Abstract
Background
Glioblastoma (GBM) is the most common primary brain tumor in adults, with a dismal prognosis. Treatment is hampered by GBM's unique biology, including differential cell response to therapy. Although several mitochondrial abnormalities have been identified, how mitochondrial DNA (mtDNA) mutations contribute to GBM biology and therapeutic response remains poorly described. We sought to determine the spectrum of functional complex III and IV mtDNA mutations in GBM.
Methods
The complete mitochondrial genomes of 10 GBM cell lines were obtained using next-generation sequencing and combined with another set obtained from 32 GBM tissues. Three-dimensional structural mapping and analysis of all the nonsynonymous mutations identified in complex III and IV proteins was then performed to investigate functional importance.
Results
Over 200 mutations were identified in the mtDNAs, including a significant proportion with very low mutational loads. Twenty-five were nonsynonymous mutations in complex III and IV, 9 of which were predicted to be functional and affect mitochondrial respiratory chain activity. Most of the functional candidates were GBM specific and not found in the general population, and 2 were present in the germ-line. Patient-specific maps reveal that 43% of tumors carry at least one functional candidate.
Conclusions
We reveal that the spectrum of GBM-associated mtDNA mutations is wider than previously thought, as well as novel structural-functional links between specific mtDNA mutations, abnormal mitochondria, and the biology of GBM. These results could provide tangible new prognostic indicators as well as targets with which to guide the development of patient-specific mitochondrially mediated chemotherapeutic approaches.
Keywords: functional prediction, glioblastoma, mitochondrial DNA (mtDNA) mutation, structural analysis, subgrouping
Glioblastoma (GBM; World Health Organization grade IV) is the most malignant and common primary brain tumor in adults.1 One percent of cases are diagnosed in patients younger than 20 years of age, 90% are primary de novo tumors, and 10% are secondary tumors that develop from lower-grade gliomas. GBM carries a dismal prognosis, with a median survival of just 14.6 months.2 Effective treatment is hampered by the unique underlying heterogeneous biology of GBM, including differential cell response to current therapies. Research over the last decade has revealed the existence of 6 main subgroups, classified on the basis of cellular origin, anatomical compartment, and various molecular alterations such as aberrations in phosphatase and tensin homolog/delta-like 3 (Drosophila), epidermal growth factor receptor, neurofibromatosis type 1, platelet derived growth factor receptor α/isocitrate dehydrogenase 1 (IDH1) gene expression, and more recently, H3 histone, family 3A and IDH1 mutations, and transcriptomic signatures, highlighting that patient-specific profiles are linked to different prognoses and/or responsiveness to therapies.3–5
Mitochondria are double-membrane organelles found in most eukaryotic cells. The inner membrane hosts the multi-subunit protein complexes of the oxidative phosphorylation system (the mitochondrial respiratory chain [MRC] complexes I–IV and ATP synthase or complex V) that produces most of the ATP required for cell metabolism. Numerous other cellular processes are also linked to MRC function, including the tricarboxylic acid cycle, reactive oxygen species (ROS) signaling, generation of nicotinamide adenine dinucleotide phosphate, mitochondrial membrane potential, and apoptosis.6 Mitochondrial DNA (mtDNA) is essential for producing 13 of the core catalytic MRC protein subunits. Consequently, mtDNA mutations often cause defects in the MRC, which in turn have dramatic and multiple consequences for the cell.
Although GBM cells contain several mitochondrial abnormalities compared with normal brain cells,7–9 and mtDNA mutations are proposed to be at the heart of the mitochondrial abnormalities observed in cancers,10 patient-specific profiles and functional roles of mtDNA mutations found in gliomas8,11–13 are poorly described. Confounding issues include the analyses of incomplete genomes, inadequate ascertainment of mutation loads, and/or the difficulties associated with proving direct links between mtDNA mutation and phenotype.
Only a proportion of the multiple mtDNA molecules resident in a cell or tissue will be mutated (heteroplasmy), influencing whether the mutation exerts a functional effect. In the absence of genetic transformation methods for the mammalian mtDNA, the usual way of proving a link is to demonstrate a corresponding biochemical defect in affected tissue. However, it is not practical to do this for every mutation, and more than one nuclear or mitochondrial mutation may be present, further confounding the interpretation.
Recently, we have demonstrated that 3D structural analysis of mitochondrial proteins in silico, which takes into account the bigenomic, quaternary structural organization of the MRC, can be used to reveal detailed mechanistic insights into the functional role of mutations in complex III and IV mtDNA genes.14
In this study, we used deep sequencing to increase the number of entire primary GBM mtDNAs available and to highlight the current known spectrum and heteroplasmies of GBM-associated mutations of complex III and IV mtDNA genes. Finally, we applied 3D structural analysis to the mutations to predict their functional significance and reveal insights into how they could contribute to the mitochondrial dysfunction observed in gliomas and individual responses to mitochondrially mediated chemotherapy.
Materials and Methods
All reagents, materials, and equipment were purchased from Sigma-Aldrich, unless stated otherwise.
Patients and Samples
Set A consisted of new mitochondrial mutation data generated from: 7 human biopsy-derived primary adult GBM cell lines and 1 matched blood sample, which were obtained from patients from Kings College Hospital, London, under ethics permission (REC reference number: 11/SC/0048); 2 primary pediatric GBM cell lines obtained from other laboratories; 1 commercial primary adult GBM tumor cell line; and finally, 1 commercial non-neoplastic adult astrocyte cell line (Supplementary Table S1). Details of the cell culture techniques are described in the Supplementary Materials and Methods. Set B consisted of mitochondrial mutational data provided by whole-genome sequencing of 32 primary adult GBM biopsies, each with a matched blood sample, gathered by various centers on behalf of The Cancer Genome Atlas network (Supplementary Table S2).15–17
Whole Mitochondrial Genome Sequence Generation
Whole mitochondrial genome libraries were generated from long PCR amplicons to avoid contaminating nuclear pseudogenes (Set A).18,19,20 Whole-exome or whole-genome libraries were generated from total DNA (Set B).16 Set A libraries were sequenced on an Illumina MiSeq,18 while the whole-exome or whole-genome libraries were sequenced on an Illumina HiSeq 2000 or GA-IIX or Illumina GAII or HiSeq, respectively.16 Library preparation and sequencing are described in detail in the Supplementary Materials and Methods.
Assembly, Mutation Identification, and Heteroplasmy Quantification
Set A and B reads were aligned using the Burrows–Wheeler Aligner,21,23 and alignments were processed using GATK Unified Genotyper24,25 (Set A) and ANNOVAR15,26 (Set B) to identify mutations and calculate heteroplasmy. More details on Set A and B mutational data generation are given in the Supplementary Materials and Methods.
3D Structural Analysis
To obtain a more comprehensive interpretation, complex III and IV mitochondrial mutational data from Set A and B mtDNAs were combined for the subsequent functional prediction and statistical analysis. The functional significance of all nonsynonymous mitochondrial mutations in complex III and IV proteins was then predicted using the 3D structural analysis method previously developed and validated on mutations associated with a broad range of human diseases, including cancer.14 Briefly, based on location in the sequence alignments, the mutations were mapped onto highly homologous bovine complex III and IV structures. Then, following the detailed analysis of the location, relative to important catalytic and/or interaction surfaces, mutations were placed into 1 of 5 structural classes: (i) frameshift, (ii) active site, (iii) binding pocket, (iv) protein interaction region, and (v) nonfunctional (Supplementary Figs. S2 and S3). In addition, ligand docking analysis was performed on the mutation F18L to determine its effect on ubiquinone binding. Further details on the 3D structural modeling (including ligand docking analysis) are provided in the Supplementary Materials and Methods.
Statistical Analyses
As the prevalences and heteroplasmies of individual mutations were varied, we used K-means and hierarchical (unweighted pair-group average method29) cluster analyses to determine the similarity of the mutations. The similarity metric (distance measure) used by the K-means module was unscaled Euclidean distance. The data matrix used to determine similarity of mutations was “3 variables × 130 cases,” where the 3 variables were (i) prevalence of each mutation in the GBM cohort (%GBM), (ii) prevalence of each mutation in the general population (%HmtDB), and (iii) heteroplasmy level of each mutation (%Het), and the 130 cases were the number of nonsynonymous and synonymous complex III and IV mutations found in the combined (Set A+B) mtDNAs. ANOVAs were used to make sure the clusters were statistically different. All analyses were performed using Statistica 10 for Windows (Statsoft UK). The HmtDB contains over 16 000 mitogenomic sequences obtained from the tissues of normal, healthy subjects (%HmtDB27,28).
Results
New Deep Coverage Mitochondrial Genomes Permit the Quantification of Heteroplasmy
We obtained 10 complete GBM cell line mtDNAs using long PCR and next-generation sequencing (Set A). The size of all 10 mtDNAs was similar to the revised Cambridge Reference Sequence (rCRS, NC_01292022), that is, 16 569 bp (Supplementary Table S4), and on average, the mitochondrial genomes were assembled from 2.6 million reads (Supplementary Table S4). The mean read lengths were 147 ± 1 nucleotides. The total number of reads with Q-score of 20 or above contributing to the assembled sequences was on average 97% ± 1 with a mean depth of coverage over the entire mitochondrial genome of 22 695 ± 5838 (Supplementary Table S4), providing the 250 reads with a minimal Q-score of at least 30 for mutation detection and heteroplasmy determination. The mean depth of coverage at mutation sites for Set B mtDNAs was >5600.15 All new mtDNAs generated have been submitted to GenBank with the following accession numbers (KJ735668-KJ735679).
Germ-line Mutations and Heteroplasmy Are Key Features
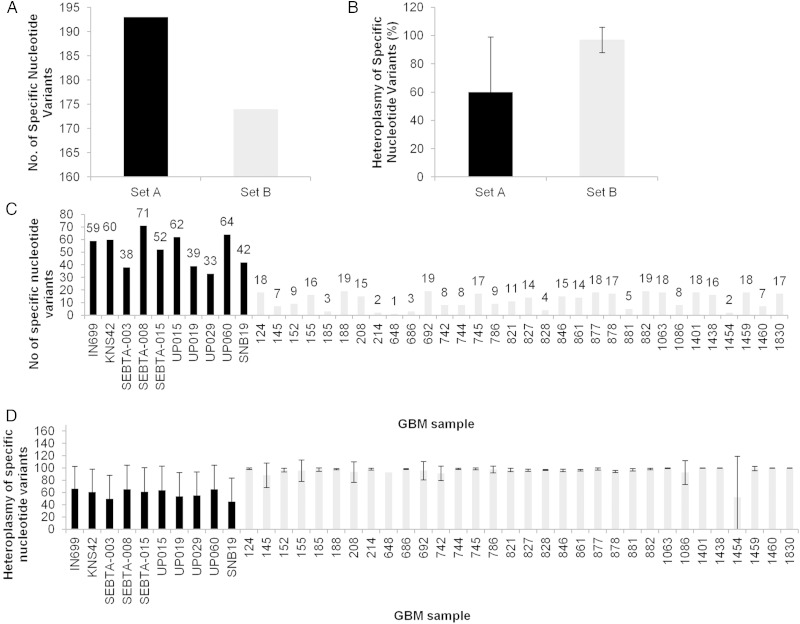
A key question is whether mutations associated with GBM are genetically inherited events (germ-line) or sporadic somatic (non-germ-line) events. Analysis of a subset of 33 GBM samples with corresponding matched bloods in the combined cohort allowed us to determine whether the mutations were germ-line or somatic. Set A mtDNAs (n = 10) contained 193 mutations relative to the rCRS, whereas Set B (n = 32) contained 174 mutations (Fig. 1A), and the average heteroplasmy of Set A mtDNAs was lower than Set B (60% ± 39 vs 97% ± 9, respectively; Fig. 1B and D). The differences between the datasets were also evident at the individual patient level, where the new data display >4-fold increase in the average number of mutations observed per sample (Set A was 52 ± 13 vs 12 ± 6 in Set B; Fig. 1C). The ratio of germ-line to somatic mutations is roughly consistent between the datasets (∼21:1 for Set A and ∼35:1 for Set B), although this is ∼2–2.5-fold higher than that previously documented for GBM studies that have focused on nuclear DNA.16
Fig. 1.
Abundance and heteroplasmy of mutations in GBM samples in 10 GBM biopsy–derived cell cultures (Set A, black bars) and 32 GBM biopsy tissues (Set B, gray bars). Combined (A and B, respectively) and in individual (C and D, respectively) profiles.
Twenty-five mutations that cause nonsynonymous amino acid substitutions in complex III and IV proteins were found in the combined GBM mtDNA dataset (Set A + B; Table 1). Thirty-two percent (8/25) have no known disease association (Set A: C9469T, T9252C, A6692del, C6237A, T15479C, A15326G, G15048A, and C14766T); 40% (10/25) were observed in GBM previously (Set B: A15218G, T15453C, G15500A, T15693C, A15758G, G6267A, G6619A, G9655A, A9667G, and G9966A); 16% (4/25) were found in both Sets A and B at very similar heteroplasmy despite the differences in sample types and methodologies used to obtain the mutational data in each case (A14793G, T14798C, C15452A, and G9477A); and 56% of the mutations (14/25) had other disease associations (A14793G, T14798C, A15218G, G15257A, C15452A, T15693C, A15758G,G15803A, G6267A, G9300A, G9477A, G9655A, A9667G, and G9966A) (Supplementary Table S5). The ratio of nonsynonymous germ-line to somatic mutations was ∼8:1, lower than that determined for all the different mutation types combined.
Table 1.
Structural classification and functional prediction, presence in control astrocytes, heritability, heteroplasmy level, and prevalence of mutations in the mtDNA complex III and IV genes identified in GBM
| Human Subunit (gene) | Human Residue (nucleotide) | %GBM* | %HmtDB** (tissue) | %Het ± SD | K-means Group | Heritability*** | Presence in Non-neoplastic Astrocytes | Structural/Functional Class | 3D Modeling Prediction |
|---|---|---|---|---|---|---|---|---|---|
| MT-CYB (mt-cyb) | T7I (C14766T) | 14.29 | 0 | 98.25 ± 3.14 | 2 | Inherited | Yes | 0 | Nonfunctional |
| H16R (A14793G) | 4.76 | 2.32 (blood) | 98.7 ± 1.27 | 2 | Inherited | No | 0 | Nonfunctional | |
| F18L (T14798C) | 30.95 | 8.66 (blood, buccal) | 98.98 ± 1.71 | 2 | Inherited | No | 3 | Functional | |
| G101D (G15048A) | 2.38 | 0 | 17 | 1 | ND | No | 2 | Functional | |
| T158A (A15218G) | 2.38 | 1.90 (blood, buccal) | 97.9 | 2 | Inherited | No | 0 | Nonfunctional | |
| D171N (G15257A) | 2.38 | 1.43 (blood, buccal) | 99.6 | 2 | ND | No | 0 | Nonfunctional | |
| T194A (A15326G) | 23.81 | 0 | 98.80 ± 0.29 | 2 | Inherited | Yes | 0 | Nonfunctional | |
| L236I (C15452A) | 30.95 | 10.18 (blood, buccal, bone) | 96.02 ± 3.76 | 2 | Inherited | Yes | 0 | Nonfunctional | |
| L236P (T15453C) | 2.38 | 0.03 (blood, buccal) | 98.5 | 2 | Inherited | No | 4 | Functional | |
| F245L (T15479C) | 2.38 | 0.38 (blood) | 16.4 | 1 | ND | No | 4 | Functional | |
| D252N (G15500A) | 2.38 | 0 | 4.99 | 1 | Inherited | No | 2 | Functional | |
| M316T (T15693C) | 2.38 | 1.25 (blood) | 99.8 | 2 | Inherited | No | 0 | Nonfunctional | |
| I338V (A15758G) | 2.38 | 0.75 (blood, buccal) | 99.8 | 2 | Inherited | No | 0 | Nonfunctional | |
| V353M (G15803A) | 2.38 | 0.1 (blood) | 93.6 | 2 | ND | No | 0 | Nonfunctional | |
| MT-CO1 (mt-co1) | L112M (C6237A) | 2.38 | 0.08 (ns) | 99.6 | 2 | ND | No | 0 | Nonfunctional |
| A122T (G6267A) | 2.38 | 0.16 (blood) | 99.9 | 2 | Inherited | No | 0 | Nonfunctional | |
| G239D (G6619A) | 2.38 | 0 | 33.7 | 1 | Somatic | No | 2 | Functional | |
| M271Ter (A6692del) | 2.38 | ND | 8 | ND | No | 1 | Functional | ||
| MT-CO3 (mt-co3) | W16R (T9252C) | 2.38 | 0 | 8.6 | 1 | ND | No | 4 | Functional |
| A32T (G9300A) | 2.38 | 0.47 (blood) | 100 | 2 | ND | No | 0 | Nonfunctional | |
| T88I (C9469T) | 4.76 | 0.06 (ns) | 99.2 | 2 | Inherited | No | 0 | Nonfunctional | |
| V91I (G9477A) | 4.76 | 4.41 (blood) | 98.55 ± 1.45 | 2 | Inherited | No | 0 | Nonfunctional | |
| S150N (G9655A) | 2.38 | 0.01 (ns) | 73.8 | 2 | Somatic | No | 4 | Functional | |
| N154S (A9667G) | 2.38 | 0.75 (blood) | 92.6 | 2 | Inherited | No | 0 | Nonfunctional | |
| V254I (G9966A) | 2.38 | 0.67 (blood, buccal) | 96.8 | 2 | Inherited | No | 0 | Nonfunctional |
Abbreviations: ND, not determined; ns, not specified; K-means group—mutations were stratified into 2 groups based on 3 variables: %GBM = prevalence of mutations in the GBM cohort, %HmtDB = prevalence of mutations found in the human mitochondrial database, and %Het = heteroplasmy.
* n = 42. **n = >16 000. ***Mutations listed as “inherited” were found in both tumor and matched blood samples and are therefore likely to occur in the germ-line, while those listed as “somatic” were found in the tumor samples only and are therefore likely to be spontaneous events.
Nine Functional Mutations Identified
Following the identification of GBM-associated mutations, and importantly their mutational load within tumor cells, we sought to determine their functional significance. We predicted that 9 mutations have significant functional impact at the level of protein structural changes, while the remaining 16 are likely to be nonfunctional (Table 1). Among the mutations classified as functional, 1 caused a frameshift in the triplet code (class 1), 3 occurred in active site regions (class 2), 1 occurred in a substrate/product binding pocket (class 3), and 4 occurred in protein interaction regions (class 4). As expected, no functional mutations were present in the non-neoplastic astrocyte control.
Functional Mutations Can Be Either Somatic or Germ-line
Among the subset of functional candidates, 2 in mitochondrially encoded (MT) cytochrome b (CYB) were germ-line (T14798C and G15500A), while 2 in MT–cytochrome c oxidase (CO) 1 and MT-CO3 were somatic (G6619A and G9655A, respectively). Contrary to what has been observed in some cancers, such as gastric cancer,30 our results suggest that both inherited and spontaneous mtDNA mutations could play a role in mitochondrial dysfunction and the biology of GBM.
A Global Structural Map
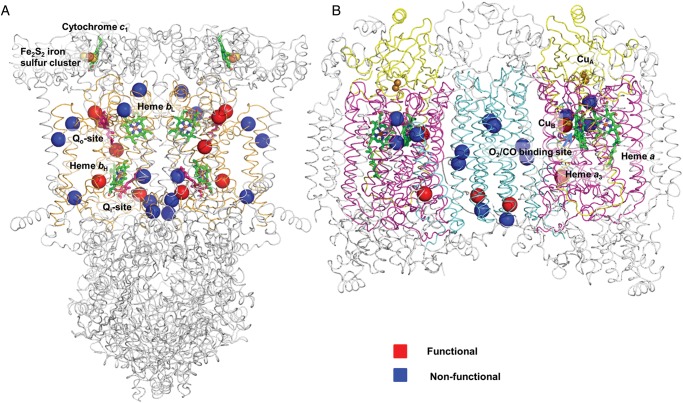
All nonsynonymous amino acid substitutions in complex III and IV genes were mapped onto their corresponding structural homologs (Fig. 2). These structures, together with decades of associated systematic biochemical analysis in the field, provide a window into the functional consequences of individual mutations. To our knowledge this is the first visual compendium of mutation sites found within the MRC proteins of GBM patients and it reveals a non-uniform distribution. Although many mitochondrial-encoded subunits act as hotspots for functional mutations, subunit MT-CO2 remains mutation free.
Fig. 2.
(A) Complexes III and (B) IV 3D crystallographic structural maps of GBM mitochondrial mutation sites. Functional sites (red spheres); nonfunctional sites (blue spheres). Both complete dimeric complexes are depicted as ribbon models, with the mitochondrial-encoded MT-CYB subunit colored as orange and MT-CO1, MT-CO2, and MT-CO3 colored as magenta, yellow, and cyan, respectively. Nuclear-encoded subunits are colored in gray, and key functional sites, such as metalloprotein and substrate binding pockets, are annotated.
Patient-Specific Structural Maps
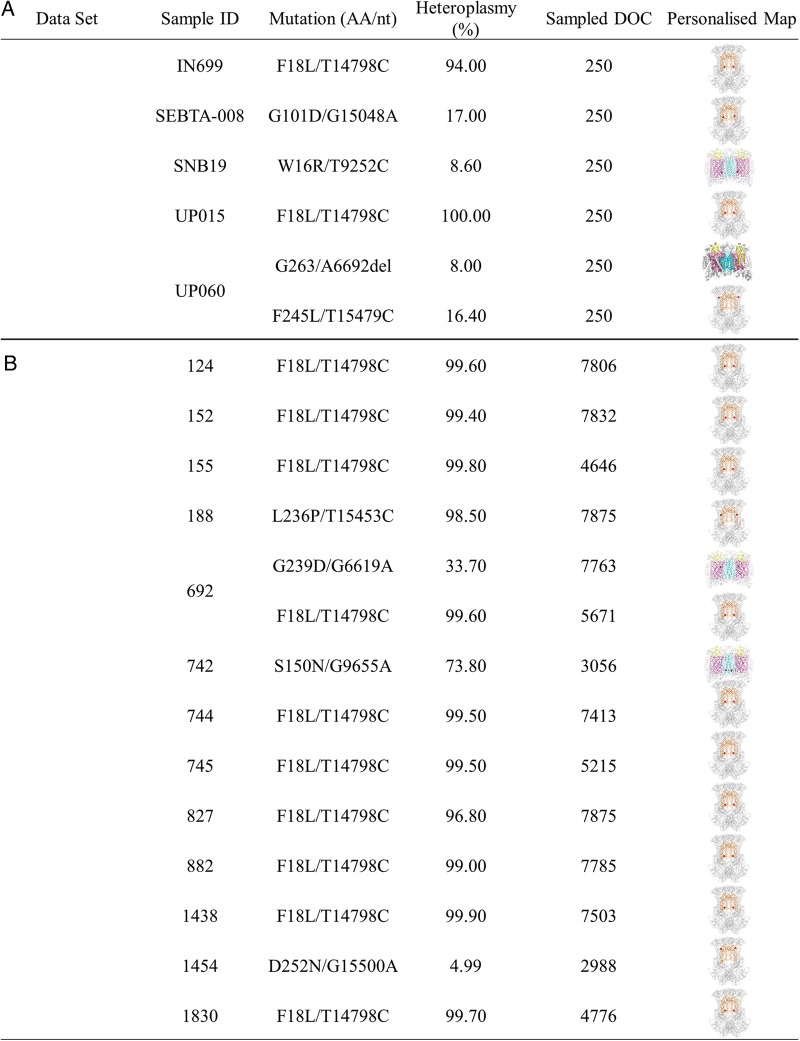
A powerful aspect of this method is the ability to easily produce maps on an individual patient basis. Structural maps (relative to the non-neoplastic astrocyte control) of the functional mitochondrial mutations (F18L, G101D, L236P, F245L, D252N, G239D, W16R, and S150N) present in individual patients were generated (Fig. 3). Altogether 16 of the 42 GBM samples (38%) carried a single functional mutation, just 2 of the 42 (∼5%) carried 2 functional mutations, while 24 GBM samples (57%) did not carry a functional mutation in either complex III or complex IV. This does not preclude the possibility that mutations in other regions of the MRC, such as in complex I or V, may be contributing to the pathology and/or drug sensitivity of GBM samples. Currently, the lack of close structural homologs to human complexes I and V limits informative mapping studies.
Fig. 3.
Functional mitochondrial mutation profiles of individual GBM biopsy–derived cell cultures (Set A) and GBM biopsy tissues (Set B). Samples 124, 152, 155, 744, 745, 827, 882, 1438, 1830, UP-015, and IN699 contained just F18L; sample SEBTA-008 contained just G101D; sample 188 contained just L236P; sample 1454 contained just D252N; SNB19 contained just W16R; and 742 contained just S150N. Sample UP-060 contained F245L and A6692del (see Fig. 5), while 692 contained both F18L and G239D. Samples 145, 185, 208, 214, 648, 686, 786, 821, 828, 846, 861, 877, 878, 881, 1063, 1086, 1401, 1459, 1460, SEBTA-003, SEBTA-015, UP-019, UP-029, and KNS42 were essentially like the SC-1800 (non-neoplastic astrocyte control) and contained no functional mutations (and so are not shown). The depth of coverage (DOC) used to calculate the percentage heteroplasmy of the functional candidates is also indicated.
Cluster Analysis Identifies 2 Distinct Groups
Across the 42 GBM patients, high variability was observed in the prevalence of individual mutations (2%–30%). A comparison with prevalence in a large group of samples from over 16 000 subjects from the general population revealed that several mutations were absent, while others were present in as much as ∼10% of the general population. The heteroplasmy revealed a more dramatic range from as little as 8% for some mutations to as much as 100% for others (Table 1).
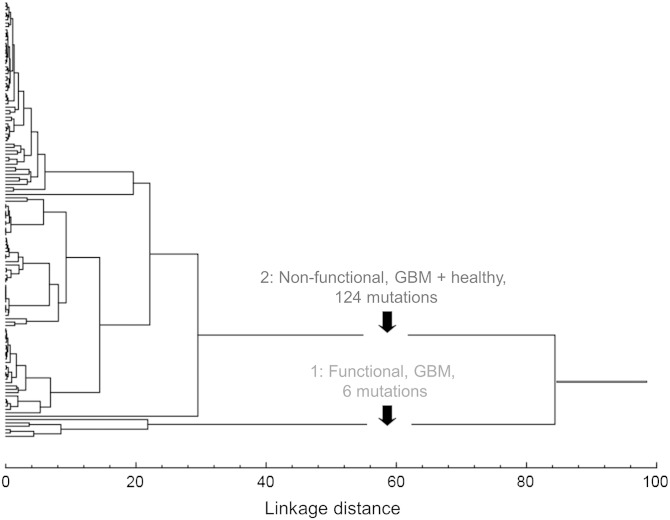
Two distinct groups of mutation were identified by integrating both the hierarchical (unweighted pair-group method with arithmetic mean) and nonhierarchical (K-means) outputs with the functional predictions (Fig. 4, Table 1):
Fig. 4.
Integration of hierarchical and nonhierarchical cluster and structural analyses of complex III and IV GBM-associated mtDNA mutations reveals 2 naturally occurring subgroups using the following variables: prevalence of each mutation in GBM; prevalence of each mutation in normal, healthy subjects; and heteroplasmy level. Group 1 mutations are significantly less frequent in healthy compared with GBM samples but are almost exclusively functional. Group 2 mutations occur in multiple healthy and GBM samples but are mostly nonfunctional.
Functional, GBM: A small group of almost exclusively functional mutations (5 of 6, ∼86%) found in single patients (2.38% ± 0.00) but 39-fold less frequently in the general human population (0.06% ± 0.16), with low mutational loads (15.52% ± 10.02)
Nonfunctional, mixed: A large group of almost exclusively nonfunctional mutations (3 of 124, 2.42%) found in multiple patients (13.94% ± 11.60) and multiple subjects in the general human population (3.42% ± 4.04), with very high heteroplasmy levels (98.13 ± 3.17%).
Post-hoc ANOVA comparisons confirmed that all 3 variables differed significantly between the 2 groups: prevalence in all the GBM samples tested (%GBM, P < .02), prevalence in the general population (%HmtDB, P < .04), and heteroplasmy (%Het, P < .0000001).
New Mechanistic Insights
We employed 3D structural analysis and docking studies to predict the effects of mutations on MRC activity and ligand binding, respectively. In order to demonstrate the wide spectrum of functional consequences that can arise from single mtDNA point mutations, we have provided graphical examples of 3 scenarios that include the dramatic loss of a large portion of protein subunits through to extremely subtle atomic rearrangements.
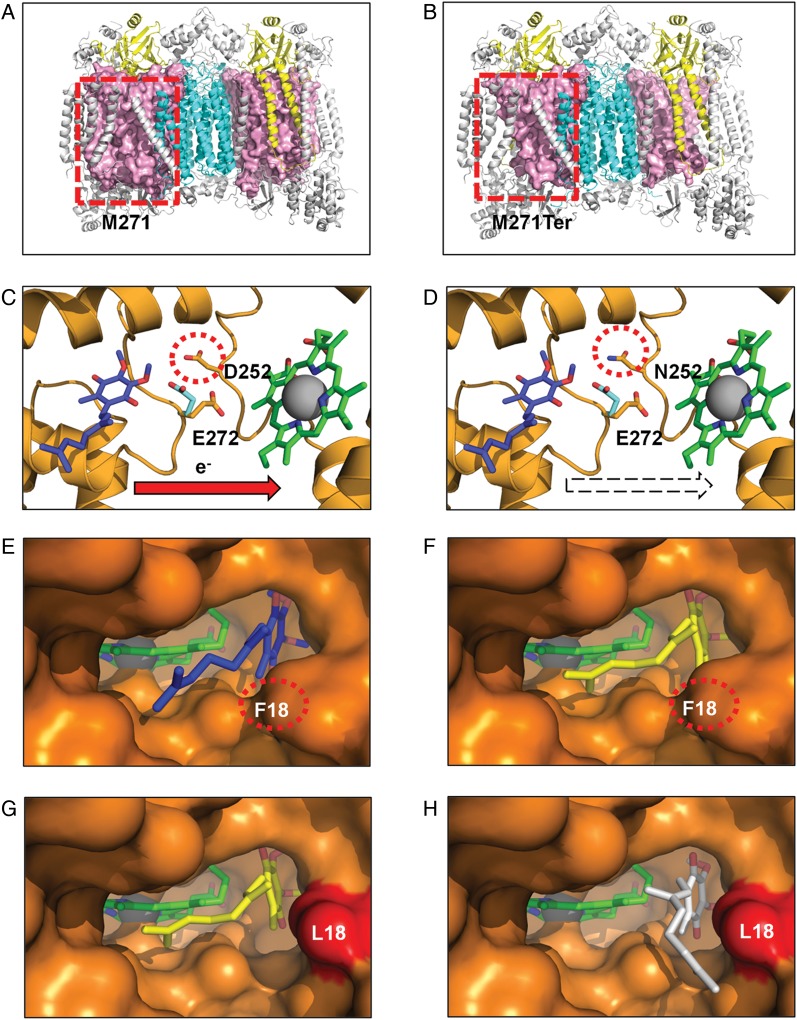
Figure 5A and B depict the structural consequence of the frameshift mutation A6692del on complex IV. In the wild type, MT-CO1 is the central and major subunit of complex IV, the terminal enzyme of the MRC that catalyzes electron transfer from cytochrome c to oxygen, conserving the released energy as coupled transmembrane proton transfers (Fig. 5A). The deletion of a single adenine nucleotide at position 6692 of the mitochondrial genome results in a change of 5 amino acids and ultimately a stop codon being introduced at position 271 of MT-CO1. This results in the deletion of 47% of the subunit and occurs in both halves of the dimeric complex (Fig. 5B). It is likely that the truncated MT-CO1 (Fig. 5B) will not have sufficient surface area to form stabilizing contacts with the redox groups (heme a, heme a3, and CuB) or the other subunits found within complex IV, whether mitochondrial (MT-CO2 and MT-CO3) or nuclear (COX4I1, COX5B, COX6A, COX7C, COX7A, COX6B, and COX6C), impairing complex IV assembly. This loss in quaternary structure will further compromise the activity of additional MRC complexes, as associations among complexes IV, I, and III have been demonstrated to be vital for MRC function, assembly, and stability.31
Fig. 5.
Structural consequences of 3 classes of functional mutation present in GBM complex III and IV mitochondrial proteins (colored as in Fig. 2). Frameshift mutation. (A) Wild-type (M271) residue in MT-CO1 is rendered as a space-filling model (light pink), highlighting its central position within each monomer. (B) The mutant A6692del (M271Ter) results in deletion of >47% of the MT-CO1 polypeptide chain, disrupting the interaction of multiple subunits that surround MT-CO1. Active site mutation. (C) Wild-type (D252) residue within the MT-CYB complex III Qo site. (D) The mutant (N252) may affect the passage of protons from the hydroxyquinone (blue) bound at the Qo site to the solvent phase. Residue E272 is shown in the 2 alternate conformations observed in x-ray crystal structures with the stigmatellin-inhibited complex (PDB ID 1KB950) (cyan) and in the HHDBT (5-n-heptyl-6-hydroxy-4, 7-dioxobenzothiazole)-inhibited complex (PDB ID 1P8432) (orange) and heme bL. The quinol binding site is inferred from the inhibitors binding site. Binding pocket mutation in complex III Qi site. (E) The wild-type Qi-site pocket is shown as a surface representation with the deeply buried heme group and bound ubiquinone (PDB ID 1NTZ34) (blue). (F) The results from in silico docking studies confirm that the predicted hydroxyquinone position (yellow) corresponds closely to that observed in the crystal structure. (G) An extra cavity is shown where the F18L GBM mutation (red) opens up the binding pocket. (H) The hydroxyquinone is observed to occupy additional binding sites (white) that are not possible in the wild-type protein due to steric constraints provided by the aromatic F18 residue. Since this highly conserved pocket is tailored precisely for hydroxyubiquinone binding, such a change is likely to interfere with its association/dissociation and ultimately efficient electron transfer from heme bH.
G15500A results in the amino acid change D252N in MT-CYB (Fig. 5C and D). MT-CYB is the central subunit of complex III, the middle enzyme of the MRC that couples the transfer of electrons from ubihydroquinone to cytochrome c while translocating protons across the inner mitochondrial membrane (Supplementary Data). On the basis of atomic structure, the D252 residue, conserved in all metazoans, is proposed to be part of a proton exit route from the catalytic site (Qo site) in complex III.32 The D252 residue occupies a key position within the Qo site, being in direct contact with the external solvent in the intermembrane space. In addition, an interaction has been proposed between residues D252 and E272 via a complex network of water-mediated hydrogen bonds. Although E272 has been proposed to be a proton acceptor, more recent mutational analysis has indicated that E272 may rather serve as a relay on the proton exit pathway.33 Figure 5C depicts the position of D252 relative to the Qo site and E272. In either case, whether a block in the protein exit pathway directly or disruption of the mobile E272 relay mechanism, the loss of a protonable amino acid residue in mutant D252N (Fig. 5D) is predicted to slow down the movement of protons from the Qo site, reduce the electron transfer rate, and thus compromise the activity of complex III.
T14798C, which results in an amino acid change F18L in MT-CYB (Fig. 5E to H), is an example of a binding pocket mutation and forms part of the ubiquinone (coenzyme Q) binding site (Qi site) of complex III (Supplementary Data). According to the structure of the wild-type Qi site in complex III, ubiquinone interacts with the buried heme in a “lock and key”-style enclosed environment (Fig. 5E). The pocket has evolved to make intimate contacts to the ubiquinone to action binding, redox reaction, and dissociation. In order to model the effect of GBM mutations on binding, we first used an in silico docking approach with the wild-type pocket and the natural substrate. We were able to simulate the precise docking of ubiquinone as observed in the crystal structure (Protein Data Bank identification [PDB ID] 1NTZ34) (Fig. 5F and Supplementary Fig. S5). We then modeled the effect of mutation F18L on this interaction and revealed that the structure of the binding cavity is opened up by the loss of a bulky aromatic group leading to alternative ubiquinone binding modes (Fig. 5G and H). It is likely that these additional modes are non-optimal for the crucial balance of association and dissociation following the redox event and are likely to influence the activity of complex III.
Discussion
Despite significant advances in our understanding and treatment of cancers, GBM patients have a very limited survival and it is clear that current treatments are woefully inadequate. Our focus here was to identify and collate specific mtDNA mutations that are likely to contribute to GBM development, progression, and/or chemosensitivity. Our results reveal that the spectrum of mtDNA mutations associated with GBM is wider than previously thought and that individual patients present a surprising diversity of mutational and heteroplasmy profiles.
New Methods Yield Subtle Signatures
Numerous mtDNA mutations have been reported in a range of brain tumor types,8,13,35–39 including GBM,8,11–13,15 although many of these studies were incomplete, as they explored small portions of the mtDNA or used nonquantitative sequencing methods of limited sensitivity (eg, Sanger sequencing). Consequently, several potentially key mtDNA mutations have almost certainly been missed and their heteroplasmies overlooked, despite the latter being an important determinant of whether an mtDNA mutation exerts a phenotype.
Using 3D structural analysis, we predicted that 9 nonsynonymous complex III/IV mutations are functional, occurring mainly in MT-CYB. This finding is consistent with a previous observation that mutations associated with neuropathies tend to occur preferentially in MT-CYB. In the only other previous study where mutations were mapped across a wide-disease cohort, cancer mutations were found solely in complex IV. It is intriguing that GBM shares a signature more closely aligned with neuropathies, rather than cancers per se. In this way GBM has an atypical profile compared with other cancers, although this may reflect the neural origins of the cells.14
Unexpectedly, 2 clearly defined subgroups of brain tumor–associated mutation were identified using a combination of statistical and structural data. Such groupings have not been previously recognized. It is generally thought that mutations that promote cancer will be selected more frequently than neutral mutations,40 so the mutations in subgroup 1 (heteroplasmy)—G101D (17%), F245L (16%), D252N (5%), G239D (34%), and W16R (9%)—are of particular interest given that they are more prevalent in GBM samples compared with those obtained from healthy individuals and are predicted to be functional. However, it is of note that 90% of the control mtDNAs were obtained from blood samples. Given that pathogenic mutations can be lost in blood over time,41 the possibility that potentially functional mtDNA mutations identified in GBM could be found in less proliferative tissues of controls cannot be ruled out and warrants further investigation.
MtDNA Mutations as Novel Diagnostic and Therapeutic Targets
The extent to which mtDNA mutations contribute to tumor aggression may depend on their specific impact on MRC activity.36,42 For example, mutations that mildly affect the activity of the MRC and stimulate ROS production36 have been shown to promote hypoxia-inducible factor 1α (HIF1-α) stabilization/hypoxic adaptation, leading to a more aggressive phenotype.42 In contrast, mutations that severely affect the activity of the MRC and abrogate ROS production have been shown to prevent HIF1-α stabilization/hypoxic adaptation and increase chromosomal stability, leading to a less aggressive tumor phenotype.36,43,44 Although severe complex IV deficiency has been linked to a less aggressive phenotype in GBM,45 whether this was a result of an underlying mutation in mtDNA was not investigated.
In this study, we identify a single germ-line point mutation, T14798C, that is present in one-third of the GBM patients tested. This is a significant finding because not only is this mutation predicted to alter the binding of the natural complex III substrate ubiquinone and thus the activity of complex III and ROS levels (complex III is a major site of ROS production), but it could alter the binding of Qi site inhibitors. Indeed, some of the most effective respiratory inhibitors, such as antimycin and stigmatellin, have been found to act directly at the ubiquinone Qi and Qo sites, respectively. There is strong biochemical and structural evidence that explains the molecular basis for the mutations at these sites that confer resistance to these compounds.34,46 This discovery raises the possibility that complex III inhibitors could be used to trigger mitochondrially mediated apoptosis in some subgroups of GBM patients more effectively than others. Similarly, such information also has the potential to inform the clinician which drugs are likely or unlikely to be effective given a particular mtDNA genetic background.
Predisposing Elements or Triggers of Tumor Formation?
Some mutations occur in the germ-line prior to tumor initiation and act as predisposing elements to neoplastic transformation but do not necessarily cause tumors independently. Other mutations occur somatically and can trigger tumor formation either alone or in combination with a predisposing element. Although both somatic and germ-line mtDNA mutations have been documented in a range of brain tumor types, studies investigating the heritability of mtDNA mutations associated with GBM are generally lacking. Gliomas are known to aggregate in some families, suggesting an inheritable genetic basis. Most of these familial cases are associated with well-known tumor syndromes (eg, neurofibromatosis, Li–Fraumeni). The discovery that 2 of the functional complex III mutations were germ-line reveals a potential genetic predisposition for a small proportion of familial gliomas that occur in the absence of known syndromes.47–49 Equally, the 2 functional complex IV mutations identified as somatic are potential candidates for triggering glioma formation. It remains to be shown whether these mutations are predisposing elements or are sufficient to drive tumor formation independently. In the absence of routine methods for the genetic manipulation of mtDNA, deciphering the contribution of single mtDNA mutations to gliomagenesis will require the development of innovative new approaches.
If the proposed functional mtDNA mutations identified are shown to contribute to gliomagenesis or correlate with key clinical parameters, such as age at diagnosis, therapeutic response, and survival outcome, this will have an impact on how the main GBM subgroups, which currently focus mainly on nuclear genome aberrancies, are treated in the future. They may also provide new targets for the development of mitochondrially mediated therapeutics. Thus future studies of GBM should focus on exploring the role of these mitogenomic elements in gliomagenesis and as indicators of prognosis and drug sensitivity, which will become possible as novel models are developed and as the size of the GBM cohorts with available molecular (particularly mitogenomic) and clinical data grows.
Funding
The authors R.E.L., Q.A., S.C.H., and H.L.F. were supported by Brain Tumour Research (to G.J.P.); K.K. was supported by the Headcase Cancer Trust (to G.J.P.) and the University of Portsmouth Research Development Fund (to J.E.M.).
Supplementary Material
Acknowledgment
We thank Dr Safa Al-Sarraj, Kings College Hospital, London, for verifying the histology of the GBM biopsies.
Conflict of interest statement. None declared.
References
- 1. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;167:896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubrow R, Darefsky AS, Jacobs DI, et al. Time trends in glioblastoma multiforme survival: the role of temozolomide. Neuro Oncol. 2013;15(12):1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 4. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. [DOI] [PubMed] [Google Scholar]
- 7. Deighton RF, Le Bihan T, Martin SF, et al. Interactions among mitochondrial proteins altered in glioblastoma. J Neurooncol. 2014;118(2):247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirches E, Krause G, Warich-Kirches M, et al. High frequency of mitochondrial DNA mutations in glioblastoma identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93(4):534–538. [DOI] [PubMed] [Google Scholar]
- 9. Ordys BB, Launay S, Deighton RF, et al. The role of mitochondria in glioma pathophysiology. Mol Neurobiol. 2010;42(1):64–75. [DOI] [PubMed] [Google Scholar]
- 10. Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeHaan C, Habibi-Nazhad B, Yan E, et al. Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. Mol Cancer. 2004;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Montanini L, Regna-Gladin C, Eoli M, et al. Instability of mitochondrial DNA and MRI and clinical correlations in malignant gliomas. J Neurooncol. 2005;74(1):87–89. [DOI] [PubMed] [Google Scholar]
- 13. Vega A, Salas A, Gamborino E, et al. mtDNA mutations in tumors of the central nervous system reflect the neutral evolution of mtDNA in populations. Oncogene. 2004;23(6):1314–1320. [DOI] [PubMed] [Google Scholar]
- 14. Lloyd RE, McGeehan JE. Structural analysis of mitochondrial mutations reveals a role for bigenomic protein interactions in human disease. PLoS One. 2013;8(7):e69003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larman TC, DePalma SR, Hadjipanayis AG, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109(35):14087–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lloyd RE, Foster PG, Guille M, et al. Next generation sequencing and comparative analyses of Xenopus mitogenomes. BMC Genomics. 2012;13:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li M, Schroeder R, Ko A, et al. Fidelity of capture-enrichment for mtDNA genome sequencing: influence of NUMTs. Nucleic Acids Res. 2012;40(18):e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic. 2004;3(2):157–167. [DOI] [PubMed] [Google Scholar]
- 21. Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. [DOI] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milne I, Stephen G, Bayer M, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14(2):193–202. [DOI] [PubMed] [Google Scholar]
- 26. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;3816:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Attimonelli M, Accetturo M, Santamaria M, et al. HmtDB, a human mitochondrial genomic resource based on variability studies supporting population genetics and biomedical research. BMC Bioinformatics. 2005;6(Suppl 4):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubino F, Piredda R, Calabrese FM, et al. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 2012;40(Database issue):D1150–D1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schlee D, Sneath PHA, Sokal RR, et al. Numerical taxonomy. The principles and practice of numerical classification. System Zool. 1975;24(2):263. [Google Scholar]
- 30. Lee HC, Huang KH, Yeh TS, et al. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol. 2014;20(14):3950–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Althoff T, Mills DJ, Popot JL, et al. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30(22):4652–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palsdottir H, Lojero CG, Trumpower BL, et al. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003;278(33):31303–31311. [DOI] [PubMed] [Google Scholar]
- 33. Seddiki N, Meunier B, Lemesle-Meunier D, et al. Is cytochrome b glutamic acid 272 a quinol binding residue in the bc1 complex of Saccharomyces cerevisiae?. Biochemistry. 2008;47(8):2357–2368. [DOI] [PubMed] [Google Scholar]
- 34. Gao X, Wen X, Esser L, et al. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003;42(30):9067–9080. [DOI] [PubMed] [Google Scholar]
- 35. Kirches E, Krause G, Weis S, et al. Comparison between mitochondrial DNA sequences in low grade astrocytomas and corresponding blood samples. Mol Pathol. 2002;55(3):204–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurelac I, MacKay A, Lambros MB, et al. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas. Hum Mol Genet. 2013;22(2):226–238. [DOI] [PubMed] [Google Scholar]
- 37. Lueth M, von Deimling A, Pietsch T, et al. Medulloblastoma harbor somatic mitochondrial DNA mutations in the D-loop region. J Pediatr Hematol Oncol. 2010;32(2):156–159. [DOI] [PubMed] [Google Scholar]
- 38. Lueth M, Wronski L, Giese A, et al. Somatic mitochondrial mutations in pilocytic astrocytoma. Cancer Genet Cytogenet. 2009;192(1):30–35. [DOI] [PubMed] [Google Scholar]
- 39. Wong LJ, Lueth M, Li XN, et al. Detection of mitochondrial DNA mutations in the tumor and cerebrospinal fluid of medulloblastoma patients. Cancer Res. 2003;63(14):3866–3871. [PubMed] [Google Scholar]
- 40. Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poulton J, Morten K. Noninvasive diagnosis of the MELAS syndrome from blood DNA. Ann Neurol. 1993;34(1):116. [DOI] [PubMed] [Google Scholar]
- 42. Iommarini L, Kurelac I, Capristo M, et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum Mol Genet. 2014;23(6):1453–1466. [DOI] [PubMed] [Google Scholar]
- 43. Gasparre G, Kurelac I, Capristo M, et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 2011;71(19):6220–6229. [DOI] [PubMed] [Google Scholar]
- 44. Park JS, Sharma LK, Li H, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18(9):1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Griguer CE, Cantor AB, Fathallah-Shaykh HM, et al. Prognostic relevance of cytochrome C oxidase in primary glioblastoma multiforme. PLoS One. 2013;8(4):e61035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fisher N, Meunier B. Molecular basis of resistance to cytochrome bc1 inhibitors. FEMS Yeast Res. 2008;8(2):183–192. [DOI] [PubMed] [Google Scholar]
- 47. Dirven CM, Tuerlings J, Molenaar WM, et al. Glioblastoma multiforme in four siblings: a cytogenetic and molecular genetic study. J Neurooncol. 1995;24(3):251–258. [DOI] [PubMed] [Google Scholar]
- 48. Sadetzki S, Bruchim R, Oberman B, et al. Description of selected characteristics of familial glioma patients - results from the Gliogene Consortium. Eur J Cancer. 2013;49(6):1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ugonabo I, Bassily N, Beier A, et al. Familial glioblastoma: a case report of glioblastoma in two brothers and review of literature. Surg Neurol Int. 2011;2:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lange C, Nett JH, Trumpower BL, et al. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20(23):6591–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.