Abstract
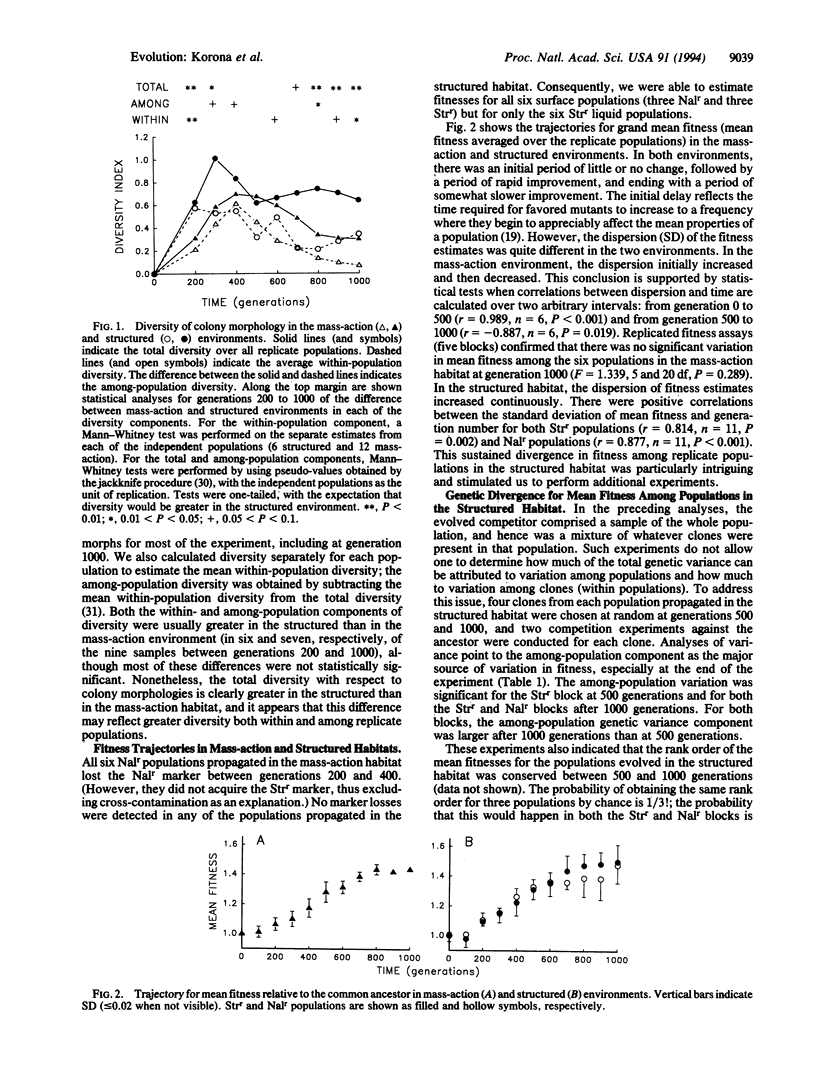
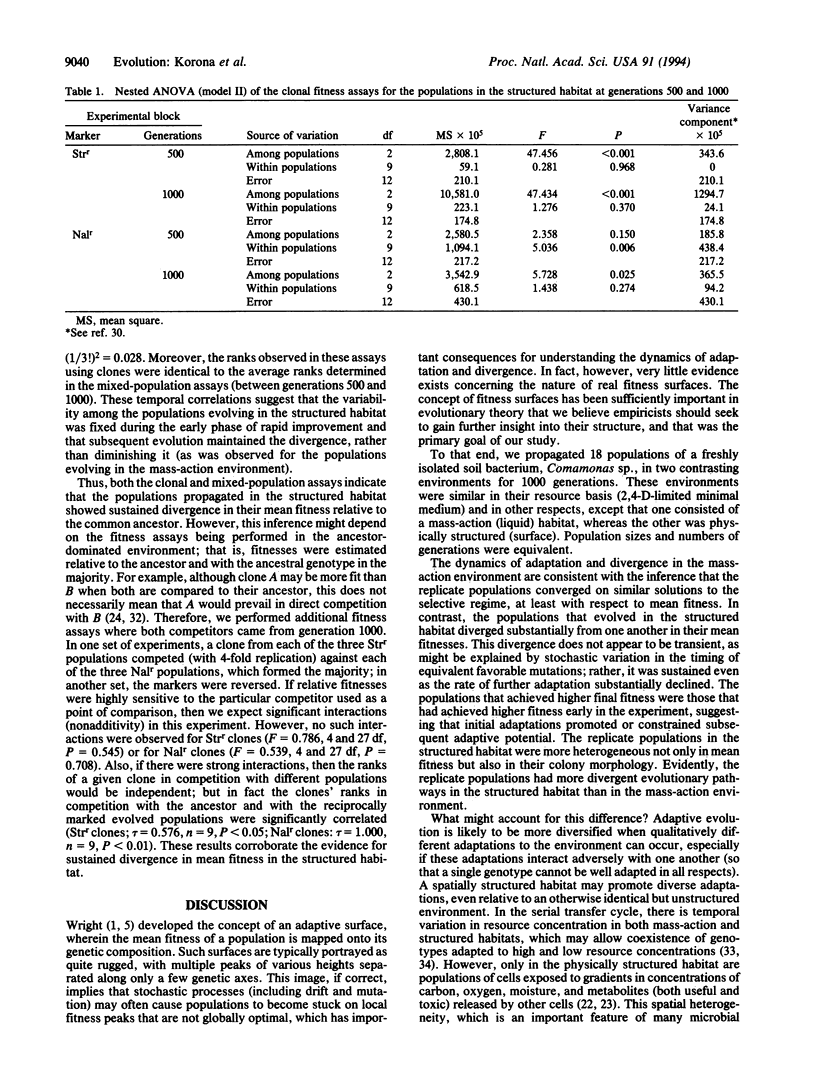
Natural selection tends to promote the divergence of populations living in different environments. Even in identical environments, however, replicate populations may diverge if they find alternative adaptive solutions. We describe the evolution of 18 bacterial populations (Comamonas sp.) founded from a single progenitor genotype and propagated separately for 1000 generations in two distinct environments, one physically unstructured (mass-action liquid) and the other structured (agar surfaces). Phenotypic diversity, as reflected in colony morphology, was greater in the structured habitat than in the unstructured habitat. More importantly, the trajectories for mean fitness, as measured by competition against the common ancestor, were more divergent for populations in the structured habitat than those in the unstructured habitat. Structured environments may accelerate evolutionary diversification by promoting genetic polymorphisms within populations, thereby increasing the complexity of genetic constraints that allow divergence among replicate populations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayala F. J. Competition between Species: Frequency Dependence. Science. 1971 Feb 26;171(3973):820–824. doi: 10.1126/science.171.3973.820. [DOI] [PubMed] [Google Scholar]
- Barton N. H., Rouhani S. The frequency of shifts between alternative equilibria. J Theor Biol. 1987 Apr 21;125(4):397–418. doi: 10.1016/s0022-5193(87)80210-2. [DOI] [PubMed] [Google Scholar]
- Chao L., Levin B. R. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry G. R., Chapalamadugu S. Biodegradation of halogenated organic compounds. Microbiol Rev. 1991 Mar;55(1):59–79. doi: 10.1128/mr.55.1.59-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. M., Hoffmann A. A. Genetic divergence under uniform selection. II. Different responses to selection for knockdown resistance to ethanol among Drosophila melanogaster populations and their replicate lines. Genetics. 1986 Sep;114(1):145–164. doi: 10.1093/genetics/114.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J Bacteriol. 1985 Jan;161(1):466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci. 1988 Jul 6;319(1196):459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- Lewin R. The uncertain perils of an invisible landscape. Science. 1988 Jun 10;240(4858):1405–1406. doi: 10.1126/science.3375828. [DOI] [PubMed] [Google Scholar]
- Paquin C. E., Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983 Nov 24;306(5941):368–370. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M. J., Goodnight C. J. Wright's shifting balance theory: an experimental study. Science. 1991 Aug 30;253(5023):1015–1018. doi: 10.1126/science.1887214. [DOI] [PubMed] [Google Scholar]
- Yin J. Evolution of bacteriophage T7 in a growing plaque. J Bacteriol. 1993 Mar;175(5):1272–1277. doi: 10.1128/jb.175.5.1272-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


