LECs extract using anti-GFP rabbit polyclonal antibody resulted in the two protein bands in GFP-IPs on membrane; an endogenous Sumoylated band at ~70kDa (Prdx6 plus EGFP-Sumo1: Fig. 4B; lanes 1 and 2, single *) and ectopically expressed GFP-Prdx6 and EGFP-Sumo1 conjugate at ~100kDa (Fig. 4B; lane 2, double **). Importantly, GFP-IPs from Prdx6−/− LECs extract revealed only one band, a conjugate of GFP-Prdx6 and EGFP-Sumo1 (Fig. 4B, lane 4). Taken together, data from these experiments indicated that both endogenous and exogenous Prdx6 were Sumoylated. It was surprising to notice that inputs as well as conjugates bands were faint in experiments with Prdx6-/-, although we used similar concentrations of DNA to transfect both Prdx6−/− and Prdx6+/+. We are currently unable to explain this discrepancy; it may be caused by the Redox-status of cells, wherein adverse ROS-induced aberrant Sumoylation signaling is prevalent and adversely affects cell physiology. Another possible explanation is that Sumoylated Prdx6 may be degraded quickly in Prdx6−/− LECs microenvironment, as Sumo-directed ubiquitination has recently emerged as a potential mechanism of protein degradation [66, 72]. We also consider possibility of low transfection efficiency, but microscopic examination ruled out this possibility.
Sumo1-ELISA assay revealed that endogenous Prdx6 is Sumoylated
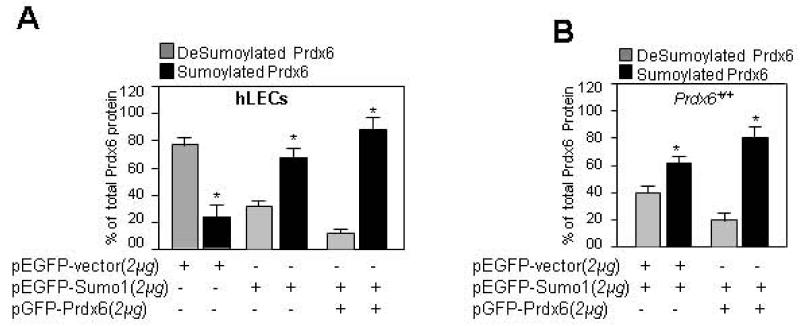
Next we analyzed levels of Sumoylated and deSumoylated forms of Prdx6 in hLECs and mLECs, to test the idea that a constant ratio between Sumoylated and deSumoylated forms of Prdx6 is genetically determined and possibly essential for normal physiology of LECs. The data may also provide a base value to assess normal functioning of LECs in relation to the Prdx6 Sumoylation/deSumoylation ratio. To measure total Prdx6 and Sumoylated Prdx6 protein, we utilized Sandwich-ELISA/Sumo-Specific ELISA assay. LECs were transiently transfected with pEGFP empty vector alone or pEGFP-Sumo1 plus pEGFP vector or pEGFP-Sumo1 plus pEGFP-Prdx6. Equal amounts of cell lysate prepared as well as normalized with GFP from these transfectants were used for assay as described in Materials and Methods. Data revealed that naturally occurring Sumoylated and deSumoylated forms of Prdx6 were present in a ratio of 1:4, respectively. Next we measured the levels of Sumoylated and deSumoylated Prdx6 in cells overexpressing Sumo1, as oxidative stress has been reported to affect Sumo conjugation aberrantly, depending on the abundance of Sumo. This experiment revealed increased Sumoylation of Prdx6 and an altered ratio between Sumoylated and deSumoylated forms of Prdx6 of approximately 1:3, (Fig. 5A). We were unable to estimate how much Sumo should be available to cells facing acute or chronic oxidative stress, but we did find that abundance of Sumo1 in cells can alter the Sumoylation status of Prdx6. Further work is required to reveal the abundance of Sumo in aging or redox cells. However, Western analysis of aging lenses, cells facing oxidative stress and Prdx6-deficient cells, (Figs. 1 and 2) showed increased Sumo conjugates, and these cells were susceptible to oxidative stress-induced cell death. Because expression and abundance of biomolecules, including Prdx6 and Sumo, are modulated during aging or stress [15, 42, 43, 73], we studied the effect of overexpression of these molecules on Sumoylation status of Prdx6. As described above, we overexpressed LECs with Prdx6 and Sumo1 and performed Sandwich ELISA assay. We found a significant increase in Sumoylated Prdx6 in pEGFP-Sumo1 plus pGFP-Prdx6 transfectants compared to pEGFP-Sumo and pEGFP-Vector ones. These experiments also indicated that abundance of Sumo and its target influenced Sumo1-specific conjugates, which in turn can affect cellular fate in cellular microenvironment.
Fig. 5.
Sensitive Sumo-ELISA/Sandwich-ELISA assays validated that both extrinsic and intrinsic Prdx6 were Sumoylated, and showed that a fraction of endogeneous Prdx6 was present in Sumoylated form. (A) hLECs were transfected with pEGFP-vector or pEGFP-vector plus pEGFP-Sumo1 or pGFP-Prdx6 plus pEGFP-Sumo1. After 48h total cell lysates were prepared and submitted to Sandwich/Sumo1-ELISA assays to check the total Prdx6 protein and Sumoylated Prdx6 protein. Sumoylated Prdx6 protein was subtracted from total Prdx6 protein, presenting as Prdx6 unSumoylated (gray bars) and Sumoylated (black bars) forms. The data represent mean ± SD from three independent experiments (*p<0.001). (B) Prdx6+/+ LECs were transfected with pEGFP-vector plus pEGFP-Sumo1 or pGFP-Prdx6 plus pEGFP-Sumo1. Total cell lysates were prepared and used to perform Sandwich/Sumo1-ELISA. Sumoylated Prdx6 protein was subtracted from total Prdx6 protein, presenting as Prdx6 unSumoylated (gray bars) and Sumoylated (black bars) forms. The data represent the mean ± SD of three independent experiments (*p<0.001 vs control).
Next we examined if the same phenomenon occurs in mLECs overexpressed with Prdx6 and Sumo1. Total cell lysate isolated from Prdx6+/+ cells transfected with pEGFP-Sumo1 plus pEGFP-vector or pEGFP-Sumo1 plus pGFP-Prdx6 were submitted for Sumo1/Prdx6-specific ELISA assay. Results in Prdx6+/+ (Fig. 5B) LECs were similar to those in hLECs, suggesting Sumoylation of Prdx6 was neither selective nor species-specific, at least in LECs. Collectively our results were consistent with in vitro observations, and revealed that Prdx6 was Sumoylated both endogenously and exogenously by Sumo1.
Sumo1 overexpression in response to oxidative stress reduced cell viability and increased ROS level in hLECs
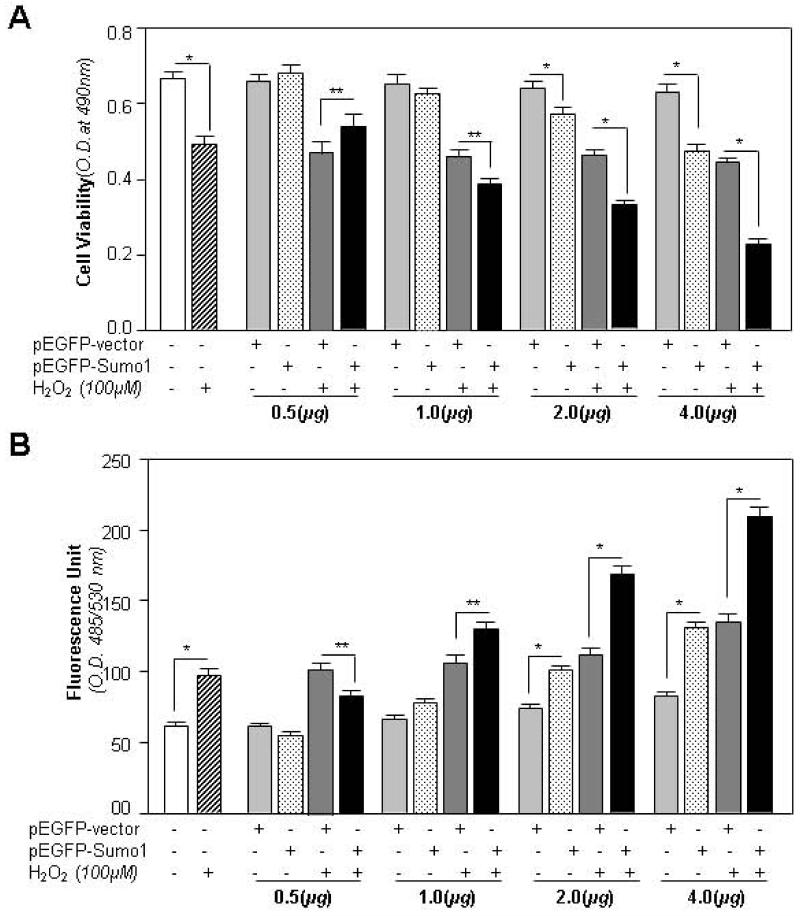
Recent reports have described internal or external environmental stress-induced aberrant Sumoylation signaling that adversely affects cellular physiology, leading to pathobiology and finally to disease [39, 74]. Because we found that an increased oxidative load directly influenced an increase in Sumo1 conjugates, we examined whether Sumo1 overexpression affects viability of cells in response to oxidative stress. After exposure of cells overexpressing Sumo1 to oxidative stress, viability assay revealed that Sumo1 overexpression did indeed negatively regulate cell viability compared to cells overexpressing pEGFP-empty vector. We also used different concentrations of pEGFP-Sumo1 to transfect cells following oxidative stress evoked by 100μM H2O2. We found that an increase in cell death (Fig. 6A, black bars vs. dark gray bars) and oxidative load (Fig. 6B, black bars vs dark gray bars) was dependent on Sumo1 concentration. Intriguingly, transfectants with empty vector (control) appeared to gain resistance (Fig. 6A, dotted bars vs light gray bars) and showed reduced oxidative load (Fig. 6A, dotted bars vs light gray bars) in response oxidative stress. These experiments indicated that aberrant Sumoylation signaling adversely affected cellular physiology. More importantly, as the levels of oxidative load were higher in cells overexpressing Sumo1 than cells containing only vector, we suggest that Sumoylation of the antioxidant protein Prdx6 may have reduced cells’ protective capacity either by reducing abundance of Prdx6 or by attenuating its transcription. Furthermore, we noticed that transfectants containing lower concentrations of pEGFP-Sumo1 (0.5μg DNA) bore reduced oxidative load and showed a tendency to increased resistance against oxidative stress (Fig. 6A and B). Fig. 6C is representative of the experiments showing photomicrographs taken after 48h of H2O2 exposure to pEGFP-Vector or pEGFP-Sumo1 transfected cells. As a whole, we concluded that cellular levels of Sumo1 were vital for normal physiological signaling and hLECs survival, while aberrant levels of Sumo1 attenuated normal physiology of cells.
Fig. 6.
Sumo1 overexpression reduced cell viability and increased the ROS level in concentration-dependent fashion at normal physiological condition, and LECs were more susceptible to oxidative stress. hLECs were overexpressed with different concentrations of pEGFP-vector or pEGFP-Sumo1 and pcDNA3 plasmid. A required amount of pcDNA3 plasmid was used to equalize DNA load during transfection. Complete media were replaced with 0.2% BSA, and cells were exposed to H2O2 (100μM). After 24h, cell viability (A) and ROS level (B) were examined. Histogram values represent mean ± SD of three independent experiments (**p<0.05; *p<0.001 vs control).
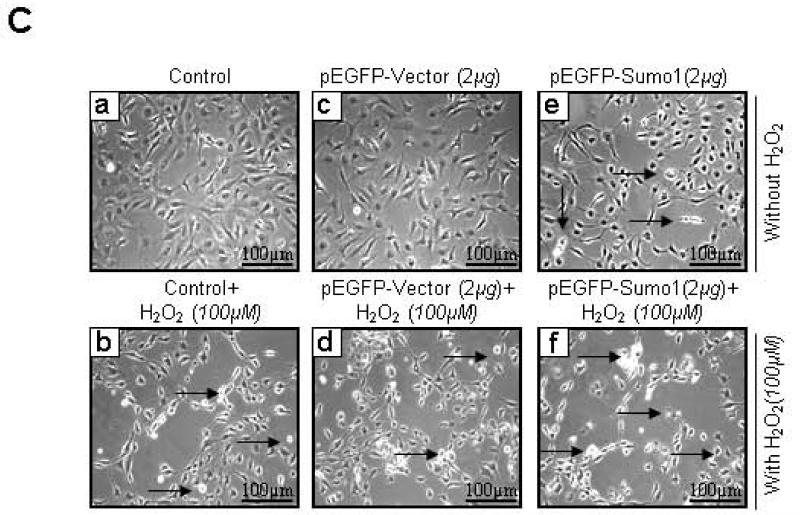
(C) Cells overexpressing Sumo1 were more susceptible to cell death in response to oxidative stress. Photomicrographs of cells showed relative cell death (white rounded cells, indicated by arrow and empty spaces where cells detached after death) in cells overexpressing Sumo1. hLECs (8 × 105) were transfected with pEGFP-vector or pEGFP-Sumo1 and exposed to H2O2 (100μM). 24h later cells were photomicrographed. a, control (no transfection); b, control+H2O2 (100μM); c, pEGFP-Vector (2μg); d, pEGFP-Vector (2μg)+H2O2 (100μM); e, pEGFP-Sumo1(2μg); f, pEGFP-Sumo1(2μg)+ H2O2 (100μM).
Overexpression of Sumo1 affected the expression of Prdx6 protein and mRNA and of its transregulator Sp1
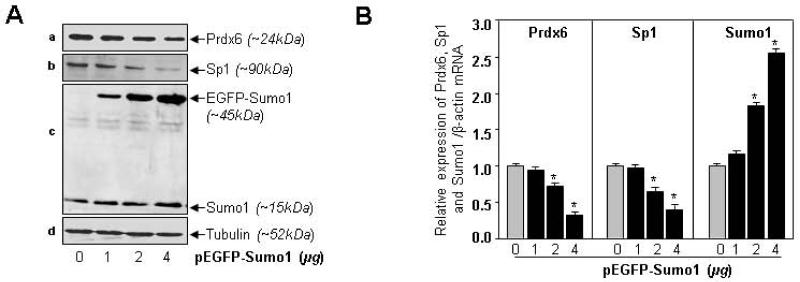
Because ROS levels were elevated in cells overexpressing Sumo1 (Fig. 6B) and these cells were highly vulnerable to oxidative stress-induced cell death, the data argued that Sumo1 may have been destabilizing/dysregulating Prdx6 activity. To address this possibility, we investigated the fate of Prdx6 and its transregulator Sp1 in cells overexpressed with variable concentrations of pEGFP-Sumo1 by assaying their protein and mRNA (Fig. 7). Expression assays, Western blot and real-time PCR by specific probes revealed that cells overexpressing Sumo1 displayed reduced expression of Prdx6 as well as its transregulator Sp1, and decline of both were linked to Sumo1 concentrations in cells (Fig. 7A, a and b vs c panel). Real-time PCR showed that expression of Prdx6 and Sp1 was not only affected at protein levels, but also that Sumo1 overexpression significantly downregulated transcripts of both, suggesting the possibility that Sumo1 dysregulated the level of Prdx6 at protein as well as transcription level.
Fig. 7.
Sumo1 suppressed expression of Prdx6 and its transregulator Sp1 protein and mRNA in dose-dependent fashion. hLECs (8X105) were transfected with different concentrations of pEGFP-Sumo1 (1, 2 and 4μg). After 48h, cells were processed for Western and real-time analysis by using specific probes to measure protein (A) and mRNA (B) expression. An inverse relation was evident between Sumo1 expression levels and Prdx6 and Sp1, suggesting aberrant expression of Sumo1 adversely affected Prdx6 and Sp1 protein and mRNA abundance. Tubulin antibody was used as an internal control. Histogram values represent mean ± SD of three independent experiments (*p<0.001 vs control).
Overexpression of Sumo1-specific proteases Senp1 upregulated Prdx6 and Sp1 expression
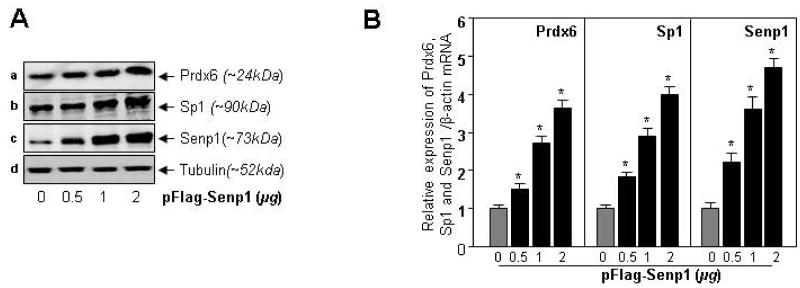
Next we examined whether cells ectopically expressed with Senp1, a Sumo1 proteases, displayed elevated expression of Prdx6 and Sp1. Protein and RNA isolated from hLECs transfected with different concentrations of pFlag-Senp1 were processed for immunoblotting and real-time-PCR. After probing to specific probe(s) of Prdx6 or Sp1, we found that Senp1 upregulated the expression of protein (Fig. 8A) and mRNA (Fig. 8B) of both molecules. Thus Prdx6 appeared to be under the control of Sumoylation and deSumoylation machinery, with effects on its protein and transcription.
Fig. 8.
Senp1 upregulated Prdx6 and Sp1 protein and mRNA expression. hLECs (8X105) were transfected with different concentrations of pFlag-Senp1 (0.5, 1 and 2). After 48h, protein and mRNA expression were measured by Western (A) and real-time PCR (B) analysis using specific probes. Senp1 increased the expression levels of both Prdx6 and its transregulator Sp1. Tubulin antibody was used as an internal control. Histogram values represent mean ± SD of three independent experiments (*p<0.001 vs control).
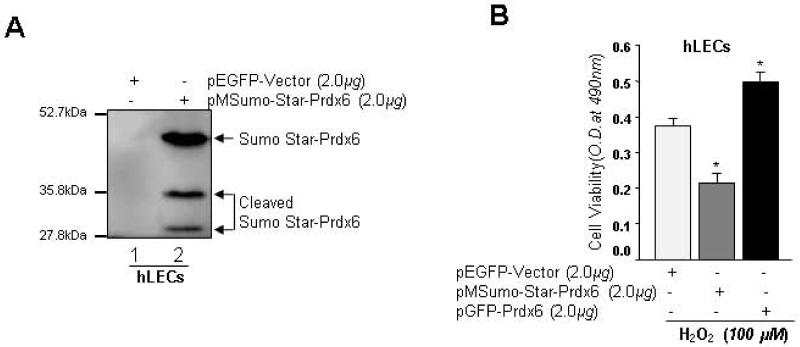
Sumoylation destabilized Prdx6 protein and its activity
Because Sumoylation is dynamical process, only a small portion of a given Sumo substrate can be detected following enrichment making it difficult to determine whether Prdx6 is degraded by Sumo1 conjugation. To resolve this issue, we used Sumo1 gene fusion technology that has been applied to other Sumoylated proteins [75-77]. Human LECs transfected with pM-Sumo-Star-Prdx6 were processed for Western analysis using anti-Prdx6 antibody. Data analysis revealed that, indeed, Sumoylation of Prdx6 enhanced its degradation (affecting its cellular stability) as seen Fig. 9A. These results suggested that Sumo1 conjugation destabilized Prdx6 activity. Next we tested the relative functionality of Sumo-Star-Prdx6 and GFP-Prdx6 in protective LECs facing H2O2-induced oxidative stress. LECs transfected with pGFP-Prdx6 showed significant resistance to oxidative stress, while LECs transfected with pMSumo-Star-Prdx6 did not show similar resistance as evidenced by viability assay. These results suggested that Sumoylation of Prdx6 destabilized Prdx6 protein and its activity.
Fig. 9.
Sumoylation of Prdx6 made it less stable and affected its protective capacity. (A) hLECs were transfected with empty vector (lane 1) and pSumo-Star-Prdx6 construct (pMSumo-Star-vector, Life sensors) (lane, 2). 48h later, cell lysates were prepared from transfectants and resolved onto SDS-PAGE and immunoblotted with anti-Prdx6 antibody. Prdx6 protein showed cleaved protein bands (lane 2). (B) hLECs were transfected with pEGFP-vector (open bar), pMSumo-Star-Prdx6 (gray bar) or pGFP-Prdx6 (black bar) and exposed to H2O2 (100μM) for 24h before being subjected to MTS analysis. Results indicated significantly reduced viability in pMSumo-Star-Prdx6 (gray bar) transfected cells, while pGFP-Prdx6 (black bar) transfected cells were protected. Histogram values represent mean ± SD of three experiments (*p<0.001 vs control).
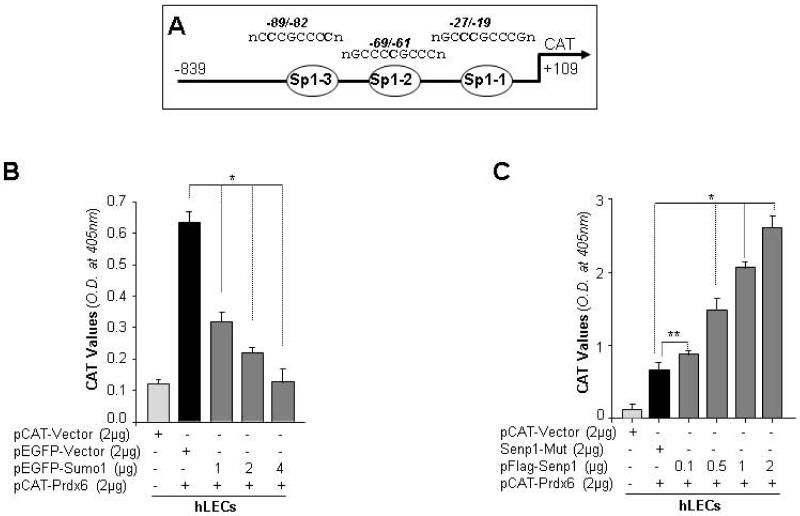
Sumo1 overexpression repressed Prdx6 transcription, while the Sumo1-proteases 1, Senp1 released the repression
Sumo modification has recently emerged as an important mechanism for transcription regulation. A number of transcription factors and coregulators are Sumoylated, which generally represses their transcription capacity, resulting in the repression of target gene transcription [21, 78, 79]. Sp1 Sumoylation leads to reduction in its transactivation capacity, is known [17, 80]. Earlier we showed that Sp1 is a regulator of Prdx6 [40]. As our data revealed Sumo1 repression of Prdx6 transcript (Fig. 7B), we studied whether suppression of Prdx6 mRNA was due to repression of its transcription. Prdx6 promoter construct-linked CAT reporter gene (-839/+109) containing all three Sp1 element sites (-839/+109) was transfected alone or with different concentrations of pEGFP-Sumo1 (Fig. 10B) or pFlag-Senp1 (Fig. 10C) in hLECs. In the presence of Sumo1, Prdx6 promoter activity significantly decreased in dose-dependent manner (Fig. 10B, gray bars vs black bars). In contrast, Senp1 overexpressing cells showed significantly increased promoter activity in a concentration-dependent manner (Fig. 10C). These data demonstrated that the reduction of Prdx6 mRNA in LECs overexpressing Sumo1 was due to repression of Prdx6 transcription.
Fig. 10.
Prdx6 gene transcription was regulated by Sumoylation and deSumoylation mechanism. (A) Schematic diagram of wild-type Prdx6 gene promoter construct (−839/+109) containing three Sp1-binding elements linked to CAT reporter plasmid vector. (B) Sumo1 significantly repressed Prdx6 promoter activity in dose-dependent manner. hLECs were transiently cotransfected with pCAT-Prdx6 (−839/+109) with different concentrations of pEGFP-Sumo1 (gray bars; 1, 2 and 4 μg). After 72h, protein was extracted and CAT-ELISA was performed to measure the effects of Sumo1 on Prdx6 promoter activity. Sumo1 downregulated (gray bars) the promoter activity. (C) Senp1 dramatically enhanced Prdx6 promoter activity in concentration-dependent fashion. hLECs were cotransfected with pCAT-Prdx6 plasmid (−839/+109) with increasing concentrations of pFlag-Senp1 (gray bars; 0.1, 0.5, 1 and 2μg). After 72h, extracted cell lysates from these transfectants were analyzed for CAT activity (shown as a histogram). The data represent the mean ± SD from three independent experiments (** p<0.05;* p<0.001). DNA concentration was maintained by cotransfecting pcDNA3 plasmid, and transfection efficiency was normalized with SEAP value as described [40, 47].
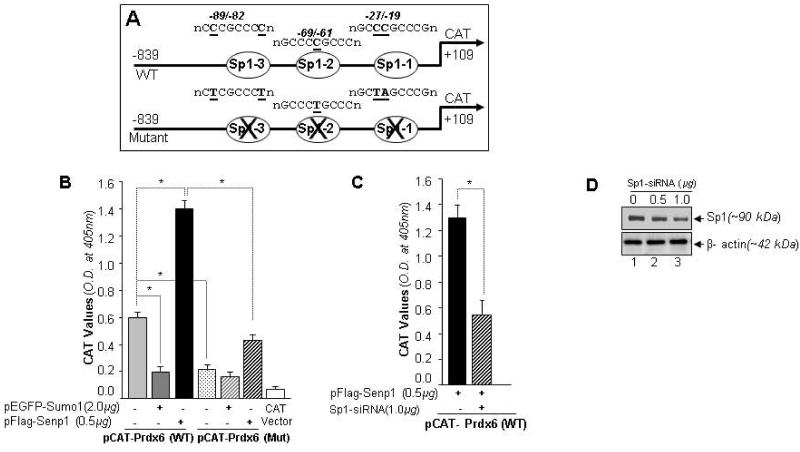
Spengler (2006) [17] reported that Sp1 is Sumoylated by Sumo1, and that Sumoylation attenuates Sp1-dependent transcription. Senp1 was found to stabilize Sp1 activity, so we predicted modulation in the transcription of Prdx6 by Sp1 Sumoylation by Sumo1 and deSumoylation by Senp1 mechanisms. To address whether repressing transcription of Prdx6 was linked to Sp1 responsive elements present in the Prdx6 promoter [40], we utilized mutant promoter disrupted at Sp1 responsive elements positioned at 27/-19, -69/-62 and -89/-82 (Fig. 11A, upper panel; diagram). In these experiments, hLECs were transfected with pCAT-Prdx6 wild-type or pCAT-Prdx6-Mutant (at all three Sp1 sites) along with either pEGFP-Sumo1 (Fig. 11B; dark gray bar) or pFlag-Senp1 (Fig. 11B; black bar), and Prdx6 promoter activity was measured. As expected, we found significantly reduced promoter activity (dark gray bar) in the presence of Sumo1 and significantly increased activity the in presence of Senp1 (black bar) in pCAT-Prdx6 wild-type construct (light gray bar). Mutant construct did not respond significantly to either Senp1 or Sumo1 (Fig. 11B). The mutant promoter showed some activity, although this was significantly lower than that of wild-type promoter and Sumo1 did not show any activity in mutant promoter. These results indicated that repression of Prdx6 promoter activity was regulated by Sumoylation/deSumoylation mechanism, and repression of Prdx6 transcription may be due to Sp1 Sumoylation, as Sumoylation of Sp1 reduces its transactivation activity [17, 80]. For further confirmation we performed Sp1 knockdown experiments; hLECs transfected with pCAT-Prdx6 along with Sp1-siRNA and Senp1 were processed to examine Prdx6 transcription activity. CAT-ELISA revealed lack of response in Prdx6 CAT promoter to Senp1, further suggesting that Sumoylation of Sp1 was a reason for repression of Prdx6 transcription (Fig. 11C).
Fig. 11.
Sumoylation and deSumoylation of Sp1 was involved in regulating Prdx6 transcription. (A) Schematic illustration of wild-type Prdx6 gene promoter construct (−839/+109) containing three Sp1-binding elements (WT) and its mutant (all three Sp1 sites disrupted using site-directed mutagenesis) linked to CAT vector. (B) Prdx6 promoter activity was down regulated in cells overexpressing Sumo1 compared to cells overexpressing Senp1. hLECs were co-transfected with wild-type Prdx6 promoter linked to CAT vector or mutated construct (at all three Sp1 sites) along with either pEGFP-Sumo1 or pFlag-Senp1. The effects of pEGFP-Sumo1 (dark gray bar) or pFlag-Senp1 (black bar) on CAT activity are shown. Empty CAT-vector served as control (open bar). We did not observe significant changes in pCAT-Prdx6 mutant coexpressed with either pEGFP-Sumo1 or pFlag-Senp1, suggesting that Prdx6 transcription was linked to Sumoylation/deSumoylation of Sp1. The transfection efficiencies were normalized using cotransfected pEGFP-vector. Data represent the mean ± SD from three independent experiments (*, p<0.001). (C) Sp1-siRNA assay confirming the involvement of Sumo1 and Senp1 in modulating Prdx6 transcriptional activity through Sp1. hLECs were transiently cotransfected with pCAT-Prdx6 reporter plasmid and pFlag-Senp1 with or without siRNA specific to Sp1, as indicated. CAT activity was monitored. The results are represented as a histogram showing that promoter activity was significantly reduced in Sp1-siRNA transfected cells compared to untransfected cells (lined bar vs black bar). Transfection efficiencies were normalized using pEGFP vector. Data represent the mean ± SD from three independent experiments (*, p<0.001). (D) Silencing of Sp1 by specific siRNA was validated by Western blotting. The membrane was striped and stained with β-actin antibody.
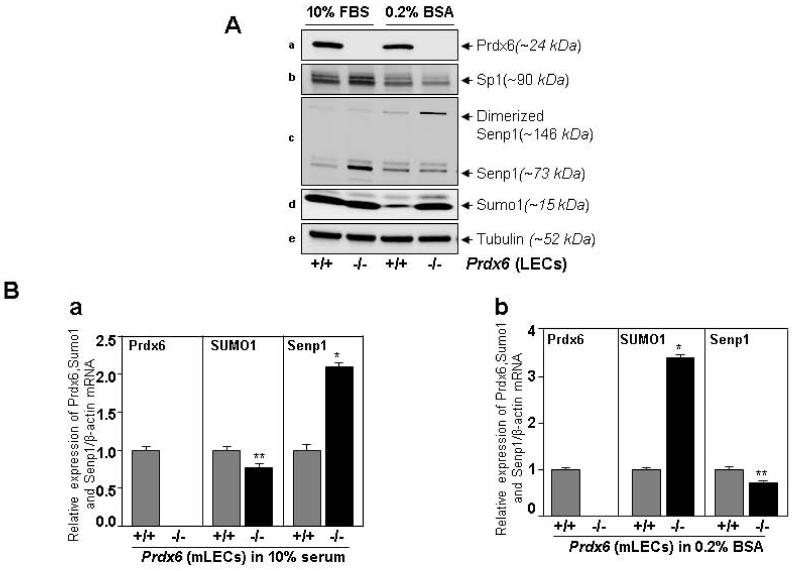
Prdx6-deficient LECs showed aberrant Sumoylation signaling resulting in increased Sumo1 and dimeric form of Senp1 (inactive form) in response to oxidative stress
A number of reports have established that Sumoylation is a critical regulator of proteins involved in cellular functions by influencing their expression and functions [2, 25, 81]. Also, recent studies have demonstrated that oxidative stress-evoked abnormal Sumoylation signaling is a cause of cell/tissue pathobiology that leads to disease [2, 15, 55, 56, 73, 74, 82-85]. Our fore mentioned current studies showed that overexpression of Sumo1 negatively regulated cellular survival by Sumoylating Prdx6 as well as its regulator Sp1 in response to oxidative stress, but the status of Sumo1 and Senp1 in redox-active cells or cells lacking antioxidant- was not clear. To address this question, we utilized Prdx6−/− LECs and measured expression levels of Senp1, Sumo1, Sp1 and Prdx6 protein and mRNA. Prdx6−/− and Prdx6+/+ LECs cultured in 0.2% BSA were examined for levels of those molecules using their corresponding probes. The level of Senp1 protein in Prdx6−/− LECs was decreased, and its dimeric form (inactive form) [29, 86, 87] was increased (Fig. 12A, panel c). Notably, the level of Sumo1 protein in these cells was dramatically increased (Fig. 12A, panel d), while level of the Sp1 was decreased (Fig. 12A, panel b), suggesting the existence of aberrant Sumoylation signaling in redox-active cells. Examining whether aberrant levels of Sumo1 and Senp1 were at protein levels only or linked to mRNA level, we found increased expression of Sumo1 mRNA and a decreased expression of Senp1 in Prdx6−/− LECs cultured in 0.2% BSA. Taken together, results showed that Senp1 and Sumo1 were abnormally regulated in redox cells, cells facing oxidative stress, and possibly aging cells, and that in turn led to oxidative stress-induced aberrant pathogenic signaling. Importantly, Prdx6+/+ and Prdx6−/− LECs cultured in 10% serum did not show any significant change in Sumo1 mRNA (although a tendency to reduced expression could be seen), while Prdx6−/− cells had significantly increased expression of Senp1 compared to Prdx6+/+ cells (Fig. 12B, a). However, we were unable to explain how Senp1 was upregulated in Prdx6−/− cells; that may require a separate line of investigation and beyond the scope of current study.
Fig. 12.
Prdx6-deficient LECs facing oxidative stress showed decreased expression and activity of Senp1, and reduced expression of Sp1. Prdx6+/+ and Prdx6−/− cells were cultured in either 10% FBS or 0.2% BSA. (A) Cell lysate was prepared and immunoblotted. Reduced Sp1 expression was observed in Prdx6−/− cell cultures in 0.2% BSA (lane 4). Also in these cells, Senp1 was found to be reduced and dimerized (lane 4) with increased expression of Sumo1. (B) RNA was isolated from Prdx6+/+ and Prdx6−/− cells cultured in either 10% FBS or 0.2% BSA and subjected to real-time PCR to measure mRNA level using specific probes corresponding to Prdx6, Sumo1 and Senp1 as indicated. The expression level of Sumo1 was increased in Prdx6−/− cells (b: Sumo1, −/−; black bar); in contrast, Senp1 expression was suppressed (b: Senp1, −/−; black bar) in cells under serum depletion stress (0.2% BSA) compared to LECs cultured at normal physiological conditions (a: Senp1, −/−; black bar and a: Sumo1, −/−; black bar). **p<0.05 and *p<0.001 showed statistically significant.
Stress-evoked aberrant Sumoylation-induced cellular injuries can be blocked by Prdx6 and Senp1
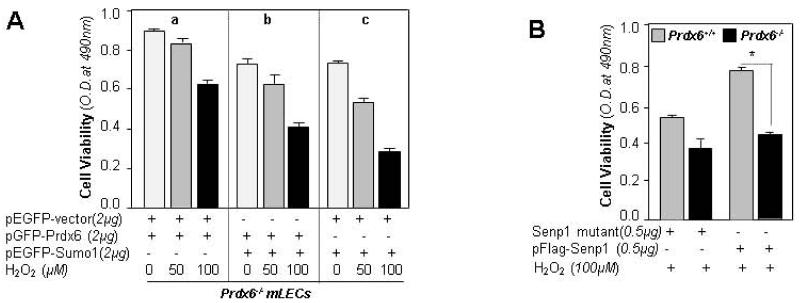
Based on results of earlier experiments (Figs. 6-12), it became evident that cells lacking Prdx6 or cells under oxidative stress displayed increased levels of Sumo1, and aberrant Sumoylation signaling dysregulates Prdx6-mediated cytoprotection by attenuating its abundance both at mRNA and protein levels. Our goals were to gain more direct evidence and measure the synergistic effects of Sumo1 overexpression and oxidative stress on cellular survival, and to determine whether an extrinsic supply of Prdx6 would attenuate the adverse process. Using Prdx6-deficient lens epithelial cells (LECs) overexpressing Sumo1 and/or Prdx6, or Prdx6 along with empty vector, with or without exposure to oxidative stress as indicated (Fig. 13A), we observed that cells cotransfected with pGFP-Prdx6 and pEGFP-Sumo1 had significantly higher survival rates (Fig. 13A; b, histograms) than cells lacking Prdx6 (Fig 13A; c, histogram) when faced with oxidative stress. Also, enhanced survival was observed in cells transfected with pGFP-Prdx6 and pEGFP-empty vector (Fig. 13A; a, histograms) which was expected, as these cells were not overwhelmed with Sumoylation signaling as were the Sumo1-transfected cells. These data argued that Prdx6 delivery can abate oxidative stress-evoked aberrant Sumoylation-mediated adverse signaling. To examine whether Senp1 overexpression protected LECs through Prdx6, we transfected Prdx6+/+ and Prdx6−/− LECs with pFlag-Senp1 or its mutant plasmid, and exposed them to oxidative stress (Fig. 13 B). Cells lacking Prdx6 had relatively little protection compared to Prdx6+/+ cells (Fig.13B, a vs b histograms), suggesting that Senp1 protected cells against oxidative stress through Prdx6. As protection was not absolute, we surmised that another type of signaling negatively regulated death pathways. Taken together, the data revealed that the aberrant Sumoylation signaling causing increased cell death due to loss or reduced expression of Prdx6 was blocked by an extrinsic supply of Prdx6, paving the way for development of Prdx6 expression-based therapeutics for blocking or preventing the progression of oxidative stress and age-associated pathobiology of cells/tissues.
Fig. 13.
Transfection and viability experiments of Prdx6-deficient LECs with Sumo1 or Senp1 or Prdx6 showed that Sumoylation of Prdx6 attenuated its protective potential. (A) Prdx6−/− cells were transfected with pGFP-Prdx6 plus pEGFP-vector (group, a) or pGFP-Prdx6 plus pEGFP-Sumo1 (group, b) or pEGFP-Vector plus pEGFP-Sumo1 (group, c). 24h later cells were exposed to H2O2 and subjected to cell viability assay as indicated. A comparison between the groups (a vs b vs c) revealed that Prdx6 was vital for cell survival and Sumoylation of Prdx6 attenuated its protective potential significantly (compare black bars in groups a, b and c). OD values were normalized with GFP OD value. (B) Prdx6-deficient cells overexpressing Senp1 revealed that Senp1 acted through Prdx6 in protecting LECs by the deSumoylating mechanism. Prdx6+/+ and Prdx6−/− cells were transfected with Senp1 mutant or pFlag-Senp1 as shown. 24h later serum-depleted LECs were exposed to H2O2 (100μM) and cell viability was measured (Prdx6−/−, black bars vs Prdx6+/+, gray bars). Data represent the mean ± SD from three independent experiments (*, p<0.001).
Discussion
Increased expression of ROS in cells in response to reduced expression and activity of antioxidant proteins or environmental/cellular stress can result in pathogenic signaling and cellular insult. Such insults are linked to the etiology and progression of many diseases, including age-related degenerative disorders, cataractogenesis, macular degeneration, neurodegenerative disorders, diabetes, cancer and so forth [41-43, 55, 56]. However, a physiological level of ROS has been shown to be involved in various cellular processes [40, 42, 61, 88, 89]. Thus ROS can either promote survival or initiate death signaling, depending upon its cellular concentration and cell background. The eye is continuously exposed to environmental stresses. These stressors induce ROS, which damages macromolecules and can alter protein integrity (by altering posttranslational modification) and function, in turn leading to etiopathology and disease states [88, 90].
Sumoylation is emerging as a critical posttranslational modification of nuclear and extranuclear proteins. It affects cellular functions by regulating protein activity [15, 55, 56, 62, 91-94]. Recently, several reports have documented that the Sumoylation process is altered by stressors such as UV radiation, heat stress and oxidative stress. Global changes in Someones have been shown under different stresses in the cellular microenvironment and in model organisms [59]. As eukaryotic gene expression is driven by a complex series of events, we believe that stress-induced Sumoylation may globally affect the LEC/lens Someones, as aging eyes are under oxidative pressure due to a decline in antioxidant defense molecules like Prdx6 [40, 42-44]. By using aging LECs/lenses to study the effects of aging and oxidative stress, and Prdx6-deficient LECs as a model for aging cells, we found that Sumoylation of most proteins was increased and the increases were linked to increased levels of ROS (Figs. 1 and 2). The key novel observation was that Prdx6 is target for Sumo1 modification. The global changes in Sumoeome were associated with Prdx6 deficiency and an increase in ROS levels and aberrant Sumoylation that results in cell damage. Using Prdx6-deficient cells, we showed that aging cells became prone to cell death due to overwhelming abnormal Sumoylation signaling (Figs. 1 and 2). It was intriguing to observe that the level of Sumo1 conjugates was significantly higher in Prdx6−/− cells, and these were much more susceptible to oxidative stress than were Prdx6+/+ cells (Fig. 2), suggesting a pivotal role for Prdx6 in abating oxidative stress-evoked aberrant Sumoylation signaling, at least in eye lens.
Recently, extranuclear roles of Sumoylation have been documented (Stephane martin (2007). Sumoylation regulates cellular transduction by modifying growth factor-mediated signaling and altering protein trafficking and protein-protein interaction [37]. Sumoylation also controls cell fate by enhancing protein stability or reducing protein degradation [3, 9]. Sumoylation has been shown to have a role in antioxidant protein modification [38, 39], leading to redox status of cells. The redox effect on the conjugation of a Sumo to its target substrate can lead to aberrant expression of Sumo. Since Sumo itself is a limiting factor for conjugation [62], its overexpression or underexpression influences the status of target substrate Sumoylation globally. Global changes in the Sumo proteome have been observed under diverse stimuli [59]. Modulation of Sumoylation status by stressors was initially observed by Saitoh and Hinchey [65], who reported that environmental stress, oxidative stress, heat shock, or osmotic stress increases Sumoylation. We found that Prdx6 is a substrate for Sumo1 conjugation (Figs. 3, 4 and 5), and importantly, a fraction of Sumoylated form of Prdx6 is present in LECs (Figs. 3 and 4). Moreover, a protein can be a target for mono- or multi-Sumoylation, depending upon the presence of a Sumo1 recognition site within the target protein. Sites are often part of a ΨKxD/E consensus motif which is recognized by the UBC9, a Sumo conjugation enzyme [95, 96]. Like ubiquitin, Sumo can attach its own lysine (K) residue(s) and can form polySumoylation. Our finding that Prdx6 is monoSumoylated was revealed by in vitro and in vivo Sumoylation assays. The molecular weight of endogenous Prdx6 on SDS-gel is ~24kDa to ~28kDa [40-44, 68]. We identified the two protein bands of high molecular weight. Analysis showed that these bands were the Sumoylated form of Prdx6 approximately ~70kDa [(endogenous Prdx6 plus EGFP-Sumo1 (~45kDa)], and 100 kDa [ectopically expressed GFP-Prdx6 (~55kDa) plus EGFP-Sumo1 (~45kDa)]. Our data showed that Prdx6 was modified by Sumo1, and Sumo1 lacks a lysine residue in consensus motif; thus, Sumo1 will not form poly chains [97], and Prdx6 cannot be polySumoylated. We propose that Sumoylation of Prdx6 is a constitutive event in regulating its function, and during stress aberrant Sumoylation signaling alters the expression and function of Prdx6. Indeed, our data revealed that overexpression of Sumo1 negatively regulates LEC survival in dose-dependent fashion by downregulating Prdx6 and its transactivator, Sp1 (Figs. 6, 7 and 8). We also found that overexpression of Sumo1 negatively regulated cell fate and downregulated the expression of both Prdx6 and Sp1 protein and mRNA. Notably, the down regulation was dependent on the concentration of Sumo1 in cells (Fig. 7). In contrast, Senp1 expression reversed the process (Fig. 8). We believe the loss of Prdx6 in cells overexpressing Sumo1 may be associated with Sumo1-mediated modulation of Prdx6 at posttranslational or transcription levels or both.
Moreover, the repression of mRNA levels argues the involvement of transcription repression as supported by downregulation of Sp1 in these cells. However, whether reduce abundance of Prdx6 was also associated with its degradation due to Sumoylation was not clear. Our experiments revealed Prdx6 Sumoylation also affects its stability. Until recently, Sumo and ubiquitin were known to have opposing effects by occupying the same lysine, thus making the proteins resistant to degradation through the ubiquitylation pathway [10, 98, 99]. Conversely, recent investigations demonstrated that Sumoylation is a directional signal for ubiquitylation and ubiquitin-dependent degradation [100-104]. We think the decrease in level of Prdx6 may also be associated with Sumoylation-mediated ubiquitin-dependent degradation. Our current study revealed that Sumo1 conjugated Prdx6 was degraded (Fig. 9A) and, importantly, Sumoylation of Prdx6 blunted its protective potential (Fig. 9B). Because Sumoylation is a dynamical process, and a fraction of protein is Sumoylated, determining whether Prdx6 Sumoylation leads to degradation would be difficult. To resolve this issue, we utilized Sumo-fusion strategy and prepared Sumo1-Prdx6 fusion cDNA eukaryotic construct. Such a strategy has been successfully applied previously [75, 77]. Our experiment revealed that Prdx6 was degraded, and we posited that the degradation might be a reason for the lower cellular abundance of Prdx6. However, it was not clear whether Sumo-1-Prdx6 was able to recruit the enzymes responsible for ubiquitin-mediated degradation; the question needs further investigation. SumoPlot analysis [25] disclosed that Prdx6 contained Sumo1 binding site, but it was diversified from consensus motif (non-consensus motif). We have started pursuing this line of research to define Sumo1 motif(s) in Prdx6. An extended Sumo consensus motif has been suggested and identified as a functional motif for Sumos [36, 105, 106]. However, our current study clearly revealed that Prdx6 is a target for Sumo1, indicating that deficiency of Prdx6 is a cause of its own aberrant Sumoylation due to increased levels of oxidative load. Therefore we believe that maintaining cellular levels of Prdx6 can abate ROS-evoked aberrant Sumoylation signaling.
Apart from the destabilization of Prdx6 by Sumoylation, Fig. 10B shows that the process also affects the transcription of Prdx6 by affecting the abundance of Prdx6’s activator Sp1. Our transactivation-based study demonstrated reduced transcription of Prdx6 promoter containing Sp1 responsive elements, which was directly associated with concentrations of Sumo1, while transfection of Senp1 restored Prdx6 transcription. The involvement of Sumo1 in repressing Prdx6 transcription through Sp1 Sumoylation came through our mutation of Sp1 sites in Prdx6 promoter and Sp1-siRNA/Senp1 experiments in which Senp1 failed to modulate Prdx6 promoter activity. The study revealed that Prdx6 was transcriptionally repressed by aberrant Sumoylation signaling involving Sp1. Sp1 is a known target for Sumo1, and Sumoylation decreases its transactivation capacity [17]. Thus the repression of Prdx6 gene can result from destabilization and degradation of Sp1 by Sumo1 signaling. Previous studies have shown that constitutively Sumo-modified Sp1 is a weak transactivator [17, 104]. When transactivation assay was followed by overexpression with Senp1, we noticed a significant increase in Prdx6 promoter activity. Our finding pointed to ROS-evoked aberrant Sumoylation signaling in LECs as a major culprit, and indicated potential rescue for the cells by expressing Senp1 or Prdx6. However, at normal physiological condition Sumo signaling is a regulatory mechanism for Prdx6. Our studies provide the proof of concept that cells in redox state or facing oxidative stress undergo an abnormal Sumoylation process due to overexpression of Sumo1 and reduced expression and inactivation of Senp1 (Fig. 12A and B). In eukaryotic cells environmental factors/oxidative stresses modify the Sumoylation process by increasing the cellular abundance of Sumo1 mRNA and protein and inactivating expression of Senp1 [15, 59, 107, 108]. Intriguingly, we observed that the level of Senp1 expression was dramatically increased in Prdx6−/− cells cultured in complete DMEM medium (Fig. 12), suggesting the involvement of serum component(s) in upregulation of Senp1. We posit that both redox-active state of Prdx6−/− cells and serum factor(s) may be responsible for increased expression of Senp1. It has been reported that Senp1 gene promoter bears androgen response element (ARE), and androgen receptor (AR) functionally binds and activates its transcription [109]. We think that AR activated by serum androgen initiated Senp1 transcription in Prdx6−/− cells, which may be more sensitive than wild-type Prdx6 cells. Furthermore, hypoxic stress has been implicated in upregulation of Senp1 transcription [110]. Hypoxia is known to be an inducer of oxidative stress. We surmise that increased expression of Senp1 can also be associated with increased oxidative load of Prdx6-deficient LECs. However, how the expression of Senp1 is increased in Prdx6−/− LECs warrants further investigation.
One of the salient conclusions of the current study was that Prdx6 is essential for protecting LECs during oxidative stress-evoked aberrant Sumoylation signaling. This was evidenced by the observation that Prdx6-deficient cells overexpressing Sumo1 showed a dramatic decrease in viability during oxidative stress, while LECs overexpressing Prdx6 had resistance against the same stress (Fig. 13A). Prdx6-deficent LECs overexpressing Senp1 did not have cytoprotection similar to that observed in wild-type Prdx6+/+ LECs (Fig. 13B), indicating that Prdx6 is vital for attenuating ROS-evoked Sumoylation-mediated adversity.
In summary, by utilizing Prdx6-deficient LECs derived from Prdx6 knockout eye lenses and human LECs facing oxidative stress, we demonstrated for the first time that Prdx6 is Sumoylated. We showed that aberrant Sumoylation signaling in response to oxidative stress caused repression of Prdx6 transcription as well as reduced abundance of Prdx6. As a whole, our current investigation demonstrates for the first time that aberrant Prdx6 Sumoylation in response to oxidative stress attenuates Prdx6 activity and perturbs the growth and survival of LECs. In contrast, Prdx6 overexpression attenuates ROS-evoked aberrant Sumoylation pathways by optimizing ROS expression and restoring cellular survival signaling. We believe that the findings will bring new perspectives for understanding the molecular mechanisms of age-related disorders, and will open new pathways in the treatment of many pathological disorders associated with environmental and cellular stresses.
Materials and methods
Cell culture
Human Lens Epithelial Cells (hLECs) (a kind gift of Dr. Venkat N. Reddy, Eye Research Institute, Oakland University, Rochester, MI, USA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 15% fetal bovine serum (FBS; Atlanta Biologicals, Inc., Flowery Branch, GA, USA), 100μg/ml streptomycin, and 100μg/ml penicillin in 5% CO2 environment at 37°C as described previously [111, 112]. Cells were harvested and cultured in 96, 24, 48 or 6 well plates and 100 mm petri dishes according to the requirements of the experiment(s).
Generation and validation of LECs isolated from lenses of Prdx6−/− and Prdx6+/+ mice
All animal experiments followed the recommendations set forth in the Statement for the Use of Animals in Ophthalmic Research by the Association for Research in Vision and Ophthalmology. Animal studies were approved by the University of Nebraska Medical Center, Omaha, NE, USA. LECs isolated from Prdx6-targeted mutants (Prdx6−/−) and wild-type (Prdx6+/+) mice were generated and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% FBS (Atlanta Biologicals, Inc., Flowery Branch, GA, USA) as described earlier [42]. Prdx6−/− 129/Sv mice were generated at Harvard Medical School under the supervision of Dr. David R. Beier. For the present study, we used Prdx6−/− mutant mice of pure 129 background, and, as controls, wild-type 129/Sv inbred mice of the same sex and age (Prdx6+/+). All animals were maintained under specific pathogen-free conditions in an animal facility. LECs were isolated from mice of identical age, and Western analysis was carried out to confirm the presence of αA-crystallin [42], a specific marker of LECs. Cells from 3-5 passages were used for the experiments.
Western blot analysis and antibodies
Total cell lysates were prepared in ice-cold radio immunoprecipitation assay (RIPA) lysis buffer, as described previously [40]. Equal amounts of protein samples were loaded onto 10%, 12% or 4-20% SDS PAGE gel, blotted onto PVDF membrane (Perkin Elmer, Waltham, MA, USA), and immunostained with primary antibodies at the appropriate dilutions. The following antibodies were used: Prdx6 monoclonal (Lab Frontier, Seoul, Korea), Prdx6 monoclonal (IP grade, Abcam, Cambridge, MA, USA), Prdx6 polyclonal (sc-134478; Santa Cruz Biotechnologies, Santa Cruz, CA, USA), Sumo1 monoclonal (sc-5308; Santa Cruz Biotechnologies, Santa Cruz, CA, USA), Sumo1 polyclonal (sc-9060; Santa Cruz Biotechnologies, Santa Cruz, CA, USA), GFP monoclonal (sc-69779; IP grade, Santa Cruz Biotechnologies, Santa Cruz, CA, USA), GFP polyclonal (Invitrogen, USA), Sp1 monoclonal (sc-17824; Santa Cruz Biotechnologies, Santa Cruz, CA, USA) and Senp1 monoclonal (sc-271360; Santa Cruz Biotechnologies, Santa Cruz, CA, USA). Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (anti-mouse, sc-2055 and anti-rabbit, sc-2054; Santa Cruz Biotechnologies, Santa Cruz, CA, USA). Specific protein bands were visualized by incubating the membrane with luminal reagent (sc-2048; Santa Cruz Biotechnologies, Santa Cruz, CA, USA) and the images were recorded with FUJIFILM-LAS-4000 luminescent image analyzer (FUJIFILM Medical Systems Inc., USA). To ascertain comparative expression and equal loading of the protein samples, the membrane stained earlier was stripped and re-probed with β-actin antibody (Sigma-Aldrich, St. Louis, MO, USA) or Tubulin monoclonal (Abcam, Cambridge, MA, USA).
Quantitation of intracellular ROS level
Intracellular ROS level was measured by use of fluorescent dye dichlorofluorescin diacetate (H2-DCF-DA), a nonpolar compound that is converted into a polar derivative (dichlorofluorescein) by cellular esterase after incorporation into cells [42, 50]. Human or mouse LECs were cultured in 96 well plates for 24h with DMEM having 15 or 10% FBS, then exposed to H2O2 at different concentrations for different time intervals determined by requirements of the experiment. hLECs were transfected with pEGFP-vector or pEGFP-Sumo1 and treated or untreated with H2O2. On the final day of the experiment, the medium was replaced with Hank’s solution containing 10mM H2-DCF-DA dye and cells were incubated. Following 30 min of incubation at room temperature, intracellular fluorescence was detected with excitation at 485 nm and emission at 530 nm as measured by a Spectra Max Gemini EM (Mol. Devices, Sunnyvale, CA).
Cell viability assay
A colorimetric MTS assay (Promega, Madison, WI, USA) was performed as described earlier [42, 50]. This assay of cellular proliferation/viability uses 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2 to 4-sulphophenyl) 2H-tetrazolium salt. When added to medium containing viable cells, MTS is reduced to a water-soluble formazan salt. The A490 nm value was measured after 4h with an ELISA reader. Results were normalized with absorbance of the untreated control(s).
TAT-HA-Prdx6 recombinant protein Purification
A full-length cDNA of Prdx6 was isolated from a human LEC cDNA library using Prdx6-specific sense (5′-GTCGCCATGGCCGGAGGTCTGCTTC-3′ contained NcoI site) and antisense primer (5′-AATTGGCAGCTGACATCCTCTGGCTC-3′). The PCR products were purified by preparative agarose gel electrophoresis. The purified products were ligated into a TA-cloning vector (Invitrogen, Carlsbad, CA, USA) and then transformed into a competent cell, and the plasmids of selected colonies were purified. The purified TA vector containing Prdx6 cDNA was digested with NcoI and EcoRI and then subcloned into a pTAT-HA expression vector (a kind gift of Dr. S. F. Dowdy) that had been digested with the same restriction enzymes. Recombinant protein was purified using QIAexpress® Ni-NTA Fast Start kit column (Qiagen Inc., Valencia, CA, USA). The host Escherichia coli BL21 (DE3) was transformed with pTAT-HA-Prdx6 and the transformants were selected on a Luria broth (LB) plate with ampicillin. The selected colonies were cultured in 10ml LB medium containing ampicillin at 37°C with shaking at 200 rpm overnight. After incubation, 10ml of the overnight cultures were combined with 250ml of pre-warmed media (with ampicillin), and then was grown at 37°C with vigorous shaking until an OD600=0.6-0.8, isopropylthiogalactoside (IPTG) was added to a concentration of 1mM and the incubation was continued for 4-5h. Cells were harvested by centrifugation at 4000g for 20min. Pellets were suspended in 10ml of lysis buffer (50mM NaH2PO4, 50mM NaCl and 10mM imidazole, pH 8.0) containing lysozyme and Benzonase® Nuclease and incubated for 30min on ice. The suspension was then centrifuged at 14000g for 30min. Supernatant was added to the Ni-NTA fast start column and allowed to drain before washing twice with 4ml of wash buffer (50mM NaH2PO4, 50mM NaCl, and 20mM imidazole, pH 8.0), followed by elution with 1ml elution buffer (50mM NaH2PO4, 50mM NaCl, and 250mM imidazole, pH 8.0). This purified protein can be either used directly for protein Sumoylation, or aliquoted and stored frozen in 10% glycerol at −80°C for further use.
Construction of DNA Plasmid
For eukaryotic expression, a full length of Sumo1 cDNA was subcloned into pEGFP-C1 vector [25]. The coding region of Sumo1 was amplified by PCR from human lens cDNA library using forward (5′-CCGTCGACATGTCTGAC CAGGAG-3′) and reverse primer (5′-TCGGATCCGT TTTGAACACCACA-3′) with restriction enzyme sites, SalI and BamHI. The PCR product was digested and ligated into pEGFP-vector. pFlag-Senp1 was a generous gift from Dr. E. Yeh (University of Texas M.D. Anderson Cancer Center, Houston, TX, USA). All the transfection experiments were carried out either with Superfactamine Reagent (Invitrogen, Carlsbad, CA, USA) or by using the Neon Transfection system (Invitrogen, Carlsbad, CA, USA).
In vitro Sumoylation Assay
Purified recombinant TAT-HA-Prdx6 was incubated with E1, E2 and Sumo1 protein for 3h at 30°C according to the manufacturers’ protocol (SUMOlink™ in vitro SUMO-1 Kit, Catalog no. 40120, Active Motif, Carlsbad, CA, USA). The reaction was stopped by adding an equal amount of 2X SDS-PAGE loading buffer, and Western analysis proceeded. Sumoylation bands were visualized by anti-Prdx6 antibody or anti-Sumo1 antibody.
In vivo Sumoylation assay
hLECs or Prdx6+/+ or Prdx6−/− were transfected with either pEGFP-vector and pEGFP-Sumo1 or pEGFP-Sumo1+ pEGFP-vector and pEGFP-Sumo1+pGFP-Prdx6. After 48h, total cell lysates were prepared in IP Lysis/Wash buffer (0.025M Tris, 0.15M NaCl, 0.001M EDTA, 1% NP-40, 5% glycerol, pH7.4 plus 5μM MG132 and 30μM NEM (N-ethylmaleimide) added) provided in the Pierce® Classic IP Kit (Catalog No. 26146) as per manufacturer’s instructions. Total cell lysates were incubated with 4μg of monoclonal anti-Prdx6/anti-GFP serum/1000 μg of protein in IP lysis buffer provided in the IP kit (Pierce, Rockford, IL, USA), and were rotated at 4°C overnight. That was followed by the addition of 20 μl of Protein A/G plus Agarose bead and further rotation for 4h at 4°C. The immunoprecipitates were collected by centrifugation and washed several times with wash buffer and 1X conditioning buffer before being boiled in SDS-sample buffer. 10% Input and IP samples were resolved on 4-20% SDS/PAGE and analyzed by Western blotting using monoclonal anti-Prdx6, anti-Sumo1 or anti-GFP sera.
Sumo1/Sandwich-ELISA (Enzyme Linked Immunosorbent Assay)
A total Prdx6 protein and its Sumoylated form was performed by sandwich-ELISA (Abnova, Taipei City, Taiwan) and EpiQuik in vivo universal protein Sumoylation assay kit following the companies’ protocols. Briefly, in sandwich-ELISA assay, hLECs or Prdx6+/+ cells were transfected with pEGFP-vector, pEGFP-Sumo1 or pEGFP-Sumo1+pGFP-Prdx6 construct. After 48 h, total cell lysates were prepared and equal amounts of protein were loaded in ELISA plate well coated with Prdx6 polyclonal antibody followed by incubation with monoclonal anti-Prdx6 antibody. After incubation with goat anti-mouse-HRP conjugated secondary Ab, OPD substrate solution was added for color development and O.D. was recorded at 490nm.
As mentioned above, cell extracts from transfectants or controls were collected in modified RIPA buffer, and Sumoylated Prdx6 was detected by ELISA using an EpiQuik in vivo universal protein Sumoylation assay kit (Epigentek, Farmingdale, NY, USA). Equal amounts of proteins from the cell extracts were added to the strip wells, which were percolated overnight with either anti-Prdx6 antibody or control IgG. They were then incubated in blocking buffer for 45min, washed three times and incubated with Sumo assay buffer for 1h at room temperature. After three washes, Sumo antibody was added and the proteins were incubated for 15 min at room temperature. Following color development by a Sumo detection system, absorbance was measured at 450nm using an ELISA plate reader. To obtain deSumoylated Prdx6, we calculated total and Sumoylated Prdx6 protein and subtracted the Sumoylated Prdx6 protein from total Prdx6 protein.
Quantitative real-time PCR
Quantitative real-time PCR was performed using LightCycler® 480II as described earlier [50, 113]. Primers shown in table 1 were purchased from Roche Applied Sciences (Indianapolis, IN, USA). The comparative Cp method was used to calculate relative fold expression levels using LightCycler ® 480 software, release 1.5.0 SP3. The Cps of target genes was normalized to the levels of β-actin as an endogenous control in each group.
Table 1.
| Gene Name | Primer Sequence |
|---|---|
| Prdx6(Human) | Forward: 5′-GCATCCGTTTCCACGACT -3′ Reverse: 5′-TGCACACTGGGGTAAAGTCC-3′ |
| Sp1(Human) | Forward: 5′-CCTGGATGAGGCACTTCTGT-3′ Reverse: 5′-GCCTGGGCTTCAAGGATT-3′ |
| Sumo1(Human) | Forward: 5′-AAGCCACCGTCATCATGTCT-3′ Reverse: 5′-TTATCCCCCAAGTCCTCAGTT-3′ |
| Senp1(Human) | Forward: 5′-TTCCTCGCTGATGACAACTG-3′ Reverse: 5′-AGTGAGTCCATAAGTAGGATACAAGGT-3′ |
| β-actin(Human) | Forward: 5′-CCAACCGCGAGAAGATGA-3′ Reverse: 5′-CCAGAGGCGTACAGGGATAG-3′ |
| Prdx6(Mouse) | Forward: 5′-TTTCAATAGACAGTGTTGAGGATCA -3′ Reverse: 5′-CGTGGGTGTTTCACCATTG-3′ |
| Sumo1(Mouse) | Forward: 5′-GTGAATCCACGTCACCATGT -3′ Reverse: 5′-TCGCCTAAGTCCTCAGTTGAA -3′ |
| Senp1(Mouse) | Forward: 5′-GCTTCCAGCCCTACGTTCTT -3′ Reverse: 5′-CGAGCTCGAGAGTCATAAACG -3′ |
| β-actin(Mouse) | Forward: 5′-CTAAGGCCAACCGTGAAAAG-3′ Reverse: 5′-ACCAGAGGCATACAGGGACA-3′ |
pM-Sumo-Star-Prdx6 Eukaryotic expression construct
Because it was difficult to detect the effect of Prdx6 Sumoylation on its stability and protective efficacy, we prepared Sumo1-Prdx6 fusion plasmid by cloning a full cDNA of Prdx6 into pM-Sumo-Star eukaryotic expression vector between BsmB1 and XhoI sites (Life Sensors) following the company’s instructions. We prepared Prdx6 forward (5′-CGTCTCTAGGTATGCCCGGAGGTCTGCTTCTCG-3′) and Reverse (5′-CTCGAGTCATCACAGCACCAGCTTCTCCAA-3′) primers containing BsmB1 and XhoI restriction sites, respectively. Products were digested with appropriate enzymes and cloned into the pM-Sumo-Star vector [114]. pM-Sumo-Star plasmid was transformed and plasmid was isolated [42, 43]. Following sequence verification plasmid was used for transfection assays.
Construction of Prdx6 Promoter-Chloramphenicol Acetyltransferase (CAT) Reporter Vector
The 5′-flanking region (-839 to +109 bp) was isolated from mouse genomic DNA and sequenced. A construct of −839 bp was prepared by ligating it to basic pCAT vector (Promega Corporation, Madison, WI, USA) using the SacI and XhoI sites. The plasmid was amplified and used for the CAT assay. Primers were as follows: 5′CTTCCTCTGGAGCTCAGAATTTAC-3′; and 5′-CAGGAACTCGAGGAAGCGGAT-3′.
Site-directed mutagenesis (SDM)
PCR-based site-directed mutagenesis was carried out with the QuikChange™ site-directed mutagenesis kit (Invitrogen, Carlsbad, CA, USA), following the company’s protocol. We made mutations in Sp1 binding sites (Mutant 1: CC changed to TA; Mutant 2: C changed to T; Mutant 3: C changed to T. Briefly, the double-stranded Prdx6 promoter construct (-839/+109) was used as template DNA with a pair of complementary primers used to mutate the Prdx6 promoter construct with PCR. The primers used for mutation are as follows:
Sp1-Mut-1for’5′-GATCTAGGTCTCCGCAGGAGCTAGCCCGCTGCTCACTG-3′;
Sp1-Mut-1rev’5′-CAGTGAGCAGCGGGCTAGCTCCGGCGGAGACCTAGATC-3′;
Sp1-Mut-2for’5′-GCCCTGCCCTAGCCCTGCCCACTCGGCCAGCACTGATC-3′;
Sp1-Mut-2rev’5′-GATCAGTGCTGGCCGAGTGGGCAGGGCTAGGGCAGGGC-3′;
Sp1-Mut-3for’5′-GCCGCCAGACTCGCGGTCGCCTCACCTCGCCCTAGC-3′;
Sp1-Mut-3rev’5′-GCTAGGGCGAGGTGAGGCGACCGCGAGTCTGGCGGC-3′.
Cotransfection and promoter activity assay
The CAT assay was performed using a CAT-ELISA kit (Roche Diagnostic Corporation, Indianapolis, IN, USA). Cells were cotransfected with reporter constructs (pCAT-Prdx6 wild-type or pCAT-Prdx6-mutant construct at all Sp1 sites) along with pEGFP-Sumo1 or pFlag-Senp1, and Prdx6 transcriptional activity was measured. To observe the effect of Sumo1 and Senp1 on Sp1’s regulation of Prdx6 transcription activity, pCAT-Prdx6-mut (mutant at all sp1 (1+2+3) sites) and Sp1-siRNA construct were used. Sp1-siRNA construct was made as described earlier [16]. After 72h of incubation, cells were harvested, extracts were prepared and the protein was normalized. CAT-ELISA was performed to monitor CAT activity in accordance with the manufacturer’s instructions. A405 was measured using a microliter plate ELISA reader. Transactivation activities were adjusted for transfection efficiencies using GFP values or SEAP values cotransfected during transfection assays.
Statistical analysis
Data are presented as mean ± S.D. of the indicated number of experiments. Data were analyzed by Student’s t-test when appropriate. A p value of **, p< 0.050 and *, p< 0.001 was defined as indicating a statistically significant difference.
Acknowledgements
Grant supports for this work provided by the National Eye Institute, NIH (EY013394 and EY017613) (to DPS) and Bridge Funding Support, University of Nebraska Medical Center, and also by Research for Preventing Blindness (RPB) are gratefully acknowledged.
Abbreviations
- SUMO
small ubiquitin-like modifier
- GFP
green fluorescent protein
- NEM
N-ethylmaleimide
- Senp
Sumo-specific proteases
- Prdx6
peroxiredoxin 6
- Sp1
Specificity protein 1
Reference
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279:27233–8. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 4.Martin SF, Hattersley N, Samuel ID, Hay RT, Tatham MH. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Anal Biochem. 2007;363:83–90. doi: 10.1016/j.ab.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8:948–59. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–5. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–70. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 9.Klenk C, Humrich J, Quitterer U, Lohse MJ. SUMO-1 controls the protein stability and the biological function of phosducin. J Biol Chem. 2006;281:8357–64. doi: 10.1074/jbc.M513703200. [DOI] [PubMed] [Google Scholar]
- 10.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–9. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 11.Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–9. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 12.Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell. 2005;16:5433–44. doi: 10.1091/mbc.E05-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichler A, Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–7. doi: 10.1034/j.1600-0854.2002.30601.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–45. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–57. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Singh DP, Bhargavan B, Chhunchha B, Kubo E, Kumar A, Fatma N. Transcriptional protein Sp1 regulates LEDGF transcription by directly interacting with its cis-elements in GC-rich region of TATA-less gene promoter. PLoS One. 2012;7:e37012. doi: 10.1371/journal.pone.0037012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spengler ML, Brattain MG. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J Biol Chem. 2006;281:5567–74. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- 18.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–59. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 19.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–42. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KI, Baek SH, Chung CH. Versatile protein tag, SUMO: its enzymology and biological function. J Cell Physiol. 2002;191:257–68. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- 21.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10:831–42. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 22.Ihara M, Yamamoto H, Kikuchi A. SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol Cell Biol. 2005;25:3506–18. doi: 10.1128/MCB.25.9.3506-3518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu YM, Huang DY, Chiu JF, Lau AT. Post-translational modification of human heat shock factors and their functions: a recent update by proteomic approach. J Proteome Res. 2012;11:2625–34. doi: 10.1021/pr201151a. [DOI] [PubMed] [Google Scholar]
- 24.Bueno MT, Garcia-Rivera JA, Kugelman JR, Morales E, Rosas-Acosta G, Llano M. SUMOylation of the lens epithelium-derived growth factor/p75 attenuates its transcriptional activity on the heat shock protein 27 promoter. J Mol Biol. 2010;399:221–39. doi: 10.1016/j.jmb.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihara K, Fatma N, Bhargavan B, Chhunchha B, Kubo E, Dey S, Takamura Y, Kumar A, Singh DP. Lens epithelium-derived growth factor deSumoylation by Sumo-specific protease-1 regulates its transcriptional activation of small heat shock protein and the cellular response. FEBS J. 2012;279:3048–70. doi: 10.1111/j.1742-4658.2012.08686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunet Simioni M, De Thonel A, Hammann A, Joly AL, Bossis G, Fourmaux E, Bouchot A, Landry J, Piechaczyk M, Garrido C. Heat shock protein 27 is involved in SUMO-2/3 modification of heat shock factor 1 and thereby modulates the transcription factor activity. Oncogene. 2009;28:3332–44. doi: 10.1038/onc.2009.188. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–9. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 28.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–9. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Li J, Zuo Y, Deng J, Wang LS, Chen GQ. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett. 2011;309:78–84. doi: 10.1016/j.canlet.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Witty J, Aguilar-Martinez E, Sharrocks AD. SENP1 participates in the dynamic regulation of Elk-1 SUMOylation. Biochem J. 2010;428:247–54. doi: 10.1042/BJ20091948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen HJ, Zhu HY, Yang C, Ji F. SENP2 regulates hepatocellular carcinoma cell growth by modulating the stability of beta-catenin. Asian Pac J Cancer Prev. 2012;13:3583–7. doi: 10.7314/apjcp.2012.13.8.3583. [DOI] [PubMed] [Google Scholar]
- 32.Perdomo J, Jiang XM, Carter DR, Khachigian LM, Chong BH. SUMOylation regulates the transcriptional repression activity of FOG-2 and its association with GATA-4. PLoS One. 2012;7:e50637. doi: 10.1371/journal.pone.0050637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Citro S, Chiocca S. Sumo paralogs: redundancy and divergencies. Front Biosci (Schol Ed) 2013;5:544–53. doi: 10.2741/s388. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Watts FZ. SUMO modification of proteins other than transcription factors. Semin Cell Dev Biol. 2004;15:211–20. doi: 10.1016/j.semcdb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–93. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dadke S, Cotteret S, Yip SC, Jaffer ZM, Haj F, Ivanov A, Rauscher F, 3rd, Shuai K, Ng T, Neel BG, Chernoff J. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol. 2007;9:80–5. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 38.Fei E, Jia N, Yan M, Ying Z, Sun Q, Wang H, Zhang T, Ma X, Ding H, Yao X, Shi Y, Wang G. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem Biophys Res Commun. 2006;347:406–12. doi: 10.1016/j.bbrc.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 39.Zhong N, Xu J. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1alpha: regulation by SUMOylation and oxidation. Hum Mol Genet. 2008;17:3357–67. doi: 10.1093/hmg/ddn230. [DOI] [PubMed] [Google Scholar]
- 40.Chhunchha B, Fatma N, Bhargavan B, Kubo E, Kumar A, Singh DP. Specificity protein, Sp1-mediated increased expression of Prdx6 as a curcumin-induced antioxidant defense in lens epithelial cells against oxidative stress. Cell Death Dis. 2011;2:e234. doi: 10.1038/cddis.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chhunchha B, Fatma N, Kubo E, Rai P, Singh SP, Singh DP. Curcumin abates hypoxia-induced oxidative stress based-ER stress-mediated cell death in mouse hippocampal cells (HT22) by controlling Prdx6 and NF-kappaB regulation. Am J Physiol Cell Physiol. 2013 doi: 10.1152/ajpcell.00345.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatma N, Kubo E, Sharma P, Beier DR, Singh DP. Impaired homeostasis and phenotypic abnormalities in Prdx6−/−mice lens epithelial cells by reactive oxygen species: increased expression and activation of TGFbeta. Cell Death Differ. 2005;12:734–50. doi: 10.1038/sj.cdd.4401597. [DOI] [PubMed] [Google Scholar]
- 43.Fatma N, Singh P, Chhunchha B, Kubo E, Shinohara T, Bhargavan B, Singh DP. Deficiency of Prdx6 in lens epithelial cells induces ER stress response-mediated impaired homeostasis and apoptosis. Am J Physiol Cell Physiol. 2011;301:C954–67. doi: 10.1152/ajpcell.00061.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubo E, Hasanova N, Tanaka Y, Fatma N, Takamura Y, Singh DP, Akagi Y. Protein expression profiling of lens epithelial cells from Prdx6-depleted mice and their vulnerability to UV radiation exposure. Am J Physiol Cell Physiol. 2010;298:C342–54. doi: 10.1152/ajpcell.00336.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury I, Fisher AB, Christofidou-Solomidou M, Gao L, Tao JQ, Sorokina EM, Lien YC, Bates SR, Feinstein SI. Keratinocyte growth factor and glucocorticoid induction of human peroxiredoxin 6 gene expression occur by independent mechanisms that are synergistic. Antioxid Redox Signal. 2014;20:391–402. doi: 10.1089/ars.2012.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fatma N, Kubo E, Sen M, Agarwal N, Thoreson WB, Camras CB, Singh DP. Peroxiredoxin 6 delivery attenuates TNF-alpha-and glutamate-induced retinal ganglion cell death by limiting ROS levels and maintaining Ca2+ homeostasis. Brain Res. 2008;1233:63–78. doi: 10.1016/j.brainres.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fatma N, Kubo E, Takamura Y, Ishihara K, Garcia C, Beebe DC, Singh DP. Loss of NF-kappaB control and repression of Prdx6 gene transcription by reactive oxygen species-driven SMAD3-mediated transforming growth factor beta signaling. J Biol Chem. 2009;284:22758–72. doi: 10.1074/jbc.M109.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fatma N, Kubo E, Toris CB, Stamer WD, Camras CB, Singh DP. PRDX6 attenuates oxidative stress- and TGFbeta-induced abnormalities of human trabecular meshwork cells. Free Radic Res. 2009;43:783–95. doi: 10.1080/10715760903062887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 50.Kubo E, Fatma N, Akagi Y, Beier DR, Singh SP, Singh DP. TAT-mediated PRDX6 protein transduction protects against eye lens epithelial cell death and delays lens opacity. Am J Physiol Cell Physiol. 2008;294:C842–55. doi: 10.1152/ajpcell.00540.2007. [DOI] [PubMed] [Google Scholar]
- 51.Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A activities. Antioxid Redox Signal. 2011;15:831–44. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–32. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 53.Park JW, Mieyal JJ, Rhee SG, Chock PB. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J Biol Chem. 2009;284:23364–74. doi: 10.1074/jbc.M109.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–34. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 55.Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009;34:200–5. doi: 10.1016/j.tibs.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarge KD, Park-Sarge OK. SUMO and its role in human diseases. Int Rev Cell Mol Biol. 2011;288:167–83. doi: 10.1016/B978-0-12-386041-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 57.Goemaere J, Knoops B. Peroxiredoxin distribution in the mouse brain with emphasis on neuronal populations affected in neurodegenerative disorders. J Comp Neurol. 2012;520:258–80. doi: 10.1002/cne.22689. [DOI] [PubMed] [Google Scholar]
- 58.Gao L, Tse SW, Conrad C, Andreadis A. Saitohin, which is nested in the tau locus and confers allele-specific susceptibility to several neurodegenerative diseases, interacts with peroxiredoxin 6. J Biol Chem. 2005;280:39268–72. doi: 10.1074/jbc.M506116200. [DOI] [PubMed] [Google Scholar]
- 59.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–8. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 60.Watts FZ. Starting and stopping SUMOylation. What regulates the regulator? Chromosoma. 2013;122:451–63. doi: 10.1007/s00412-013-0422-0. [DOI] [PubMed] [Google Scholar]
- 61.Sang J, Yang K, Sun Y, Han Y, Cang H, Chen Y, Shi G, Wang K, Zhou J, Wang X, Yi J. SUMO2 and SUMO3 transcription is differentially regulated by oxidative stress in an Sp1-dependent manner. Biochem J. 2011;435:489–98. doi: 10.1042/BJ20101474. [DOI] [PubMed] [Google Scholar]
- 62.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–85. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 63.Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6:361–70. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones DP, Mody VC, Jr., Carlson JL, Lynn MJ, Sternberg P., Jr. Redox analysis of human plasma allows separation of pro-oxidant events of aging from decline in antioxidant defenses. Free Radic Biol Med. 2002;33:1290–300. doi: 10.1016/s0891-5849(02)01040-7. [DOI] [PubMed] [Google Scholar]
- 65.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–8. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 66.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–15. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 67.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–8. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fatma N, Kubo E, Sen M, Agarwal N, Thoreson WB, Camras CB, Singh DP. Peroxiredoxin 6 delivery attenuates TNF-α-and glutamate-induced retinal ganglion cell death by limiting ROS levels and maintaining Ca2+ homeostasis. Brain Res. 2008;1233:63–78. doi: 10.1016/j.brainres.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubo E, Singh DP, Fatma N, Akagi Y. TAT-mediated peroxiredoxin 5 and 6 protein transduction protects against high-glucose-induced cytotoxicity in retinal pericytes. Life Sci. 2009;84:857–864. doi: 10.1016/j.lfs.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 70.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PloS one. 2010;5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28:269–79. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 73.Bischof O, Dejean A. SUMO is growing senescent. Cell Cycle. 2007;6:677–81. doi: 10.4161/cc.6.6.4021. [DOI] [PubMed] [Google Scholar]
- 74.Luciani A, Villella VR, Vasaturo A, Giardino I, Raia V, Pettoello-Mantovani M, D’Apolito M, Guido S, Leal T, Quaratino S, Maiuri L. SUMOylation of tissue transglutaminase as link between oxidative stress and inflammation. J Immunol. 2009;183:2775–84. doi: 10.4049/jimmunol.0900993. [DOI] [PubMed] [Google Scholar]
- 75.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–35. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 76.Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng E.-c., Chen H, Krause DS, Min W. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J Exp Med. 2010;207:1183–1195. doi: 10.1084/jem.20092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J Biol Chem. 2004;279:49091–8. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 78.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–54. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 79.Freiman RN, Tjian R. Regulating the regulators: lysine modifications make their mark. Cell. 2003;112:11–7. doi: 10.1016/s0092-8674(02)01278-3. [DOI] [PubMed] [Google Scholar]
- 80.Hsu TI, Wang MC, Chen SY, Huang ST, Yeh YM, Su WC, Chang WC, Hung JJ. Betulinic acid decreases specificity protein 1 (Sp1) level via increasing the sumoylation of sp1 to inhibit lung cancer growth. Mol Pharmacol. 2012;82:1115–28. doi: 10.1124/mol.112.078485. [DOI] [PubMed] [Google Scholar]
- 81.Krumova P, Weishaupt JH. Sumoylation in neurodegenerative diseases. Cell Mol Life Sci. 2013;70:2123–38. doi: 10.1007/s00018-012-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feligioni M, Brambilla E, Camassa A, Sclip A, Arnaboldi A, Morelli F, Antoniou X, Borsello T. Crosstalk between JNK and SUMO signaling pathways: deSUMOylation is protective against H2O2-induced cell injury. PLoS One. 2011;6:e28185. doi: 10.1371/journal.pone.0028185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7:609–21. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X, Ding B, LeJeune D, Ruggiero C, Li S. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One. 2009;4:e5267. doi: 10.1371/journal.pone.0005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grant MM. Identification of SUMOylated proteins in neuroblastoma cells after treatment with hydrogen peroxide or ascorbate. BMB Rep. 2010;43:720–5. doi: 10.5483/BMBRep.2010.43.11.720. [DOI] [PubMed] [Google Scholar]
- 86.Yeh ETH. SUMOylation and De-SUMOylation: Wrestling with Life’s Processes. Journal of Biological Chemistry. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 88.Wang CH, Wu SB, Wu YT, Wei YH. Oxidative stress response elicited by mitochondrial dysfunction: implication in the pathophysiology of aging. Exp Biol Med (Maywood) 2013;238:450–60. doi: 10.1177/1535370213493069. [DOI] [PubMed] [Google Scholar]
- 89.Erol A. Insulin resistance is an evolutionarily conserved physiological mechanism at the cellular level for protection against increased oxidative stress. Bioessays. 2007;29:811–8. doi: 10.1002/bies.20618. [DOI] [PubMed] [Google Scholar]
- 90.Cerutti PA, Trump BF. Inflammation and oxidative stress in carcinogenesis. Cancer Cells. 1991;3:1–7. [PubMed] [Google Scholar]
- 91.Wei F, Scholer HR, Atchison ML. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J Biol Chem. 2007;282:21551–60. doi: 10.1074/jbc.M611041200. [DOI] [PubMed] [Google Scholar]
- 92.Hoppe JB, Rattray M, Tu H, Salbego CG, Cimarosti H. SUMO-1 conjugation blocks beta-amyloid-induced astrocyte reactivity. Neurosci Lett. 2013;546:51–6. doi: 10.1016/j.neulet.2013.04.050. [DOI] [PubMed] [Google Scholar]
- 93.Konopacki FA, Jaafari N, Rocca DL, Wilkinson KA, Chamberlain S, Rubin P, Kantamneni S, Mellor JR, Henley JM. Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc Natl Acad Sci U S A. 2011;108:19772–7. doi: 10.1073/pnas.1111575108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simpson-Lavy KJ, Johnston M. SUMOylation regulates the SNF1 protein kinase. Proc Natl Acad Sci U S A. 2013;110:17432–7. doi: 10.1073/pnas.1304839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–94. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 97.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–74. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 98.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–32. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 99.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 100.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–12. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–33. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 102.Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–75. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 103.Denuc A, Marfany G. SUMO and ubiquitin paths converge. Biochem Soc Trans. 2010;38:34–9. doi: 10.1042/BST0380034. [DOI] [PubMed] [Google Scholar]
- 104.Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J Mol Biol. 2008;380:869–85. doi: 10.1016/j.jmb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 105.Subramanian L, Benson MD, Iniguez-Lluhi JA. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J Biol Chem. 2003;278:9134–41. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- 106.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shinbo Y, Niki T, Taira T, Ooe H, Takahashi-Niki K, Maita C, Seino C, Iguchi-Ariga SM, Ariga H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13:96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- 108.Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8:667–76. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bawa-Khalfe T, Cheng J, Wang Z, Yeh ET. Induction of the SUMO-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J Biol Chem. 2007;282:37341–9. doi: 10.1074/jbc.M706978200. [DOI] [PubMed] [Google Scholar]
- 110.Xu Y, Zuo Y, Zhang H, Kang X, Yue F, Yi Z, Liu M, Yeh ET, Chen G, Cheng J. Induction of SENP1 in endothelial cells contributes to hypoxia-driven VEGF expression and angiogenesis. J Biol Chem. 2010;285:36682–8. doi: 10.1074/jbc.M110.164236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ibaraki N, Chen SC, Lin LR, Okamoto H, Pipas JM, Reddy VN. Human lens epithelial cell line. Exp Eye Res. 1998;67:577–85. doi: 10.1006/exer.1998.0551. [DOI] [PubMed] [Google Scholar]
- 112.Singh DP, Ohguro N, Chylack LT, Jr., Shinohara T. Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Invest Ophthalmol Vis Sci. 1999;40:1444–51. [PubMed] [Google Scholar]
- 113.Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, Lerner CP, Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278:25179–90. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 114.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]