ABSTRACT
Multiple host molecules are known to be involved in the cellular entry of filoviruses, including Ebola virus (EBOV); T-cell immunoglobulin and mucin domain 1 (TIM-1) and Niemann-Pick C1 (NPC1) have been identified as attachment and fusion receptors, respectively. However, the molecular mechanisms underlying the entry process have not been fully understood. We found that TIM-1 and NPC1 colocalized and interacted in the intracellular vesicles where EBOV glycoprotein (GP)-mediated membrane fusion occurred. Interestingly, a TIM-1-specific monoclonal antibody (MAb), M224/1, prevented GP-mediated membrane fusion and also interfered with the binding of TIM-1 to NPC1, suggesting that the interaction between TIM-1 and NPC1 is important for filovirus membrane fusion. Moreover, MAb M224/1 efficiently inhibited the cellular entry of viruses from all known filovirus species. These data suggest a novel mechanism underlying filovirus membrane fusion and provide a potential cellular target for antiviral compounds that can be universally used against filovirus infections.
IMPORTANCE Filoviruses, including Ebola and Marburg viruses, cause rapidly fatal diseases in humans and nonhuman primates. There are currently no approved vaccines or therapeutics for filovirus diseases. In general, the cellular entry step of viruses is one of the key mechanisms to develop antiviral strategies. However, the molecular mechanisms underlying the entry process of filoviruses have not been fully understood. In this study, we demonstrate that TIM-1 and NPC1, which serve as attachment and fusion receptors for filovirus entry, interact in the intracellular vesicles where Ebola virus GP-mediated membrane fusion occurs and that this interaction is important for filovirus infection. We found that filovirus infection and GP-mediated membrane fusion in cultured cells were remarkably suppressed by treatment with a TIM-1-specific monoclonal antibody that interfered with the interaction between TIM-1 and NPC1. Our data provide new insights for the development of antiviral compounds that can be universally used against filovirus infections.
INTRODUCTION
Viruses in the family Filoviridae are filamentous, enveloped, nonsegmented negative-strand RNA viruses that are divided into three genera: Marburgvirus, Ebolavirus, and Cuevavirus. Members of the genera Marburgvirus and Ebolavirus are known to cause severe hemorrhagic fever in humans and nonhuman primates, whereas nothing is known about the pathogenicity of the not yet isolated Cuevavirus (1, 2). There is one known species of Marburgvirus, Marburg marburgvirus, consisting of two viruses, Marburg virus (MARV) and Ravn virus. In contrast, five distinct species are known in the genus Ebolavirus: Zaire ebolavirus, Sudan ebolavirus, Taï forest ebolavirus, Bundibugyo ebolavirus, and Reston ebolavirus, represented by EBOV, Sudan virus (SUDV), Taï forest virus (TAFV), Bundibugyo virus (BDBV), and Reston virus (RESTV), respectively (3). The genus Cuevavirus has one species with one known virus named Lloviu virus (LLOV). In the last decade, the frequency of filovirus hemorrhagic fever outbreaks increased, with the latest one currently ongoing in the neighboring countries Guinea, Liberia, and Sierra Leone (4). Although filoviruses pose a significant threat to public health in western and central Africa and are of worldwide concern with regard to imported cases and potential bioterrorism, there are currently no approved vaccines or therapeutics available.
Filovirus particles consist of at least seven structural proteins, including a glycoprotein (GP), a nucleoprotein (NP), viral proteins (VP) 24, VP30, VP35, VP40, and an RNA-dependent RNA polymerase. The envelope GP is the only viral surface protein and mediates both receptor binding and fusion of the viral envelope with the host cell endosomal membrane during the entry process into cells (5, 6). In particular, EBOV GP is known to interact with membrane-anchored cellular C-type lectins (e.g., DC-SIGN) mainly through its mucin-like domain, which contains a number of N- and O-linked glycosylation sites (7–13). Infection is initiated by binding of GP to attachment factors, such as C-type lectins, followed by internalization of the virus particle into endosomes via macropinocytosis (14–16). Vesicles containing virus particles mature to late endosomes and/or lysosomes, in which low pH leads to proteolytic processing of GPs by cysteine proteases, such as cathepsins (17–19). Although the initiation of the conformational change in GP leading to membrane fusion is not fully understood, it has been suggested that the proteolytically digested GP exposes the putative receptor-binding region, which then interacts with the NPC1 (Niemann-Pick C1) molecule. NPC1 is a large cholesterol transporter protein that localizes in late endosomes and lysosomes (20–22) and has been shown to serve as a fusion receptor for filovirus entry (23–25).
TIM-1 (T-cell immunoglobulin and mucin domain 1) was identified as a filovirus receptor candidate using a bioinformatics approach by performing correlation analysis between gene expression profiles of cells and their permissiveness to viral infection (26). It has been demonstrated that TIM-1 directly interacts with phosphatidylserine (PtdSer) on the viral envelope, suggesting that this molecule is important for the GP-independent attachment of viral particles to cells (27–29). TIM-1 and related PtdSer-binding proteins, such as TIM-4 and Axl (a receptor tyrosine kinase), have subsequently been shown to promote infection of several different enveloped viruses in a manner independent of specific receptor recognition by their envelope glycoproteins (27–29). However, TIM-1 contributes in different ways to virus infection: for filoviruses, alphaviruses, flaviviruses, and arenaviruses, TIM-1 enhances infection, whereas for Lassa virus, herpes simplex virus 1, influenza A virus (H7N1), and severe acute respiratory syndrome (SARS) coronavirus it does not (27, 28, 30).
These cellular attachment factors show distinct expression patterns, depending on the tissue or cell type. Filovirus infection of primary target cells, such as monocytes, macrophages, dendritic cells, hepatocytes, and endothelial cells (31), is likely dependent on the expression of C-type lectins, whereas infection of other cell types later in infection (32) seems to rely on multiple attachment factors. However, NPC1 is required for infection and is ubiquitously expressed. To better understand filovirus entry into cells, we used a monoclonal antibody (MAb), M224/1, against TIM-1 that efficiently inhibited filovirus infection of highly susceptible Vero E6 cells. Interestingly, MAb M224/1 primarily prevented membrane fusion rather than virus attachment. In this study, we demonstrate that the interaction between TIM-1 and NPC1 is important for filovirus entry, suggesting that this interaction could be a novel cellular target for antiviral strategies.
MATERIALS AND METHODS
Viruses and cells.
EBOV (Mayinga), SUDV (Boniface), TAFV (Taï Forest), BDBV (Bundibugyo), RESV (Pennsylvania), MARV (Angola), and vesicular stomatitis virus (VSV) (Indiana serotype) were propagated on Vero E6 cells and stored at −80°C. All infectious work with filoviruses was performed in the biosafety level 4 laboratories at the Integrated Research Facility of the Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT.
Replication-incompetent pseudotyped VSVs containing the green fluorescent protein (GFP) and luciferase genes instead of the VSV G gene were produced as described previously (5, 33). Infectivities of VSV bearing filovirus GPs or VSV G were determined by counting the number of Vero E6 cells expressing GFP under a fluorescence microscope or by measuring luciferase activity in infected cell lysates using a luciferase assay kit (Promega).
Human embryonic kidney 293T cells, African green monkey kidney Vero E6 cells, and human leukemic Jurkat T cells from the repository of our laboratory were used for virus infection and host protein expression. 293T-derived Platinum-GP (Plat-GP) cells (Cell Biolabs) were used for the production of retrovirus vectors. 293T, Plat-GP, and Vero E6 cells were grown in Dulbecco's modified Eagle's medium (DMEM). Jurkat T cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium. Both media were supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin.
Generation of M224/1 and virus entry-inhibition assays.
Five-week-old female BALB/c mice (Japan SLC) were immunized via the intraperitoneal route with formalin-fixed Vero E6 cells. Subsequently, splenocytes were fused with B-cell myeloma cells according to standard procedures (34). Hybridomas were then screened for the ability of secreted MAbs to inhibit the infection of VSV pseudotyped with EBOV GP in Vero E6 cells. MAb M224/1 was obtained screening approximately 1,000 different clones. We confirmed that this antibody did not bind to EBOV GP in immunoblotting and enzyme-linked immunosorbent assays (data not shown). MAb M224/1 and a control MAb (anti-influenza virus hemagglutinin [HA]–IgG1, WZ83 59-6-1, from the repository of our laboratory) were purified from mouse ascites using the Affi-Gel protein A MAPS II kit (Bio-Rad). MAb M224/1 was cleaved with papain, and its Fab fragments were purified using a protein G separation column (ImmunoPure Fab preparation kit; Pierce). To evaluate the inhibitory effect of MAb M224/1 on virus infection, Vero E6 cells were pretreated with MAb M224/1 or its Fab fragments for 30 min at 37°C and then infected with filoviruses, VSV, or pseudotyped VSVs at a multiplicity of infection (MOI) of 0.02 to 0.04 in the presence of the antibody. After incubation for 24 to 48 h, cells infected with filoviruses were fixed and stained with a mixture of anti-GP (anti-EBOV 42/3.7 or anti-MARV FS0505) (35, 36) and anti-NP (anti-EBOV 74/7 or anti-MARV FS0609) (37, 38) as primary antibodies and anti-mouse IgG–Alexa Fluor 488 (A11029; Invitrogen) and anti-rabbit IgG–Alexa Fluor 488 (A11034; Invitrogen) as secondary antibodies. Cells infected with VSV were fixed 8 h postinfection and stained with a mixture of anti-VSV G(N)1-9 (33) and anti-VSV M 195-2 (12) as primary antibodies, and anti-mouse IgG–Alexa Fluor 488 (A11029; Invitrogen) as the secondary antibody. Virus infection was quantified by measuring the number of fluorescent cells. Filovirus GP-mediated infection was quantified 18 h post-VSV pseudotype inoculation by counting the number of GFP-expressing cells under a fluorescence microscope. The relative percentage of infectivity was calculated by setting the number of cells infected in the absence of MAb M224/1 to 100%. Animal studies were carried out in strict accordance with the Guidelines for Proper Conduct of Animal Experiments of the Science Council of Japan. The animal experiments were conducted in strict compliance with animal husbandry and welfare regulations. The mouse study was approved by the Hokkaido University Animal Care and Use Committee (permit no. 08-0235).
Expression cloning.
A full-length cDNA library was prepared from Vero E6 cells (In-Fusion SMARTer directional cDNA library construction kit; Clontech Laboratories) and cloned into a murine leukemia virus (MLV)-based retroviral vector, pMX (39). Plat-GP cells were cotransfected with pMX containing cDNA library genes and a VSV G-expressing plasmid using Lipofectamine 2000 (Invitrogen). Two days later, culture supernatants were collected and cleared of cell debris by centrifugation at 300 × g for 5 min and by passing through 0.45-μm-pore filters. The clean supernatant was then used for infection of Jurkat T cells at an MOI of 0.15, followed by incubation with MAb M224/1 for 1 h at 4°C. After washes with cold phosphate-buffered saline (PBS), the cells were incubated with anti-mouse IgG–Alexa Fluor 488 (A11029; Invitrogen), and positively stained cells were collected using a MoFlo Astrios cell sorter (Beckman Coulter). The collected cells were propagated until 1 × 106 cells were obtained for the next cell sorting. These processes were repeated several times until a cluster of cells expressing the putative target molecule for MAb M224/1 was obtained. Finally, the genomic DNA was extracted from the sorted cell population, and library genes inserted into genomic DNA of the sorted cells were amplified by PCR using library-specific primers and then sequenced.
Generation of TIM-1- and DC-SIGN-expressing cell lines.
Coding sequences of Vero E6 TIM-1 (DDBJ accession no. AB969733) and human DC-SIGN (GenBank accession no. NM021155) (12) were inserted into the MLV retroviral vector pMXs-IRES-GFP (pMXs-IG) (39). To generate the retrovirus, Plat-GP cells were cotransfected with pMXs-IG encoding Vero E6 TIM-1, its IgV domain deletion mutant, or DC-SIGN cDNAs and an expression plasmid for VSV G using Lipofectamine 2000 (Invitrogen). Two days later, the retroviruses were collected in the culture supernatants, clarified through 0.45-μm-pore filters, and then used to infect 293T and Jurkat T cells. Transduced GFP-positive cells were collected using a MoFlo Astrios cell sorter (Beckman Coulter) and used for experiments.
Flow cytometric analysis.
Parental and transduced 293T cells were detached using 0.25% trypsin, washed with cold PBS–2% FCS, and incubated with MAb M224/1 or the goat anti-TIM-1 polyclonal antibody (AF1750; R&D Systems). Primary antibody binding was detected with anti-goat IgG–Alexa Fluor 647 (305-606-047; Jackson ImmunoResearch Laboratories). After several washes, the cells were analyzed employing a FACSCanto flow cytometer (BD Biosciences) and FlowJo software (Tree Star). 293T cells cotransfected with the expression plasmids containing genes encoding NPC1 fused to the N-terminal fragment of monomeric Kusabira-Green [mKG(N)] and/or TIM-1 fused to the C-terminal fragment of mKG [mKG(C)] were analyzed using a FACSCanto flow cytometer (BD Biosciences) and FlowJo software (Tree Star).
Purification and fluorescence labeling of Ebola VLPs.
Ebola virus-like particles (VLPs) were generated by transfection of 293T cells with the expression plasmids for EBOV VP40, NP, and GP using TransIT LT-1 (Mirus). The culture supernatant was harvested 48 h posttransfection and cleared of cell debris by centrifugation at 3,500 rpm for 15 min. VLPs were precipitated through a 30% sucrose cushion by centrifugation at 11,000 rpm for 1 h at 4°C using an SW28 rotor (Beckman Coulter). Pelleted VLPs were then resuspended in TNE buffer (10 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM EDTA) and fractionated through a 20 to 60% sucrose gradient in TNE buffer at 27,000 rpm for 2.5 h at 4°C using an SW40 rotor (Beckman Coulter). The membranes of purified VLPs were then labeled with a lipophilic tracer, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) (Invitrogen), by incubation of the VLPs with a 100 μM solution of DiI in the dark for 1 h at room temperature with gentle agitation (15).
Real-time imaging of the DiI-labeled VLPs in living cells.
Parental Vero E6 cells and Vero E6 cells expressing GFP-Rab7 (15) were grown in 35-mm-diameter glass-bottom culture dishes (Matsunami Glass). Thirty minutes prior to the experiment, the cells were incubated with phenol red-free MEM (Invitrogen)–2% FBS–4% bovine serum albumin (BSA) containing either 50 μg/ml MAb M224/1, 50 μg/ml anti-TIM-1 polyclonal antibody (AF1750; R&D Systems), 20 mM NH4Cl, or 100 μM 5-(N-ethyl-N-isopropyl) amiloride (EIPA), a macropinocytosis inhibitor. The cells were then incubated with DiI-labeled VLPs in the same medium in the presence or absence of the antibodies or inhibitors at room temperature for 30 min. The effect of the different temperatures (i.e., 4°C, room temperature, and 37°C) on the VLP attachment had been previously assessed in the same assay, and there were no appreciable difference in the total numbers of internalized VLPs across the conditions (15). Unbound VLPs were removed by washing with medium, and finally the cells were incubated with or without antibodies or inhibitors for various times at 37°C. For the analysis of attachment of DiI-VLPs on the cell surface, cells were fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature. Nuclei of cells were visualized by Hoechst 33342 staining (Cell Signaling Technology). The cells were analyzed using a Fluoview FV10i confocal laser scanning microscope (Olympus). For measurement of adsorbed DiI-VLPs on the cell surface, images of 8 to 10 optical sections were acquired in 1-μm steps. The number of DiI signals was determined in 50 individual cells (approximately 1 to 10 dots/cell), and the average number per cell was calculated for each condition. The size and fluorescence intensity of DiI dots were analyzed in 50 individual cells with MetaMorph software (Molecular Devices).
Immunofluorescence assay.
Vero E6 or 293T cells were fixed with 4% PFA for 15 min at room temperature and permeabilized with 0.05% Triton X-100 for 15 min. After being blocked with 1% BSA in PBS, the cells were stained with the goat anti-TIM-1 polyclonal antibody (AF1750; R&D Systems), a mouse anti-NPC1 MAb (ab55706; Abcam), or the control antibodies mouse IgG (557273; BD Biosciences) and goat IgG (731635; Beckman Coulter) for 1 h. Cells were washed 3 times with PBS and first stained with a donkey anti-goat IgG–Alexa Fluor 594 (A11058; Invitrogen). After being washed 3 times, cells were then stained with goat anti-mouse IgG–Alexa Fluor 488 (A11029; Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI) for 1 h. The cellular localization of TIM-1 and NPC1 was analyzed using an LSM 780 confocal microscope (Carl Zeiss) with ZEN 2009 software (Carl Zeiss). We confirmed that cross-reactivities of the fluorescent-labeled secondary antibodies to the respective primary antibodies were not appreciably detected, and nonspecific reaction of the secondary antibodies to cellular components was also minimal (data not shown).
Coimmunoprecipitation assay.
The Vero E6-derived TIM-1 gene was C-terminally tagged with FLAG and inserted into a pCAGGS expression vector. The full-length cDNA encoding NPC1 (DDBJ accession no. AB971140) was amplified from total RNA extracted from 293T cells by reverse transcription-PCR (RT-PCR) with specific primer pairs, C-terminally tagged with hemagglutinin (HA), and cloned into the same expression vector. 293T cells were then transfected with plasmids expressing C-terminally FLAG-tagged TIM-1 (TIM-1–FLAG) and/or C-terminally HA-tagged NPC1 (NPC1-HA) using polyethylenimine. Two days after transfection, cells were washed with PBS and treated with lysis buffer (0.05% NP-40, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10% glycerol) containing protease inhibitors (Halt protease inhibitor cocktail; Thermo Scientific). Cell extracts were mixed with protein G Sepharose (GE Healthcare) coupled with an anti-FLAG M2 antibody (Sigma) or anti-HA 3F10 antibody (Roche) and incubated on a rotator at 4°C overnight. The next day, the Sepharose beads were washed three times with wash buffer (0.05% NP-40, 20 mM Tris-HCl [pH 7.4], 250 mM NaCl, 10% glycerol), and the bound proteins were eluted in SDS sample buffer, followed by analysis by SDS-PAGE and immunoblotting with anti-FLAG or HA antibodies.
BiFC assay.
For bimolecular fluorescence complementation (BiFC), expression plasmids for TIM-1–mKG(C) and NPC1-mKG(N) were generated by inserting TIM-1 and NPC1 into fragmented mKG vectors (CoralHue Fluo-chase system; Amalgaam). 293T cells were cotransfected with the complementary pair of the mKG fusion plasmids. After 24 h, cells with fluorescent signals derived from complemented mKG molecules accumulated in cells were counted using flow cytometry. Complemented mKG molecules have been shown to be little dissociated in cells (40, 41). For visualization of early endosomes and Golgi compartments, 293T cells transfected with the mKG fusion plasmids were incubated with CellLight Early Endosomes-RFP (red fluorescent protein) reagents (C10587; Life Technologies) or CellLight Golgi-RFP reagents (C10593; Life Technologies) for 16 h at 37°C. For visualization of lysosomes, cells were incubated with 50 nM LysoTracker Red DND-99 (L-7528; Life Technologies) for 30 min at 37°C. After incubation, cells were fixed with 4% PFA for 10 min at room temperature. The cellular localizations of these organelles and mKG molecules were analyzed using an LSM 780 confocal microscope (Carl Zeiss) with ZEN 2009 software (Carl Zeiss).
RESULTS
Anti-TIM-1 MAb M224/1 blocks filovirus entry into Vero E6 cells.
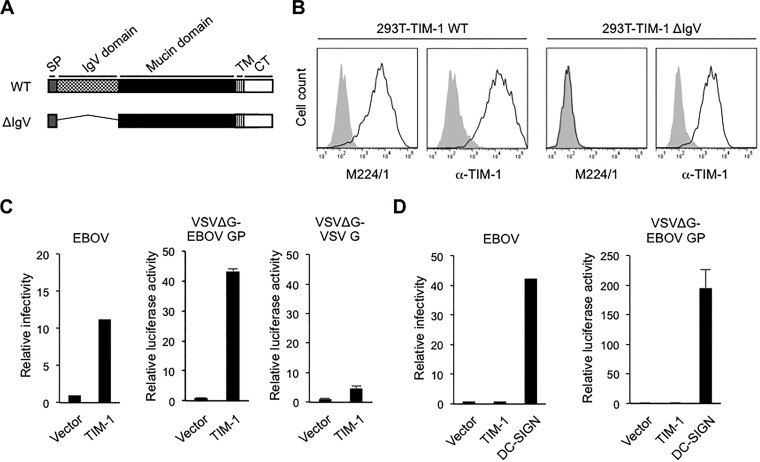
To determine any potential receptor and/or coreceptor molecules required for filovirus entry, we first generated mouse MAbs against surface molecules of Vero E6 cells. Assessment of the antifiloviral activity of the MAbs by studying their ability to block the infectivity of VSV pseudotyped with EBOV GP (VSVΔG-EBOV GP) identified MAb M224/1 (IgG1) as a potent entry inhibitor. Expression cloning using a Vero E6 cDNA library identified TIM-1 as the target molecule of MAb M224/1. TIM-1 consists of two domains: an N-terminal IgV domain that forms a PtdSer-binding pocket and a highly glycosylated mucin domain (42) (Fig. 1A). MAb M224/1 recognized full-length TIM-1 on 293T cells but not a mutant lacking its IgV domain (ΔIgV), indicating that the IgV domain is important for the interaction with MAb M224/1 (Fig. 1B). Ectopic expression of this Vero E6 cell-derived TIM-1 in 293T cells, which naturally lack cell surface TIM-1 (26), dramatically enhanced susceptibility of cells to EBOV and VSVΔG-EBOV GP infection, whereas only a limited effect was observed on infection with VSV bearing VSV G protein (VSVΔG-VSV G) (Fig. 1C). Interestingly, while expression of DC-SIGN led to productive EBOV and VSVΔG-EBOV GP infection of Jurkat T cells that are poorly permissive for filovirus infections (6), expression of TIM-1 did not (Fig. 1D). These observations were consistent with previous studies (7, 43), suggesting that there might be a distinct mechanism underlying EBOV GP-mediated entry, depending on which attachment factor is used, TIM-1 or DC-SIGN.
FIG 1.
Binding of M224/1 to the TIM-1 IgV domain and effects of TIM-1 expression on virus infectivity. (A) Schematic representation of wild-type (WT) TIM-1 and its IgV deletion mutant (ΔIgV). SP, signal peptide; TM, transmembrane region; CT, cytoplasmic tail. (B) MLV-transduced 293T cells stably expressing the Vero E6 TIM-1 WT and ΔIgV mutant were stained with MAb M224/1 or the anti-TIM-1 polyclonal antibody and analyzed by flow cytometry. Black lines represent TIM-1-expressing cells. Gray shading represents vector-transduced control cells. (C) 293T cells stably expressing Vero E6-derived TIM-1 and vector-transduced control cells were infected with EBOV (left panel), VSVΔG-EBOV GP (middle panel), or VSVΔG-VSV G (right panel). (D) Jurkat T cells stably expressing Vero E6-derived TIM-1 or DC-SIGN and vector-transduced control cells were infected with EBOV (left panel) or VSVΔG-EBOV GP (right panel). EBOV supernatant was collected 48 h postinfection, and viral titers were determined by performing the 50% tissue-culture-infective dose (TCID50) assay (52). Luciferase activities of pseudotyped VSVs were measured 24 h postinfection. The relative infectivity was determined by setting the value (TCID50 or luciferase activity) of infected control cells to 1.0. Each experiment was performed three times. One representative experiment for EBOV is shown. For VSV, the mean of three independent experiments is shown. Error bars represent standard deviation (SD).
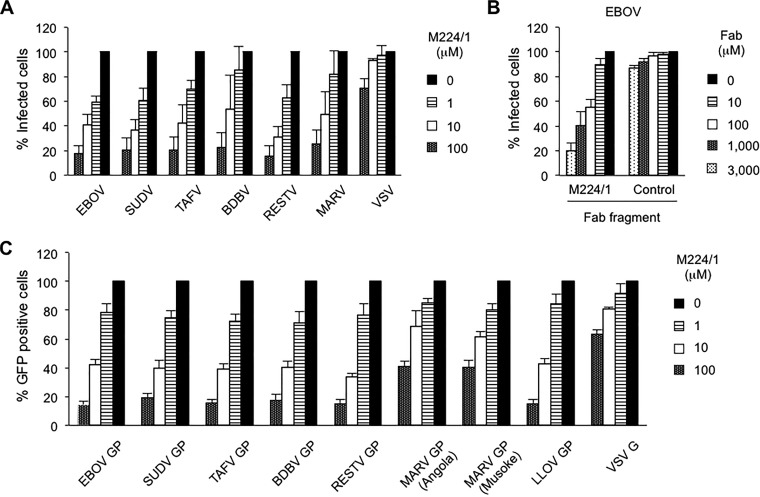
Next, we investigated the ability of MAb M224/1 to inhibit infection and replication of representatives from all known filovirus species (Fig. 2A). We observed that replication of all tested filoviruses was dramatically decreased in MAb M224/1-treated cells in a dose-dependent manner. Replication of VSV, which served as a control in this assay, was not substantially affected by MAb M224/1 (Fig. 2A), suggesting that this antibody has a filovirus-specific inhibitory effect. Notably, EBOV replication was reduced even when cells were treated with the M224/1 Fab fragment, suggesting that cross-linking by the entire IgG molecule is not required for the inhibitory activity of MAb M224/1 (Fig. 2B). Filovirus GP-mediated entry analyzed using VSVΔG-EBOV GP was also considerably inhibited by MAb M224/1 (Fig. 2C). Treatment with a control MAb did not show any inhibitory effects, even at the highest concentration tested (100 μM) (data not shown). These results indicated that this antibody blocked the entry step of filovirus infections (i.e., attachment, internalization, or membrane fusion).
FIG 2.
Inhibition of filovirus GP-mediated infection by MAb M224/1. (A) Vero E6 cells were pretreated with the indicated concentrations of MAb M224/1 for 30 min at 37°C and infected with filoviruses or VSV in the presence of MAb M224/1. Infected cells were stained with virus-specific antibodies and counted. The relative percentage of infected cells was determined by setting the number of untreated infected cells to 100% (approximately 50 to 100 fluorescent cells per microscopic field). (B) Vero E6 cells were infected with EBOV in the presence of M224/1 Fab fragments or a control antibody (WZ83 59-6-1). The relative percentage of EBOV-infected cells was determined as described above. (C) In the presence of different MAb M224/1 concentrations, Vero E6 cells were infected with VSV pseudotypes bearing the indicated filovirus GP or VSV G. GFP-positive cells were counted, and the relative percentage of infected cells was determined as described above. The mean of three independent experiments is shown. Error bars represent SD.
M224/1 primarily inhibits viral membrane fusion rather than virus attachment.
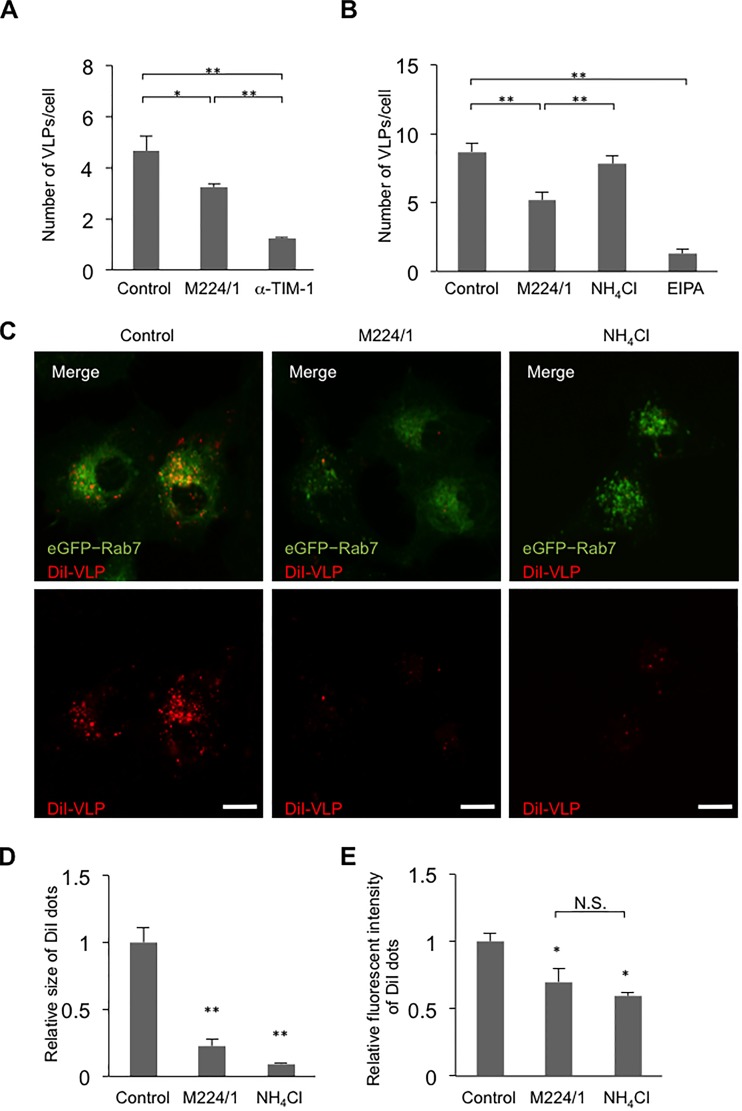
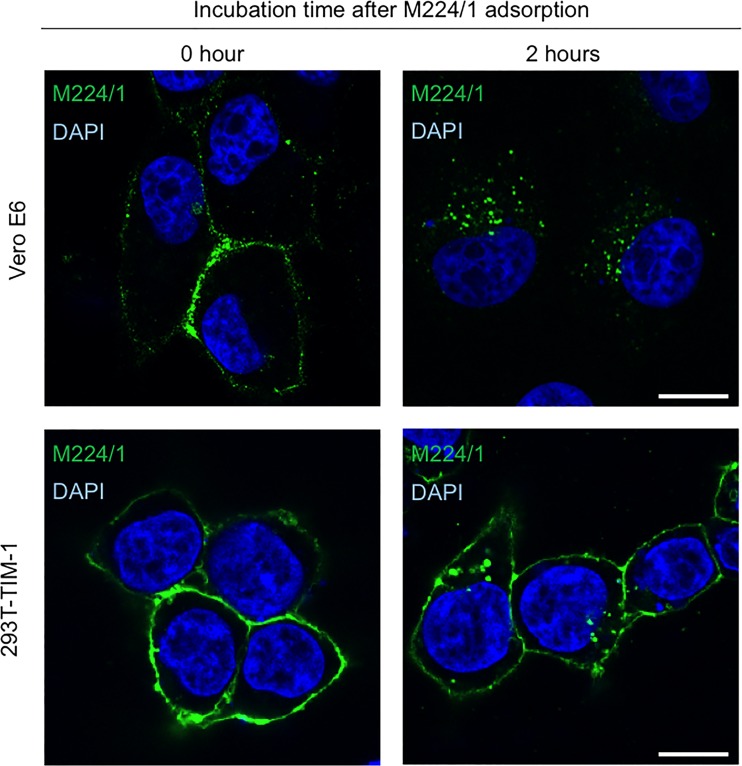
Since TIM-1 is proposed to serve as an attachment factor for filovirus infection, we first hypothesized that MAb M224/1 inhibits virus adsorption to cells by competing with the PtdSer-binding activity of TIM-1. To analyze this, we generated fluorescent (DiI)-labeled VLPs consisting of EBOV GP, VP40, and NP (44, 45) and compared the number of bound particles on cell surface in the presence and absence of MAb M224/1. Interestingly, we found that the number of VLPs attached to the surface of Vero E6 cells was only slightly decreased in the presence of MAb M224/1, whereas an anti-TIM-1 polyclonal antibody efficiently reduced VLP binding to cells (Fig. 3A). Consistent with the reduced VLP attachment, a slight reduction of VLP internalization was observed in MAb M224/1-treated cells, whereas many fewer VLPs were detected in cells treated with 5-(N-ethyl-N-isopropyl) amiloride (EIPA), a macropinocytosis inhibitor (Fig. 3B). Trypsinization of cells after 37°C incubation did not affect the number of visible VLPs (data not shown), confirming that the internalized VLPs, but not cell-surface-bound VLPs, were indeed counted in this assay. The number of VLPs internalized into untreated cells was similar to that measured for cells treated with NH4Cl, an inhibitor of endosomal acidification, confirming that VLPs detected under the assay conditions were likely in a prefusion state. We then analyzed membrane fusion by detection of dequenched DiI fluorescence (Fig. 3C to E). In this assay, once the DiI-labeled VLP envelopes fuse with the endosomal membrane, the fluorescent signal is enhanced (15). In Vero E6 cells expressing GFP-fused Rab7, a late endosome marker, we observed remarkably enlarged and enhanced VLP fusion signals (15) after cells were incubated with DiI-labeled VLPs for 5 h at 37°C, indicating that membrane fusion occurred in the endosomes (Fig. 3C, left panels). In contrast, the DiI signals in MAb M224/1-treated cells were considerably weaker than those in untreated cells and similar to those in NH4Cl-treated cells (Fig. 3C, middle and right panels). Quantification of the size and intensity of the DiI fluorescent dots confirmed that their average size and intensity were reduced in MAb M224/1-treated cells compared to untreated cells (Fig. 3D and E). These results suggest that MAb M224/1 primarily prevents membrane fusion. To confirm intracellular delivery of M224/1, Vero E6 cells and 293T cells transfected with a plasmid expressing TIM-1 were incubated with MAb M224/1 for 30 min on ice and subsequently incubated for 2 h at 37°C, and localization of the antibody was analyzed by confocal laser scanning microscopy. We found that cell-bound MAb M224/1 was indeed internalized into intracellular vesicles (Fig. 4).
FIG 3.
Inhibition of Ebola virus VLP-induced membrane fusion by MAb M224/1. (A and B) Vero E6 cells were pretreated with MAb M224/1, anti-TIM-1 polyclonal antibody, NH4Cl, or EIPA for 30 min and incubated with DiI-labeled Ebola virus VLPs for 30 min at room temperature. (B) After VLP adsorption, the cells were shifted to 37°C incubation for 30 min in the presence of antibodies or inhibitors. DiI signals on the cell surface (A) and in the cytoplasm (B) were quantified by confocal laser scanning microscopy. (C to E) After VLP adsorption (red), Vero E6 cells expressing GFP-Rab7 (green) were treated with MAb M224/1 or NH4Cl and incubated for 5 h at 37°C. (C) DiI signals representing VLPs in the cytoplasm were observed in the same focal plane by confocal laser scanning microscopy. Scale bars represent 20 μm. (D and E) Fluorescent intensities of the DiI signals were quantified. Their relative size and the intensity of DiI dots were determined by defining the value of untreated infected cells as 1. The mean of three independent experiments is shown. We confirmed that the control MAb (WZ83 59-6-1) had no significant effect on the membrane fusion activity (data not shown). Error bars represent SD. Statistical analysis was performed using Student's t test (*, P < 0.05; **, P < 0.01; N.S., not significant).
FIG 4.
Internalization of TIM-1-bound MAb M224/1 into intracellular vesicles. Vero E6 cells (upper panels) and 293T cells transfected with a plasmid expressing TIM-1 (lower panels) were incubated with MAb M224/1 for 30 min on ice (0 h) and subsequently incubated for 2 h at 37°C (2 h). MAb M224/1 was visualized with anti-mouse IgG–Alexa Fluor 488 and analyzed by confocal laser scanning microscopy. Nuclei of cells were stained with DAPI (blue). Scale bars represent 20 μm.
TIM-1 interacts with NPC1.
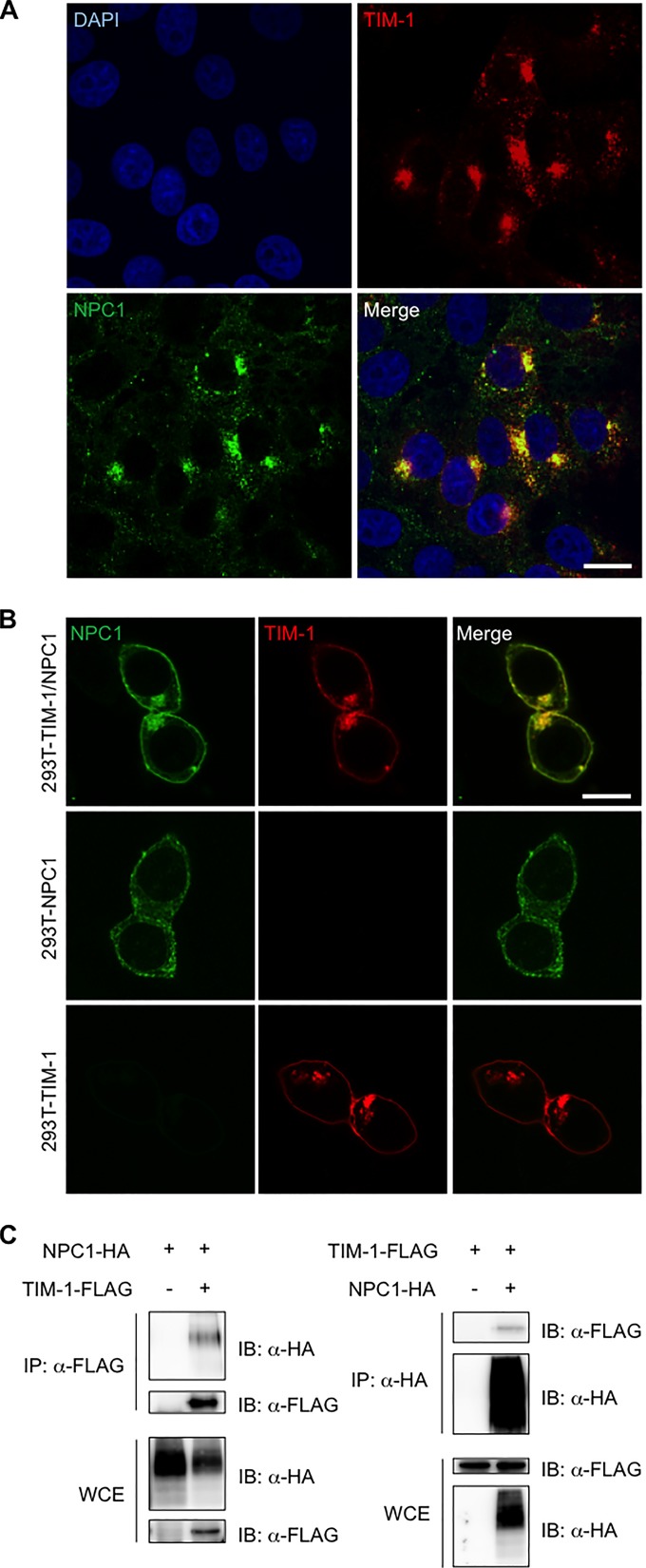
It has been postulated that EBOV membrane fusion occurs in late endosomes and/or lysosomes following the interaction of GP with NPC1, an essential molecule for infection, as the fusion receptor (23–25). The fact that anti-TIM-1 MAb M224/1 inhibited endosomal membrane fusion led us to hypothesize that TIM-1 might directly interact with NPC1. To confirm this, we first examined the localization of TIM-1 and NPC1 in Vero E6 cells. Consistent with a previous study showing that TIM-1 clusters mostly in the cytoplasm (46), our immunofluorescent assay demonstrated that TIM-1 localized mainly in the cytoplasm. As previously reported, NPC1 was primarily located in the endosomal and lysosomal membranes in Vero E6 cells. Notably, we found that TIM-1 and NPC1 were mostly colocalized in intracellular vesicles in the cytoplasm of Vero E6 cells (Fig. 5A), while these proteins were also detected and colocalized in plasma membranes when they were overexpressed in 293T cells (Fig. 5B). We next examined direct interaction between TIM-1 and NPC1 by an immunoprecipitation assay (Fig. 5C). TIM-1–FLAG and NPC1-HA were expressed in 293T cells and immunoprecipitated using either anti-FLAG or anti-HA antibodies. We found that NPC1-HA was coprecipitated with TIM-1–FLAG captured by the anti-FLAG antibody. Vice versa, TIM-1–FLAG was coprecipitated with NPC1-HA bound by the anti-HA antibody. We further found that while TIM-1 localized in the plasma membrane and perinuclear vesicles in cells transfected with the plasmid, NPC1 was diffusely present in the cytoplasm when expressed alone but shifted to expression in perinuclear vesicles, depending on the presence of TIM-1 (Fig. 5B). These results strongly suggested that TIM-1 directly bound to NPC1.
FIG 5.
Colocalization and interaction of TIM-1 with NPC1 in Vero E6 cells. (A) Intracellular TIM-1 (red) and NPC1 (green) in Vero E6 cells were visualized with specific antibodies and analyzed by confocal laser scanning microscopy. Nuclei of cells were visualized with DAPI (blue). Scale bars represent 20 μm. (B) Intracellular TIM-1 (red) and NPC1 (green) in 293T cells transfected with plasmids encoding TIM-1-FLAG and/or NPC1-HA were visualized with specific antibodies and analyzed by confocal laser scanning microscopy. Scale bars represent 20 μm. (C) 293T cells were transfected with plasmids encoding TIM-1–FLAG and/or NPC1-HA and subjected to immunoprecipitation (IP) with anti-FLAG (left panel) or anti-HA (right panel) antibodies followed by immunoblotting (IB). WCE, whole-cell extract.
M224/1 inhibits the binding of TIM-1 to NPC1.
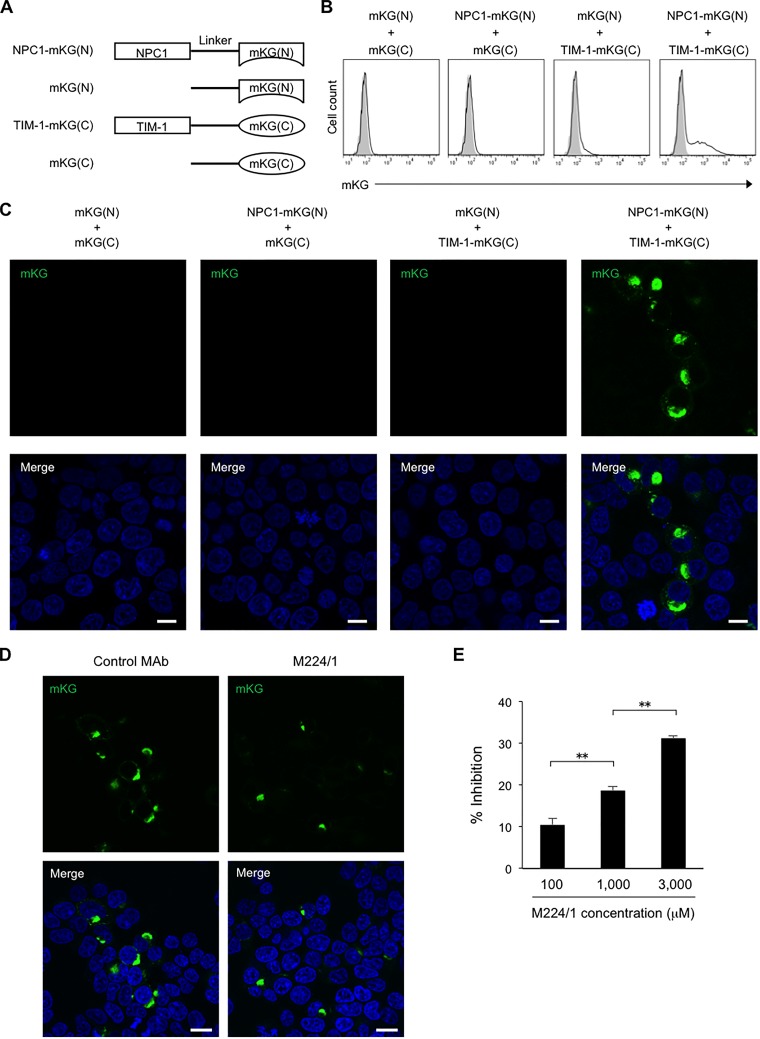
To further characterize the intracellular interaction between TIM-1 and NPC1, a BiFC assay was performed. In this assay, TIM-1 and NPC1 were fused to two inactive fragments of mKG (Fig. 6A), and the fluorescence of reconstructed mKG molecules accumulating in cells could be observed only upon complementation of the two molecules. We detected the fluorescence of mKG in 293T cells cotransfected with TIM-1–mKG(C) and NPC1-mKG(N) by flow cytometry, indicating that the direct binding between TIM-1 and NPC1 occurs intracellularly (Fig. 6B). Furthermore, fluorescence microscopy showed that the fluorescence signal was mainly located in intracellular vesicles not on the cell surface (Fig. 6C). Next, we investigated the effect of MAb M224/1 on intracellular interaction between TIM-1 and NPC1. We found that the mKG expression was remarkably decreased in MAb M224/1-treated cells and that the number of mKG-positive cells was reduced by the antibody treatment in a dose-dependent manner (Fig. 6D and E).
FIG 6.
Visualization of the intracellular interaction between TIM-1 and NPC1. (A) Schematic representation of truncated mKG fused to either TIM-1 or NPC1. mKG(N), N-terminal fragment of mKG; mKG(C), C-terminal fragment of mKG. (B) 293T cells cotransfected with the indicated plasmid pairs were analyzed for mKG signals by flow cytometry 24 h posttransfection. Open and shaded histograms indicate the cells transfected with the indicated plasmid pairs and the nontransfected cells, respectively. (C) 293T cells were cotransfected with the indicated plasmid pairs. Cytoplasmic localization of mKG (green) representing the TIM-1–NPC1 binding was visualized by confocal laser scanning microscopy 24 h posttransfection. Nuclei of cells were stained with DAPI (blue). Scale bars represent 20 μm. (D and E) 293T cells were cotransfected with plasmids encoding NPC1-mKG(N) and TIM-1–mKG(C). (D) At 8 h posttransfection the cell culture medium was replaced with fresh medium containing 3,000 μM MAb M224/1 or a control antibody. After a 16-h incubation, mKG signals were detected using confocal laser scanning microscopy. Nuclei of cells were stained with DAPI (blue). Scale bars represent 20 μm. (E) 293T cells were incubated with different concentrations of MAb M224/1 or the control antibody. The number of mKG-positive cells was counted by flow cytometry. The reduced numbers of mKG-positive cells in MAb M224/1-treated cells are presented as percentage of inhibition compared to the number of mKG-positive cells treated with the control antibody. Each experiment was performed four times, and the results are presented as the mean. Error bars represent SD. Significance was calculated using Student's t test (**, P < 0.01).
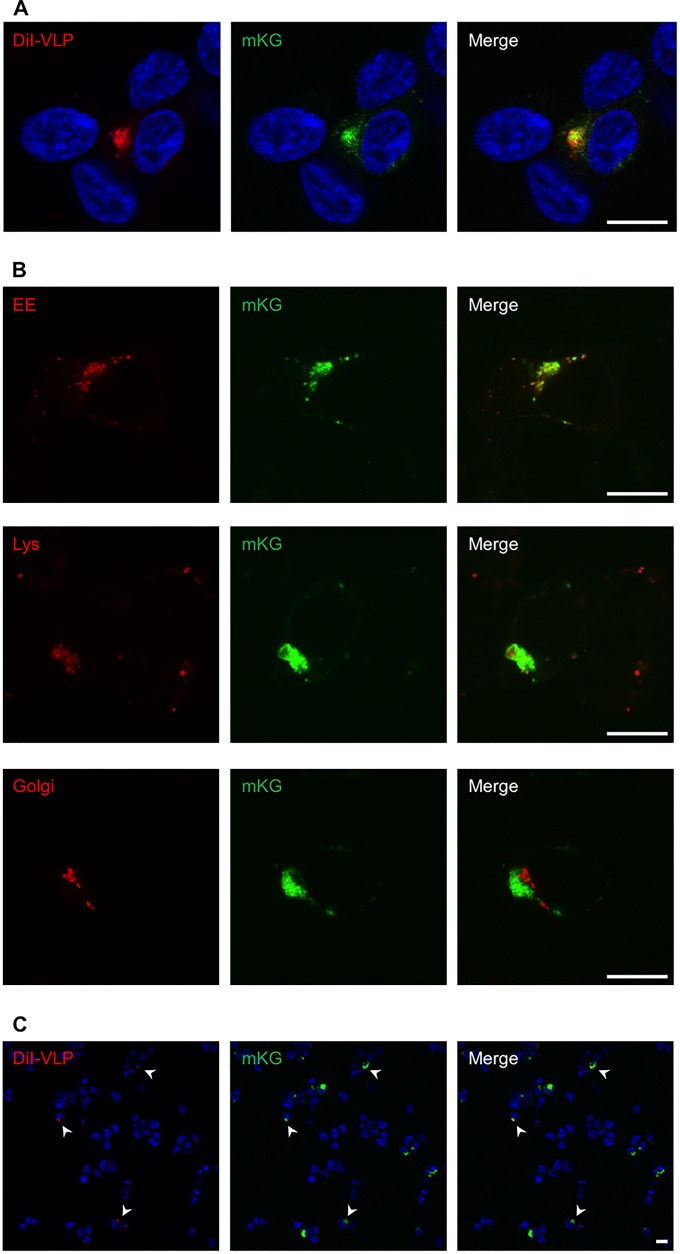
VLP-induced membrane fusion occurs in intracellular vesicles where TIM-1 and NPC1 colocalize and interact.
Finally, we investigated the colocalization of TIM-1/NPC1 complex with intracellular vesicles (likely late endosomes/lysosomes) in which VLP-induced membrane fusion occurs. We found that the enhanced or enlarged DiI signals representing VLP-induced membrane fusion colocalized with the mKG signals, the sites of TIM-1/NPC1 interaction in endosomes (Fig. 7A). We found that the TIM-1/NPC1 complex colocalized with endosomes/lysosomes but not Golgi compartments (Fig. 7B). Interestingly, most of the enhanced or enlarged DiI signals representing VLP-induced membrane fusion were associated with mKG-positive cells, and only a limited number of DiI signals were found in mKG-negative cells (Fig. 7C). This might be consistent with the observation that overexpression of TIM-1 enhanced virus infection, as shown in Fig. 2, whereas it is also possible that membrane fusion between the VLP envelope and the endosomal membrane could occur more efficiently in cells where the TIM-1/NPC1 interaction takes place (mKG positive) than in mKG-negative cells. Taken together, these results implied that the interaction between TIM-1 and NPC1 was important for EBOV GP-mediated membrane fusion.
FIG 7.
The TIM-1/NPC1 interaction required for EBOV membrane fusion. (A) 293T cells cotransfected with plasmids encoding NPC1-mKG(N) and TIM-1-mKG(C) were incubated with DiI-labeled VLPs for 30 min on ice. The cells were subsequently incubated for 5 h at 37°C and fixed with 4% PFA. DiI (red) and mKG (green) signals in the cytoplasm in the same focal plane were observed by confocal laser scanning microscopy. Nuclei of cells were visualized by Hoechst 33342 staining (blue). Scale bars represent 20 μm. (B) Early endosomes (EE), lysosomes (Lys), and Golgi compartments in 293T cells cotransfected with the plasmids were visualized as described in Materials and Methods. Scale bars represent 20 μm. (C) Lower-magnification images of cells described in panel A are shown. Arrowheads indicate the cells in which both mKG (TIM-1/NPC1) and enlarged DiI signals (membrane fusion) were detected. Scale bars represent 20 μm.
DISCUSSION
It is well established that filoviruses utilize multiple host cell molecules for attachment and entry into cells, but the molecular mechanisms underlying this process are not fully understood. While filovirus envelope GPs have been shown to mediate both receptor binding and fusion of the viral envelope with the host cell membrane, a bioinformatics approach previously identified TIM-1 as a GP-independent attachment factor promoting filovirus infection (26–29). Using a more classical approach to discover cell surface molecules serving as viral receptors, we also detected TIM-1 and demonstrated its important role in filovirus infection. However, the present study suggests that although TIM-1 promotes filovirus entry, the process is not as simple as just enhanced attachment of the virus particle to cell surface.
Although TIM-1 is mainly expressed on the plasma membrane, intracellular TIM-1 can be detected in early endosomes and lysosomes and has been shown to cycle dynamically to and from the cell surface (46). Our data also show that TIM-1 is present in intracellular vesicles as well as on the cell surface. Importantly, we found that TIM-1 colocalized and directly interacted with NPC1 in endosomes where membrane fusion of Ebola VLPs occurred. Although NPC1 has been shown to be essential for GP-mediated membrane fusion in late endosomes/lysosomes during filovirus infection (23–25, 27), the contribution of intracellular TIM-1 to a postattachment step remained unclear. Our data suggest that TIM-1 is involved not only in filovirus attachment but also in efficient membrane fusion through the interaction with NPC1. It appears that after internalization of TIM-1-captured virus particles via macropinocytosis, the binding of TIM-1 to NPC1 in late endosomes/lysosomes is required for GP-mediated membrane fusion (Fig. 8). This hypothesis is strongly supported by the fact that filovirus infection and EBOV GP-mediated membrane fusion in Vero E6 cells were considerably decreased in the presence of TIM-1-specific MAb M224/1, which interfered with the binding of TIM-1 to NPC1 in late endosomes/lysosomes.
FIG 8.
Proposed model of EBOV GP-mediated cellular entry into TIM-1-expressing cells. (Part 1) EBOV attaches to the cell surface via binding of TIM-1 to PtdSer on the viral envelope. (Part 2) After internalization of TIM-1-bound virus particles into early endosomes via macropinocytosis, the virus particles are transported to late endosomes/lysosomes, in which the GP is proteolytically processed. (Part 3) NPC1 interacts with TIM-1 and processed GP, leading to membrane fusion and RNP release.
The proposed model for EBOV entry into cells includes several steps: The first step is GP-dependent (e.g., C-type lectins) or -independent (e.g., TIM-1) virus attachment, which is then followed by the internalization of attached particles via macropinocytosis. Next, GP is cleaved in late endosomes by cysteine proteases, such as cathepsins, resulting in the exposure of the receptor-binding region and the final binding of GP to NPC1, triggering membrane fusion. Our results suggest an additional step in this process, the binding of TIM-1 to NPC1, which is important for membrane fusion. We hypothesize that TIM-1 may facilitate the binding of NPC1 to the receptor-binding region in GP by adjacently bridging the endosomal membrane and viral envelope. This event could be important for membrane fusion triggered by the conformational change in GP. Another possibility is that the interaction between TIM-1 and NPC1 directly initiates the conformational change in GP following the binding to NPC1, which then triggers membrane fusion.
However, it is known that filoviruses infect macrophages and dendritic cells, which do not express TIM-1. We hypothesize that previously identified attachment factors, such as the TAM (i.e., Tyro3, Axl, and Mer) receptor family, or yet unknown cellular molecule(s) may have the potential to function as fusion facilitators. Members of the TAM receptor family are expressed on the surfaces of macrophages and dendritic cells (47) and can indeed facilitate PtdSer-mediated virus uptake via indirect binding to PtdSer on the viral envelope (30, 48). In addition, activated Axl was shown to be internalized by endocytosis and degraded in lysosomes (49). Therefore, we propose that some other attachment factors may play a similar role in cells lacking TIM-1.
Under natural physiological conditions, the TIM-1 IgV domain recognizes PtdSer exposed on apoptotic cells and facilitates the clearance of these cells by phagocytosis (50, 51). PtdSer is also present on the outer membrane of the viral envelope and facilitates virus attachment and uptake via a process known as apoptotic mimicry (27–29). Our results demonstrated that the Fab fragment of MAb M224/1 recognizing the IgV domain in TIM-1 inhibited filovirus entry. Taken together, our data suggest that MAb M224/1 directly binds to the pivotal site of the IgV domain, which is important for the TIM-1 functions to promote filovirus entry (i.e., PtdSer recognition and interaction with NPC1). Notably, MAb M224/1 significantly prevented membrane fusion (approximately 80%), while its inhibitory effect on virus attachment was limited (30 to 40%), suggesting that the inhibition of membrane fusion is the principal mechanism of M224/1-mediated reduction of filovirus infections. Therefore, TIM-1 may be primarily involved in the EBOV GP-mediated membrane fusion process rather than virus attachment. We further demonstrated that MAb M224/1 interfered with the binding of TIM-1 to NPC1, suggesting that this novel interaction is likely required for filovirus membrane fusion and may serve as an attractive target for antiviral strategies. Thus, deciphering the detailed structures of the intermolecular interface between MAb M224/1 and TIM-1 or TIM-1 and NPC1 may provide new insights into the development of antivirals, such as low-molecular-weight compounds that can be universally used against filovirus infections.
ACKNOWLEDGMENTS
We thank Mari Ishijima for technical assistance and Kim Barrymore for editing the manuscript.
This work was supported by KAKENHI, a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Japan Society for the Promotion of Science (JSPS), Japan. Funding was also provided by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) and the Japan Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA), within the framework of the Science and Technology Research Partnership for Sustainable Development (SATREPS). This study was partly funded by the Program for Leading Graduate Schools from MEXT, Japan, and by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, United States.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Maruyama J, Miyamoto H, Kajihara M, Ogawa H, Maeda K, Sakoda Y, Yoshida R, Takada A. 2014. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J Virol 88:99–109. doi: 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, Herrera JE, Pizarro M, Hutchison SK, Echevarria JE, Lipkin WI, Tenorio A. 2011. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukreyev AA, Chandran K, Dolnik O, Dye JM, Ebihara H, Leroy EM, Muhlberger E, Netesov SV, Patterson JL, Paweska JT, Saphire EO, Smither SJ, Takada A, Towner JS, Volchkov VE, Warren TK, Kuhn JH. 2014. Discussions and decisions of the 2012-2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012–June 2013. Arch Virol 159:821–830. doi: 10.1007/s00705-013-1846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keita S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traore A, Kolie M, Malano ER, Heleze E, Bocquin A, Mely S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Gunther S. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 5.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A 94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wool-Lewis RJ, Bates P. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol 72:3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker S, Spiess M, Klenk HD. 1995. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J Gen Virol 76:393–399. doi: 10.1099/0022-1317-76-2-393. [DOI] [PubMed] [Google Scholar]
- 9.Gramberg T, Hofmann H, Moller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH, Becker S, Munch J, Pohlmann S. 2005. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin G, Simmons G, Pohlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA, Doms RW. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol 77:1337–1346. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzi A, Gramberg T, Simmons G, Moller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, Steinkasserer A, Becker S, Bates P, Hofmann H, Pohlmann S. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol 78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuno K, Kishida N, Usami K, Igarashi M, Yoshida R, Nakayama E, Shimojima M, Feldmann H, Irimura T, Kawaoka Y, Takada A. 2010. Different potential of C-type lectin-mediated entry between Marburg virus strains. J Virol 84:5140–5147. doi: 10.1128/JVI.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada A, Fujioka K, Tsuiji M, Morikawa A, Higashi N, Ebihara H, Kobasa D, Feldmann H, Irimura T, Kawaoka Y. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol 78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler HJ. 2011. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 204(Suppl 3):S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. 2010. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misasi J, Chandran K, Yang JY, Considine B, Filone CM, Cote M, Sullivan N, Fabozzi G, Hensley L, Cunningham J. 2012. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol 86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins ME, Davies JP, Chen FW, Ioannou YA. 1999. Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol Genet Metab 68:1–13. doi: 10.1006/mgme.1999.2882. [DOI] [PubMed] [Google Scholar]
- 21.Neufeld EB, Wastney M, Patel S, Suresh S, Cooney AM, Dwyer NK, Roff CF, Ohno K, Morris JA, Carstea ED, Incardona JP, Strauss JF III, Vanier MT, Patterson MC, Brady RO, Pentchev PG, Blanchette-Mackie EJ. 1999. The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J Biol Chem 274:9627–9635. doi: 10.1074/jbc.274.14.9627. [DOI] [PubMed] [Google Scholar]
- 22.Patel SC, Suresh S, Kumar U, Hu CY, Cooney A, Blanchette-Mackie EJ, Neufeld EB, Patel RC, Brady RO, Patel YC, Pentchev PG, Ong WY. 1999. Localization of Niemann-Pick C1 protein in astrocytes: implications for neuronal degeneration in Niemann-Pick type C disease. Proc Natl Acad Sci U S A 96:1657–1662. doi: 10.1073/pnas.96.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye J M, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB Jr, Chiorini J, Maury W. 2011. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci U S A 108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. 2013. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog 9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. 2013. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 87:8327–8341. doi: 10.1128/JVI.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morizono K, Chen IS. 2014. Role of phosphatidylserine receptors in enveloped virus infection. J Virol 88:4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisbert TW, Hensley LE. 2004. Ebola virus: new insights into disease aetiopathology and possible therapeutic interventions. Expert Rev Mol Med 6:1–24. doi: 10.1017/S1462399404008300. [DOI] [PubMed] [Google Scholar]
- 32.Feldmann H, Klenk HD. 1996. Filoviruses. Chapter 72 In Baron S. (ed), Medical microbiology, 4th ed. University of Texas Medical Branch, Galveston, TX: http://www.ncbi.nlm.nih.gov/books/NBK8129/. [PubMed] [Google Scholar]
- 33.Nakayama E, Tomabechi D, Matsuno K, Kishida N, Yoshida R, Feldmann H, Takada A. 2011. Antibody-dependent enhancement of Marburg virus infection. J Infect Dis 204(Suppl 3):S978–S985. doi: 10.1093/infdis/jir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahhosseini S, Das D, Qiu X, Feldmann H, Jones SM, Suresh MR. 2007. Production and characterization of monoclonal antibodies against different epitopes of Ebola virus antigens. J Virol Methods 143:29–37. doi: 10.1016/j.jviromet.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Kajihara M, Nakayama E, Marzi A, Igarashi M, Feldmann H, Takada A. 2013. Novel mutations in Marburg virus glycoprotein associated with viral evasion from antibody mediated immune pressure. J Gen Virol 94:876–883. doi: 10.1099/vir.0.049114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada A, Watanabe S, Okazaki K, Kida H, Kawaoka Y. 2001. Infectivity-enhancing antibodies to Ebola virus glycoprotein. J Virol 75:2324–2330. doi: 10.1128/JVI.75.5.2324-2330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Changula K, Yoshida R, Noyori O, Marzi A, Miyamoto H, Ishijima M, Yokoyama A, Kajihara M, Feldmann H, Mweene AS, Takada A. 2013. Mapping of conserved and species-specific antibody epitopes on the Ebola virus nucleoprotein. Virus Res 176:83–90. doi: 10.1016/j.virusres.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kajihara M, Marzi A, Nakayama E, Noda T, Kuroda M, Manzoor R, Matsuno K, Feldmann H, Yoshida R, Kawaoka Y, Takada A. 2012. Inhibition of Marburg virus budding by nonneutralizing antibodies to the envelope glycoprotein. J Virol 86:13467–13474. doi: 10.1128/JVI.01896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 31:1007–1014. doi: 10.1016/j.exphem.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto J, Watanabe T, Seki T, Karasawa S, Izumikawa M, Seki T, Iemura S, Natsume T, Nomura N, Goshima N, Miyawaki A, Takagi M, Shin-Ya K. 2009. Novel in vitro protein fragment complementation assay applicable to high-throughput screening in a 1536-well format. J Biomol Screen 14:970–979. doi: 10.1177/1087057109341406. [DOI] [PubMed] [Google Scholar]
- 41.Ueyama T, Kusakabe T, Karasawa S, Kawasaki T, Shimizu A, Son J, Leto TL, Miyawaki A, Saito N. 2008. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: evaluation using the novel monomeric Kusabira-Green system and live imaging of phagocytosis. J Immunol 181:629–640. doi: 10.4049/jimmunol.181.1.629. [DOI] [PubMed] [Google Scholar]
- 42.McIntire J J, Umetsu DT, DeKruyff RH. 2004. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin Immunopathol 25:335–348. doi: 10.1007/s00281-003-0141-3. [DOI] [PubMed] [Google Scholar]
- 43.Lasala F, Arce E, Otero JR, Rojo J, Delgado R. 2003. Mannosyl glycodendritic structure inhibits DC-SIGN-mediated Ebola virus infection in cis and in trans. Antimicrob Agents Chemother 47:3970–3972. doi: 10.1128/AAC.47.12.3970-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Licata JM, Johnson RF, Han Z, Harty RN. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol 78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol 76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balasubramanian S, Kota SK, Kuchroo VK, Humphreys BD, Strom TB. 2012. TIM family proteins promote the lysosomal degradation of the nuclear receptor NUR77. Sci Signal 5:ra90. doi: 10.1126/scisignal.2003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. 2007. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J Immunol 178:5635–5642. doi: 10.4049/jimmunol.178.9.5635. [DOI] [PubMed] [Google Scholar]
- 48.Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen IS. 2011. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe 9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valverde P. 2005. Effects of Gas6 and hydrogen peroxide in Axl ubiquitination and downregulation. Biochem Biophys Res Commun 333:180–185. doi: 10.1016/j.bbrc.2005.05.086. [DOI] [PubMed] [Google Scholar]
- 50.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. 2008. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest 118:1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, Sharpe AH, Ito S, Dranoff G, Kaplan GG, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27:927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoint. Am J Epidemiol 27:493–497. [Google Scholar]