ABSTRACT
The duplication of the poxvirus double-stranded DNA genome occurs in cytoplasmic membrane-delimited factories. This physical autonomy from the host nucleus suggests that poxvirus genomes encode the full repertoire of proteins committed for genome replication. Biochemical and genetic analyses have confirmed that six viral proteins are required for efficient DNA synthesis; indirect evidence has suggested that the multifunctional H5 protein may also have a role. Here we show that H5 localizes to replication factories, as visualized by immunofluorescence and immunoelectron microscopy, and can be retrieved upon purification of the viral polymerase holoenzyme complex. The temperature-sensitive (ts) mutant Dts57, which was generated by chemical mutagenesis and has a lesion in H5, exhibits defects in DNA replication and morphogenesis under nonpermissive conditions, depending upon the experimental protocol. The H5 variant encoded by the genome of this mutant is ts for function but not stability. For a more precise investigation of how H5 contributes to DNA synthesis, we placed the ts57 H5 allele in an otherwise wild-type viral background and also performed small interfering RNA-mediated depletion of H5. Finally, we generated a complementing cell line, CV-1–H5, which allowed us to generate a viral recombinant in which the H5 open reading frame was deleted and replaced with mCherry (vΔH5). Analysis of vΔH5 allowed us to demonstrate conclusively that viral DNA replication is abrogated in the absence of H5. The loss of H5 does not compromise the accumulation of other early viral replication proteins or the uncoating of the virion core, suggesting that H5 plays a direct and essential role in facilitating DNA synthesis.
IMPORTANCE Variola virus, the causative agent of smallpox, is the most notorious member of the Poxviridae family. Poxviruses are unique among DNA viruses that infect mammalian cells, in that their replication is restricted to the cytoplasm of the cell. This physical autonomy from the nucleus has both cell biological and genetic ramifications. Poxviruses must establish cytoplasmic niches that support replication, and the genomes must encode the repertoire of proteins necessary for genome synthesis. Here we focus on H5, a multifunctional and abundant viral protein. We confirm that H5 associates with the DNA polymerase holoenzyme and localizes to the sites of DNA synthesis. By generating an H5-expressing cell line, we were able to isolate a deletion virus that lacks the H5 gene and show definitively that genome synthesis does not occur in the absence of H5. These data support the hypothesis that H5 is a crucial participant in cytoplasmic poxvirus genome replication.
INTRODUCTION
Smallpox has plagued humans throughout history. The etiological agent of this deadly disease is variola virus, a member of the Poxviridae family of viruses. Smallpox was declared eradicated as a natural pathogen in 1980 after a global vaccination campaign that utilized a closely related poxvirus, vaccinia virus. Vaccinia virus is now the prototypic poxvirus for experimental study. Vaccinia virus possesses a large double-stranded DNA (dsDNA) genome (∼195 kb) which is replicated in the cytoplasm of the host cell, exhibiting both physical and genetic autonomy from the cell nucleus. The duplication of the viral genome takes place in cytosolic, membrane-delimited compartments (1) known as replication factories. Genetic, genomic, and biochemical analyses have revealed that the vaccinia virus genome encodes a core set of six proteins that are directly involved in and required for DNA replication in cultured cells. These include a catalytic DNA polymerase (Pol; E9), a heterodimeric processivity factor comprised of the viral uracil DNA glycosylase (UDG; D4) and a nonenzymatic bridging protein (A20), a single-stranded DNA binding (SSB) protein (I3), and a nucleoside triphosphatase/primase predicted to have helicase activity (D5) (2–14). A viral serine/threonine protein kinase (B1) is also required for viral DNA replication; it functions to combat the antiviral action of the cellular dsDNA binding protein BAF (15). Additional virus genome-encoded enzymes that are predicted to play roles in viral replication, recombination, and/or genome maturation include the DNA ligase (A50); a putative FEN-1 like endonuclease (G5); the precursor biosynthetic enzymes thymidine kinase (J2), thymidylate kinase (A48), and ribonucleotide reductase (F4, I4); and a Holliday junction resolvase (A22) (16–22). Lastly, the abundant, multifunctional phosphoprotein H5, which is discussed herein, has been postulated to participate in DNA replication. Whether H5 is in fact important for genome replication and, if so, how has remained unknown.
H5 is expressed throughout infection and has been implicated as playing roles in DNA replication, transcription, and morphogenesis (1, 23–30). Furthermore, it has been reported to be encapsidated within the virion core (31–35). H5 has a predicted molecular weight (MW) of 22,300 but migrates anomalously on SDS-polyacrylamide gels (apparent MW, 35,000) due the presence of an amino-terminal proline-rich region (36). The H5 protein is present in the genomes of all chordopoxviruses but is absent in the genomes of entomopoxviruses; its amino acid sequence is highly conserved in members of the Orthopoxviridae family. The intracellular localization of H5 has been monitored by immunofluorescence, and it is present in replication factories (1, 23, 27, 29). Yeast two-hybrid assay analysis has revealed an interaction with the A20 subunit of the DNA polymerase processivity factor as well as the viral kinase B1 (30). In 2010, D'Costa et al. published their survey study of the Dales collection of temperature-sensitive (ts) mutants of the IHD-W strain of vaccinia virus, revealing for the first time that strain Dts57 has a lesion in the H5 open reading frame (ORF) (37). Phenotypic characterization of this virus under nonpermissive growth conditions revealed a severe restriction in viral DNA replication. In combination with the intracellular localization of H5 to replication factories and its ability to interact ectopically with A20 and B1, these genetic data provided a compelling reason to further investigate the contribution of H5 to viral genome replication.
In this study, we began with the construction and analysis of a vaccinia virus recombinant carrying the same H5 mutation as Dts57 in a clean, unmutagenized strain WR background. Because this variant of H5 is impaired in function but is nevertheless stable, we took a second approach involving the administration of small interfering RNAs (siRNAs) specific for H5 to evaluate the impact of H5 depletion on viral replication. Finally, we generated an H5-expressing cell line in order to generate a recombinant virus lacking the H5 allele in which the H5 open reading frame was deleted and replaced with mCherry (vΔH5). Analysis of vΔH5 infections in noncomplementing cell lines allowed us to demonstrate incontrovertibly that H5 is an essential gene whose expression is required for DNA replication.
MATERIALS AND METHODS
Reagents.
The Flp-In T-Rex system, Lipofectamine 2000, Geneticin (G418), blasticidin, hygromycin, zeocin, Alexa Fluor 488 goat anti-rabbit (GAR) IgG, and Alexa Fluor 594 goat anti-mouse (GAM) IgG were purchased from Life Technologies (Grand Island, NY). A codon-optimized H5 (coH5) ORF was synthesized by GeneArt Gene Synthesis (now part of Life Technologies). Paraformaldehyde (PFA) was purchased from Electron Microscopy Sciences (Hatfield, PA). A zeta probe blotting membrane was purchased from Bio-Rad (Hercules, CA). [32P]dATP and [32P]TTP were purchased from PerkinElmer Life Sciences (Boston, MA). The TransIT-TKO (TKO) transfection reagent was purchased from Mirus Bio LLC (Madison, WI). siRNA duplexes were designed by and purchased from Dharmacon RNAi Technologies (Lafayette, CO). Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA).
Cells and virus.
African green monkey BSC40 and human thymidine kinase-minus (TK−) 143B cells were maintained in Dulbecco modified Eagle medium (DMEM; Life Technologies) containing 5% fetal calf serum (FCS). Flp–In–CV-1 fibroblast cells were obtained from Life Technologies and maintained in DMEM containing 10% FCS. Wild-type (WT) vaccinia virus of the IHD-W (generously provided by Richard Condit, University of Florida, Gainesville, FL) and WR strains, Dts57 (generously provided by Richard Condit), and WRts57 (see below for a description of the method of construction) were propagated in BSC40 cells. vΔH5 was propagated in CV-1 cells expressing a doxycycline (Dox)-inducible, codon-optimized H5 protein (see below for the method used for the generation of both). vTF7.3 (38) was a gift from Bernard Moss (NIH, Bethesda, MD). vTM-3×FLAG-UDG, vTM-A20, and vTM-Pol have been previously described (9, 10, 13). Viral stocks were prepared by ultracentrifugation of cytoplasmic lysates through 36% sucrose; titers were determined by plaque assays performed on BSC40 cells. vΔH5 titers were determined on the H5-expressing, complementing CV-1 cell line. For studies with temperature-sensitive virus, 31.5°C and 39.7°C were used as the permissive and nonpermissive temperatures, respectively.
Virion purification.
BSC40 or H5-expressing CV-1 cells treated with Dox (8 × 107 cells) were infected with WT virus or vΔH5 (multiplicity of infection [MOI] = 2), respectively. At 24 h postinfection (hpi), virions were purified on sucrose gradients and quantitated as previously detailed (39). The infectious yield was determined by titration on the appropriate cell line.
Prepermeabilization experiments.
BSC40 cells were infected with WT virus (MOI = 3) for 7 h. For samples denoted “prepremeabilized,” the cells were chilled, incubated for 30 s on ice with 0.1% Triton X-100, and fixed with 4% PFA in phosphate-buffered saline (PBS). For slides that were not prepermeabilized, the cells were chilled and fixed with 4% PFA–PBS. All samples were incubated on ice with 0.1% Triton X-100 for 5 min and then stained with polyclonal anti-H5 (1:400) (26) serum, followed by Alexa Fluor 488 GAR IgG. Pictures were captured at a magnification of ×20 using a Nikon Eclipse TE2000-U microscope and NIS-Elements BR3.2 software (Tokyo, Japan).
Generation and localization of H5-mCherry. (i) Generation of H5-mCherry.
To generate the pInt-H5-mCherry construct, the mCherry open reading frame was amplified from the pRSET-mCherry vector (40) using primers mCherry 5′ (5′-GCAGATCTCATATGGGATCCGTGAGCAAGGGCGAGG-3′) and mCherry 3′ (5′-GTAGATCTTCACTTGTACAGCTCGTCC-3′), where restriction sites are in bold nonitalic type, the stop codon is underlined, and bold italic type indicates a unique BamHI site that was engineered immediately upstream of the mCherry-coding sequence. The PCR product was digested and positioned behind the vaccinia virus G8 promoter in the pInt vector (41). The H5 open reading frame lacking a termination codon was amplified from the WT virus genome using an upstream H5 cloning primer (5′-GCGGATCCATGGCGTGGTCAATTAC-3′) and a downstream H5 no-stop 3′ primer (5′-GCGGATCCCTTCTTACAAG-3′) (restriction sites are in bold type). This H5 no-stop fragment was positioned upstream of the mCherry sequence via a unique BamHI site (indicated in bold and italics in the primer mCherry 5′ sequence above) that was engineered immediately upstream of the mCherry-coding sequence. Sequence analysis was used to verify the pInt-H5-mCherry construct.
(ii) Visualization of H5-mCherry.
BSC40 cells were infected with WT virus (MOI = 3) and transfected with the pInt-H5-mCherry construct at feed using Lipofectamine 2000. One set of infections was maintained in the presence of cytosine arabinoside (araC; 20 μM). At 7 hpi, live-cell images were acquired at a magnification of ×20 using a Nikon Eclipse Ti microscope and NIS-Elements BR3.2 software.
Expression and purification of the DNA polymerase holoenzyme complex.
The trimeric DNA polymerase complex was overexpressed by coinfecting BSC40 cells with vTF7.3, vTM-3×FLAG-UDG, vTM-A20, and vTM-Pol (each at an MOI of 2) (9, 10, 13, 38). The holoenzyme complex was retrieved as previously described (13). Fractions were analyzed by SDS-PAGE/silver stain analysis.
Construction of WRts57.
To generate a ts virus carrying the single G189R substitution in the H5 gene (previously reported by Condit and colleagues [37, 42]) in an otherwise wild-type background, we employed an overlap PCR strategy. Genomic viral DNA from the WR laboratory strain was used as a template for two PCRs for sequences that overlapped in the region of the mutation. The first amplicon (obtained by PCR with primer 5′ WR:Dts57 [5′-GAGGATCCCCATCTCCTGGAGTC-3′] and primer 3′ G189R [5′-CAACTTTAGATTTTCTCTCGGTG-3′]) initiated 297 nucleotides (nt) upstream of the GGA → AGA transition, and the second amplicon (obtained by PCR with primer 5′ G189R [5′-GATCTAGTCACCGAGAGAAAATC-3′] and primer 3′ WR:Dts57 [5′-GAGGATCCCGATCTCTTTTAGCG-3′]) extended 304 nt downstream of the intended mutation. The latter product included the first 257 nt of the adjacent H6 ORF. The bold residues represent the restriction enzyme sites, while the underlined residues mark the altered nucleotides. The resultant PCR products were used as the templates for another round of PCR using primers 5′ WRts57 and 3′ WRts57. The final PCR product was digested and cloned into the pUCNeo vector (39). The resultant plasmid, pUCNeo-G189R H5, was validated by DNA sequence analysis.
Insertion of the H5 G189R allele into the WT virus was accomplished using transient dominant selection. Briefly, BSC40 cells were infected with WR (MOI = 0.03); at 3 hpi, 5 μg of supercoiled pUCNeo-G189R H5 DNA was introduced by transfection and cells were shifted to 31.5°C. Geneticin (G418) was added to a final concentration of 3 mg/ml at 16 hpi to select for viral recombinants, and transfections were harvested at 48 hpi. G418-resistant viruses were isolated by two rounds of plaque purification in the presence of G418 on human TK− 143B cells. Integration of the neomycin resistance (neo) cassette was confirmed by PCR analysis. Additional rounds of plaque purifications were performed at 31.5°C in the absence of G418 to enable recombinational resolution of the tandem repeats and the subsequent loss of the neo gene. To identify the plaques that had lost neo and retained the G189R allele, DNA sequencing was performed. Plaques of the appropriate genotype were subjected to iterative plaque purification until all (>20) progeny plaques had lost the neo gene and contained the G189R lesion.
siRNA-mediated interference.
RNA duplexes of 21 nucleotides were comprised of the sense H5-specific siRNA (si-H5) sequence 5′-GAUAGAAGCGAUGGAUACUtt-3′ and the antisense si-H5 sequence 5′-AGUAUCCAUCGCUUCUAUCtt-3′, where the lowercase nucleotides indicate a terminal 3′ dT dinucleotide. BSC40 cells were seeded at a density of 2.25 × 105 cells. At 18 h postseeding, each siRNA duplex (20 nM) was transfected using the TransIT-TKO reagent. At 18 h posttransfection, cells were infected with WT virus (MOI = 3); at the time points indicated below, cells were harvested and analyzed for viral yield, protein accumulation, and bulk viral DNA accumulation.
Immunofluorescence microscopy of siRNA-treated infected cells.
BSC40 cells mock transfected (treated with TKO alone) or transfected with si-H5 were infected with WT virus in the presence of bromodeoxyuridine (BrdU; 25 μg/ml). At 7 hpi, cells were fixed and stained with anti-BrdU (5 μg/ml; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and anti-H5 (1:400) (26) antibodies, followed by Alexa Fluor 594 GAM IgG or Alexa Fluor 488 GAR IgG. Images were acquired at a magnification of ×20 using a Nikon Eclipse Ti microscope and NIS-Elements BR3.2 software.
Determination of 24-h viral yield.
BSC40 or CV-1–coH5 cells (treated or not treated with Dox) were infected (MOI = 2) and harvested at 24 hpi. Cells were collected by centrifugation, resuspended in 1 mM Tris (pH 9.0), and disrupted by freeze-thawing and sonication. Viral yields were assessed in plaque assays under the appropriate permissive conditions.
Southern dot blot hybridization to assess bulk viral DNA accumulation.
Infected (MOI = 3) BSC40 or CV-1–coH5 cells (not treated with Dox) were harvested at the time points (hpi) indicated below and subjected to Southern dot blot analysis as previously described (43). Biological replicates were spotted in quadruplicate and visualized by autoradiography and phosphorimager analysis.
Pulsed-field gel electrophoresis.
BSC40 or CV-1–coH5 cells (not treated with Dox) were infected (MOI = 3) and harvested at 10 hpi. The washed cell pellet was processed as previously detailed (7). The DNA was resolved on a CHEF Mapper XA apparatus (Bio-Rad) at 6 V/cm for 13 h at 14°C, using a switching time gradient of 1 to 25 s, a linear ramping factor, and a 120° angle (this program resolved DNA species of 5 to 500 kb). The DNA was visualized by ethidium bromide staining, and the image was captured and quantified using AlphaView software (ProteinSimple, Santa Clara, CA).
Immunofluorescence-based viral uncoating assay.
BSC40 cells were infected (MOI = 15) in the absence or presence of cycloheximide (cyclo; 25 μg/ml) or araC (20 μM). At 3 hpi, cells were fixed in 4% PFA–PBS on ice. Cells were stained with an anti-A5 antibody (44), followed by visualization with an Alexa Fluor 488 GAR secondary antibody. Pictures were captured at a magnification of ×40 using a Nikon Eclipse TE2000-U microscope and NIS-Elements BR3.2 software.
Immunoblot analysis of viral protein accumulation.
Whole-cell lysates (from cells treated with siRNA and CV-1 cells expressing coH5 [CV-1–coH5 cells]), postnuclear supernatants (from vΔH5 infections), or purified WT and vΔH5 virions (0.5, 1.0, and 2.5 μg virions) were resolved on SDS-acrylamide gels and transferred electrophoretically to nitrocellulose membranes. The membranes were probed with antisera directed toward E9 (45), A20 (11), D5 (4), I3 (12), H5 (26), L4 (39), and F17 (39). The immunoreactive proteins were detected using horseradish peroxidase-conjugated secondary antiserum (Bio-Rad) and chemiluminescent SuperSignal West Pico reagents (Pierce, Rockford, IL). Proteins were visualized by exposure on a FluorChem HD2 documentation system (ProteinSimple) and quantified using AlphaView software.
Construction of the CV-1–coH5 stable cell line. (i) Generation of the doxycycline-inducible CV-1 stable cell line.
We engineered Flp–In–CV-1 cells to constitutively express the tetracycline repressor (TetR). Briefly, the pcDNA6/TR plasmid (Life Technologies) containing the cytomegalovirus (CMV)-driven TetR ORF was linearized with FspI and transfected into the Flp–In–CV-1 cells. Flp–In–CV-1–TetR cells were clonally selected with 30 μg/ml blasticidin. TetR expression was verified by immunoblot analysis using a TetR antibody (Clontech, Mountain View, CA). The clone with the highest level of TetR expression was chosen for the subsequent integration of the coH5 gene.
(ii) Generation of CV-1–coH5 cells.
H5 was codon optimized for expression in mammalian cells by GeneArt and cloned into pcDNA5/FRT/TO (Life Technologies). Flp–In–CV-1–TetR cells were cotransfected with pcDNA/FRT/TO-coH5 and pOG44, a plasmid containing the Saccharomyces cerevisiae Flp recombinase. Flp recombinase mediates the integration of the coH5 ORF into the single FLP recombination target (FRT) site present in the Flp–In–CV-1 parental cell line. Codon-optimized H5 expression will be regulated by the CMV protomer in which two copies of the tet operator (TO) sequence have been inserted in tandem. Integration of the coH5-expressing plasmid was selected for by growth in medium containing 75 μg/ml hygromycin and verified by immunoblot analysis. In parallel, a Flp–In–CV-1–TetR cell line in which a single chloramphenicol acetyltransferase (CAT) transgene was integrated (the CV-1–CAT cell line) was generated as a control.
Construction of an H5 deletion mutant (vΔH5).
The vΔH5 construct was designed such that more than one-half of the H5 ORF was replaced with the mCherry-coding sequence. The endogenous H5 promoter and the first 36 nt and last 51 nt of the H5 ORF were retained to mediate mCherry expression and leave H4 and H6 expression unperturbed. PCRs were performed with the 5′ XhoI H4/H5 primer (5′-CCGCTCGAGCGTTATAAAGGTTGAT-3′) and the 3′ SpeI H5 primer (5′-GCGACTAGTATCCGCTTTA-3′) to amplify a 360-nt product consisting of the first 139 nt of H4L, 185 nt of H4/H5 intergenic DNA, and the first 36 nt of H5R or with the 5′ SpeI/ClaI H5 primer (5′-GGCACTAGTCTCATCGATGAGGGAAAATCTAAAG-3′) and the 3′ XbaI H6 primer (5′-GCTCTAGAGGAATCTGTGGACGAT-3′) to amplify a 390-nt product containing the last 51 nt of H5R and the first 339 nt of H6R. The PCR products were digested with XhoI/SpeI or SpeI/XbaI (identified in bold in the sequences provided above) and used in a three-part ligation with pBSIIKS that had been digested with XhoI/XbaI. The resultant vector (pBSIIKS-H4H5H6) was digested with SpeI and ClaI and ligated with an mCherry insert amplified from the pRSET-mCherry vector (40) using the 5′ SpeI mCherry primer (5′-GCGACTAGTGTGAGCAAGGGCGAG-3′) and the 3′ ClaI mCherry primer (5′-CCATCGATTTACTTGTACAGCTCG-3′). The resultant plasmid was designated mCherry-ΔH5, and its DNA sequence was verified.
BSC40 cells were infected with WT virus (MOI = 3) and transfected with linearized mCherry-ΔH5 DNA at 3 hpi. Cells were harvested at 24 hpi, and mCherry-expressing virus was isolated by iterative plaque purification on complementing CV-1–coH5 cells (preinduced with 50 ng/ml Dox for 18 to 24 h). Four rounds of plaque purification were performed until all of the progeny plaques expressed mCherry, and then one additional round of purification was performed. The replacement of the H5 ORF with mCherry and the purity of the viral stock were confirmed by DNA sequence analysis. The resulting virus was named vΔH5.
Preparation of digital figures.
Original data were scanned on an Epson Perfection scanner (Long Beach, CA). Images from immunoblot analysis were acquired using a FluorChem HD2 documentation system (ProteinSimple, Santa Clara, CA). Statistical analysis and graph preparation were performed using SigmaPlot software (Systat Software, Chicago, IL). Final figures were assembled and labeled with Canvas software (Deneba Systems, Miami, FL). The dashed lines separating the lanes in Fig. 1D, 3A (inset), and 4C represent the juxtaposition of samples that were analyzed on the same gel but that were not originally adjacent to each other.
FIG 1.
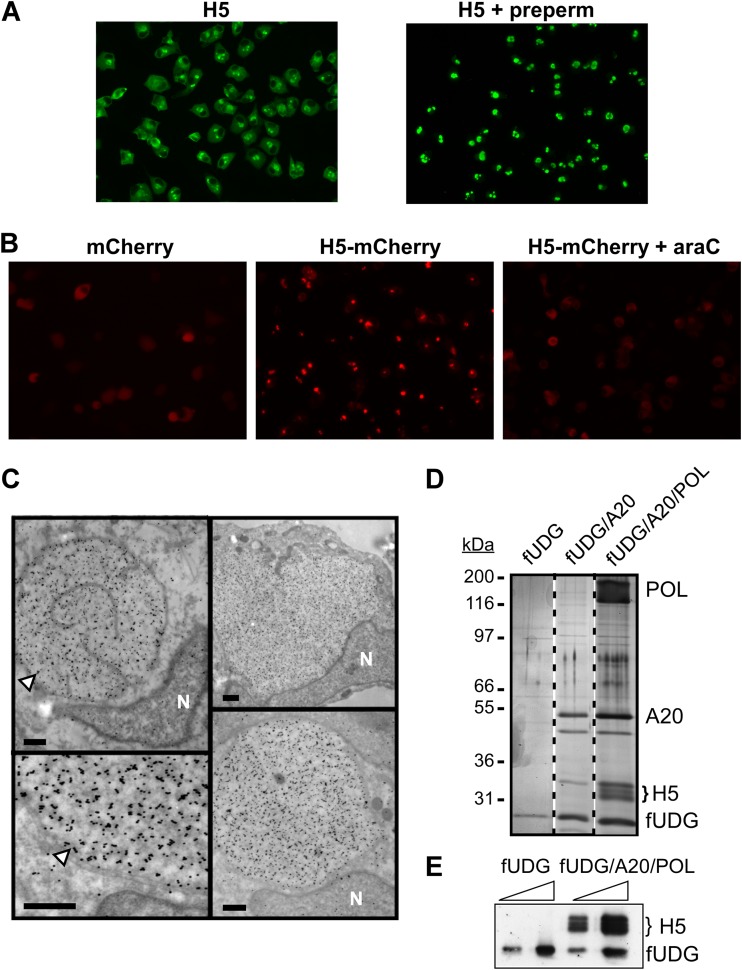
Evidence in support of a role for H5 in vaccinia virus DNA replication: intracellular localization and association with the polymerase holoenzyme. (A) H5 is anchored within replication foci. BSC40 cells were infected with WT virus for 7 h and then processed for immunofluorescence microscopy; one set of samples (right) was prepermeabilized (preperm) with 0.1% Triton X-100 for 30 s prior to fixation. H5 was visualized using a polyclonal anti-H5 antibody. (B) H5-mCherry, but not mCherry, localizes to active viral DNA replication sites. BSC40 cells were infected with WT virus and transfected with plasmids mediating the expression of mCherry or an H5-mCherry fusion protein from a viral intermediate promoter. Expression of the H5-mCherry fusion protein was also assessed in cells infected in the presence of the DNA replication inhibitor araC (right). At 7 hpi, the proteins were visualized by live-cell imaging. (C) H5 is concentrated in replication paddocks. BSC40 cells were infected with WT virus (in the presence of BrdU) and processed for immunoelectron microscopy at 7 hpi. Sections were incubated with anti-H5 serum, followed by a 10-nm gold particle-labeled secondary antibody. Arrowheads, areas of H5 staining. The nucleus (N) is noted. Bars, 0.5 μm. (D) H5 is specifically retrieved with the trimeric DNA polymerase holoenzyme complex. BSC40 cells were coinfected with vTF7.3 and a combination of viruses that direct the overexpression of components of the vaccinia virus DNA polymerase holoenzyme (3×FLAG-tagged UDG [fUDG], A20, Pol). These proteins were purified by sequential affinity and anion-exchange chromatography. Peak fractions were pooled and analyzed by silver staining. The brace identifies the migration of the H5 protein. Protein standards are marked on the left, and their molecular masses (in kilodaltons) are indicated. (E) Confirmation by immunoblot analysis that H5 associates with the trimeric DNA polymerase holoenzyme. Increasing concentrations of purified 3×FLAG-tagged UDG and the holoenzyme complex (fUDG/A20/Pol) were analyzed using polyclonal anti-H5 and monoclonal anti-FLAG antibodies.
FIG 3.
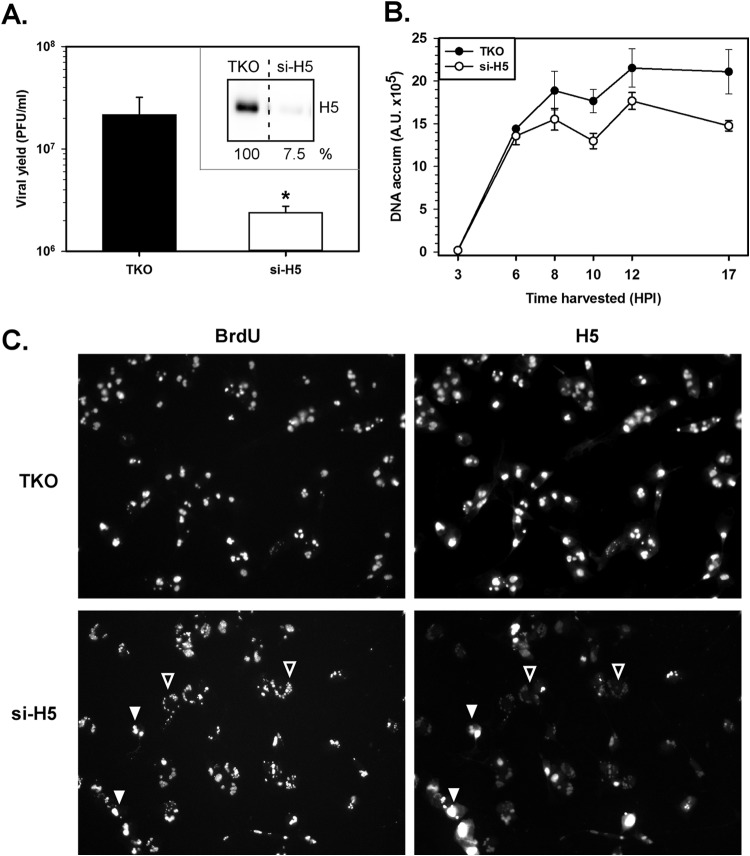
Impact of siRNA-mediated depletion of H5 on the viral life cycle. (A) BSC40 cells (2.25 × 105) were seeded and preloaded with transfection reagent alone (TKO) or with si-H5 for 18 h. Cells were then infected with WT virus for 17 h, and the infectious viral yield was determined. Statistical analysis was performed using a Student t test. *, P ≤ 0.05. The experiment was repeated at least 3 times, and standard errors are shown. (Inset) Whole-cell lysates were examined by immunoblot analysis, and the levels of H5 were quantified. The percentage of H5 detected compared to the amount detected with TKO treatment alone is shown below. (B) BSC40 cells preloaded with si-H5 or treated with TKO alone were infected with WT virus and harvested at various times (in hours) postinfection. Viral DNA was quantified by Southern dot blot hybridization. Two biological replicates were assayed in quadruplicate; the average values are plotted, and standard errors are shown. (C) Visualization of viral DNA replication factories by immunofluorescence microscopy. BSC40 cells treated and infected as described in the legend to panel A (with BrdU included in the growth medium) were harvested at 7 hpi. Cells were fixed, permeabilized, and incubated with anti-BrdU or anti-H5 antibodies, followed by incubation with Alexa Fluor-conjugated secondary antibodies. Open arrowheads, pinpoint BrdU+ H5lo DNA replication foci; filled arrowheads, larger WT BrdU+ H5+ replication factories.
RESULTS
H5 is localized to viral replication factories and is retrieved upon purification of the viral DNA polymerase holoenzyme complex.
H5 has been hypothesized to participate in viral DNA replication, in part because it can associate with a component of the DNA polymerase processivity factor, the A20 protein (28, 30), and is a substrate for the viral B1 kinase, which is also important for viral replication (46). H5 has been used as a marker for replication factories in numerous immunofluorescence analyses (1, 23, 27, 29), and we have extended the assessment of H5's localization during the early phases of viral infection. At 7 hpi, the majority of H5 localized to discrete foci within the infected cell known as replication factories or paddocks (Fig. 1A, left). Some diffuse, cytoplasmic H5 staining could be observed. To determine if H5 is a stably anchored component of the replication factories, we subjected infected cells to a mild prepermeabilization step (exposure to 0.1% Triton X-100 for 30 s on ice) prior to fixation and staining for immunofluorescence. This protocol releases any cytosolic soluble proteins, while proteins that are tethered to nucleic acids or cytoskeletal elements are retained (47). Indeed, as shown in Fig. 1A (right), the diffuse cytosolic H5 was released during prepermeabilization, but replication factory-associated H5 was anchored and its staining was not diminished. We also introduced plasmids expressing the mCherry protein or an H5-mCherry fusion protein into infected cells and examined the localization of these fluorescent proteins by live-cell imaging. Whereas mCherry was dispersed throughout the cell (Fig. 1B, left), the H5-mCherry protein localized to discrete foci (Fig. 1B, middle). These foci were evocative of replication factories, and indeed, in the presence of an inhibitor of DNA replication, araC, the H5-mCherry protein demonstrated a diffuse cytoplasmic staining (Fig. 1B, right). Lastly, cells were infected with WT vaccinia virus and exposed to BrdU from 4 to 7 hpi, at which point the cells were processed for immunoelectron microscopy. Sections were stained with an anti-H5 antibody, and abundant staining was seen throughout the interior of the membrane-delimited replication paddocks (Fig. 1C). These foci also showed strong staining for the I3 SSB protein and for BrdU, a marker of nascent DNA (data not shown) (7). Taken together, these data provide strong evidence that H5 localizes to replication sites.
We have previously described a protocol for overexpression and purification of vaccinia virus UDG, UDG/A20 (processivity factor), or UDG/A20/DNA Pol (holoenzyme) from infected cells. A 3× FLAG tag on UDG at the N terminus of D4 (fUDG) (13, 48) enables an initial affinity purification, after which the sample is resolved on an anion-exchange (Mono Q) column to separate the trimeric holoenzyme (UDG/A20/Pol) or dimeric processivity factor (UDG/A20) from excess, monomeric fUDG. We noted that the trimeric complex, but not monomeric fUDG or dimeric fUDG/A20, often contained an additional cluster of proteins with an electrophoretic migration of 30 kDa to 35 kDa (Fig. 1D, bracket). Immunoblot analysis allowed us to conclude that this cluster of proteins was indeed H5 (Fig. 1E). An interaction between H5 and A20 was previously shown by yeast two-hybrid and coimmunoprecipitation analyses (28, 30), but in our case, the stable association of H5 during Mono Q chromatography required the presence of the complete polymerase holoenzyme. Of note, the holoenzyme elutes from the Mono Q resin at 750 mM NaCl, indicating either that the association of H5 with the complex is mediated by hydrophobic interactions or that there are strong electrostatic interactions that persist under stringent conditions.
Further investigation of Dts57: does this mutant provide strong genetic evidence for a key role for H5 in vaccinia DNA replication?
D'Costa and colleagues presented a phenotypic characterization of Dts57 (37, 42, 49) as part of their reevaluation of the Dales collection of temperature-sensitive (ts) mutants (42, 49). The Dales library of ts mutants (50), including Dts57, was created by mutagenizing the wild-type IHD-W strain of vaccinia virus with nitrosoguanidine, followed by propagation in the presence of bromodeoxyuridine. Marker rescue analysis revealed that the temperature sensitivity of Dts57 mapped to the H5 gene (42), and sequence analysis revealed a single nucleotide transition leading to a Gly → Arg (G189R) substitution near the C terminus of the protein (37). Preliminary phenotypic characterization of Dts57 revealed a strict DNA-negative phenotype, accompanied by a block to postreplicative gene transcription and virion morphogenesis (37). These data were the first genetic evidence that H5 might indeed have an essential role during viral DNA replication.
To further characterize H5's role in DNA replication, we were interested in reconstructing the Dts57 lesion in an otherwise wild-type genetic background of the WR strain, which is the virus that we routinely use. It is important to note that the predicted amino acid sequences of the H5 proteins encoded by the genomes of the wild-type WR and IHD-W strains are identical. We therefore used site-directed mutagenesis and transient dominant selection to introduce the G189R lesion into the endogenous H5 allele of our WR strain. This new recombinant, WRts57, was subjected to phenotypic characterization.
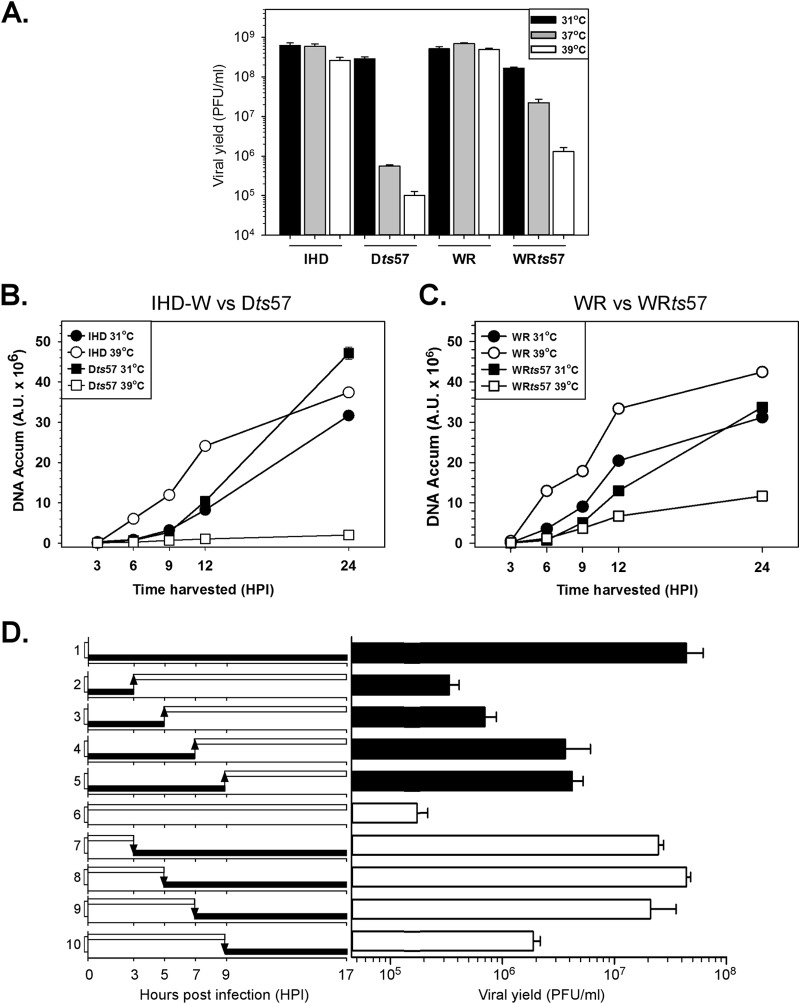
One-step growth analysis was performed to compare the growth kinetics of both ts57 viruses (Dts57 and WRts57) to those of their wild-type parents (IHD-W and WR, respectively) at 31°C, 37°C, and 39°C. While both sets of WT virus infections produced comparable levels of infectious virus under all three temperature conditions (Fig. 2A), both sets of ts57 infections demonstrated clear temperature sensitivity. Relative to the yields from Dts57 infections performed at 31°C, the yields from infections performed at 37°C and 39°C were severely diminished by 527-fold and 2,843-fold, respectively. For the reconstructed WRts57 virus, the yields at 37°C and 39°C were reduced 7-fold and 230-fold, respectively, relative to those at 31°C. Thus, although the WRts57 virus retains temperature sensitivity, the defect is not as severe as that in the original IHD-W isolate.
FIG 2.
Phenotypic characterization of WRts57. (A) The ts57 lesion confers a temperature-sensitive growth defect. BSC40 cells were infected with WT virus (IHD-W or WR) or Dts57 or WRts57 and incubated at 31°C, 37°C, or 39°C for 17 h (MOI = 3). The average from two biological replicates, each of which was titrated in duplicate, is plotted, and standard errors are shown. (B) Dts57 exhibits a strict block of DNA replication. BSC40 cells were infected with WT virus (IHD-W) or Dts57 and incubated at the permissive temperature (31°C) or the nonpermissive temperature (39°C). Viral DNA was harvested at various times (in hours) postinfection and analyzed by Southern dot blot hybridization to quantify the amount of accumulated viral DNA (DNA Accum) at each time point. Samples were analyzed in quadruplicate, and the average values are plotted. A.U., arbitrary units. (C) Viral DNA accumulation is reduced in WRts57 infections at the nonpermissive temperature. BSC40 cells were infected with WT virus (WR) or WRts57 and incubated at the permissive temperature (31°C) or the nonpermissive temperature (39°C). Viral DNA was quantified as described in the legend to panel B. (D) Execution point analysis of WRts57 reveals a postreplicative block in the viral life cycle. BSC40 cells were infected with WRts57 and incubated at either the permissive temperature (31°C; black bars) or the nonpermissive temperature (39°C; white bars) for the length of the study or shifted to the opposite temperature at 3, 5, 7, or 9 hpi. The experimental strategy is depicted on the left, and the numbers 1 to 10 on the y axis indicate the experimental set. All samples were harvested at 17 hpi, and the viral yield was determined; the average from two biological replicates (titrated in duplicate) is plotted (right), and standard errors are shown.
To assess if the block in the production of viral progeny was at the stage of DNA replication, we performed a comparative analysis of the WT and ts57 viruses using a Southern dot blot hybridization assay. IHD-W and Dts57 infections were performed at a permissive temperature (31°C) and a nonpermissive temperature (39°C), virus was harvested at 3 to 24 hpi, and the levels of viral DNA were quantified (Fig. 2B). In cells infected with IHD-W, viral DNA accumulated in a linear fashion over time at both temperatures, with higher levels of DNA being seen at 39°C than at 31°C (Fig. 2B, open versus filled circles). In contrast, whereas Dts57 DNA accumulated normally at 31°C (Fig. 2B, filled squares), a severe diminution in DNA accumulation was seen at 39°C (Fig. 2B, open squares); at 24 hpi, 24-fold less DNA had accumulated at 39°C than at 31°C. This block in DNA replication was not due to the absence of key replication proteins, since both the DNA polymerase and A20 protein accumulated to wild-type levels during infections at the nonpermissive temperature (data not shown). Lastly, consistent with the observation of D'Costa et al. (37), we verified that the Dts57 H5 protein accumulates to wild-type levels at the nonpermissive temperature (data not shown) and is thus ts for function but not for stability. Interestingly, we noted that the Dts57 H5 protein was hyperphosphorylated under nonpermissive conditions, as assessed after immunoprecipitation of extracts prepared from 32PPi-labeled cells (data not shown); hyperphosphorylated, slower-migrating species of H5 were also evident in our immunoblot analysis (data not shown).
When infections were performed with strains WR and WRts57 under the same experimental conditions detailed above, the trends were similar but less extreme (Fig. 2C). Again, during WR infections, more DNA accumulated when infections were performed at 39°C (Fig. 2C, open circles) than when they were performed 31°C (Fig. 2C, filled circles). However, during WRts57 infections, this ratio was inverted, with more viral DNA being synthesized at the low temperature than at the high temperature. WRts57 therefore remains ts for DNA synthesis, albeit only moderately so. The size and integrity of the viral DNA synthesized during WRts57 infections at the nonpermissive temperature were also monitored using pulsed-field gel electrophoresis. We observed a modest reduction in the levels of the mature, monomeric genome under nonpermissive conditions without noting any increase in the accumulation of subgenomic or supragenomic DNA species (not shown).
The major execution point of WRts57 is at late times postinfection.
While DNA accumulation was impacted by the WRts57-H5 allele, the modest diminution that was observed would certainly not account for the 2-log-unit reduction in viral yield that was seen. Thus, we were interested in performing execution point studies to define which stages of infection were most affected under nonpermissive conditions. WRts57 infection of BSC40 cells was therefore initiated at the permissive temperature (Fig. 2D, black bars) or the nonpermissive temperature (Fig. 2D, white bars), and cultures were either left at those temperatures or shifted to the opposite temperature at 3, 5, 7, or 9 hpi. All samples were harvested at 17 hpi, and the viral yield was determined. As demonstrated earlier, the viral yield from WRts57 infections at the nonpermissive temperature was ∼240-fold less than that from infections at the permissive temperature (Fig. 2D; compare the result for set 1 with that for set 6). Furthermore, the WRts57 temperature restriction was reversible. Cells in which infections were initiated at the nonpermissive temperature and then shifted to the permissive temperature at various times postinfection were able to produce infectious virus. As can be noted in Fig. 2D for sets 7, 8, and 9, as long as cells infected at the nonpermissive temperature were shifted to the permissive temperature by 7 hpi, the amount of infectious virus produced was comparable to that generated when infections were performed entirely at the permissive temperature (Fig. 2D, set 1). Only when infections were shifted at 9 hpi (Fig. 2D, set 10), in the midst of virion morphogenesis, was there an ∼10-fold reduction in the amount of infectious virus produced. Viral yield decreased another ∼10-fold if the infections were left at the nonpermissive temperature until 17 hpi (Fig. 2D, set 6). Clearly, the H5 mutation in this virus has the greatest impact on the late stages of infection (after 7 hpi).
Conversely, infections in which the initial 3 h of infection occurred at the permissive temperature produced only minimally more virus than those performed solely at the nonpermissive temperature. Even allowing the initial 7 or 9 h of infection to occur at the permissive temperature (Fig. 2D, sets 4 and 5) led to an ∼10-fold reduction in viral yield compared to the yields from cells maintained at the permissive temperature for the full 17 hpi (Fig. 2D, set 1). Again, these data reveal that the major execution point of WRts57 is after 7 to 9 hpi. These data are consistent with our observation that although DNA replication is modestly impacted during WRts57 infections at the nonpermissive temperature, genome replication is not the stage that is the most vulnerable to impairment by H5.
siRNA-mediated depletion of H5.
Since the ts57 H5 protein (37) and the previously characterized tsH5-4 protein (26) are both stable at the nonpermissive temperature and since H5 is likely to be multifunctional, our analysis of H5 has been limited to studying the first step at which the ts proteins are incapable of executing their function. It is probable that the ts proteins retain some of the capabilities of WT H5 at the nonpermissive temperature. This begged the question of how the absence of H5 would impact the progression of the viral life cycle: where would infection arrest? To approach this question, we treated cells with siRNA directed against H5 (or the transfection reagent [TKO] alone) for ∼18 h and then infected them with WT virus. Cells were harvested at 12 hpi for immunoblot analysis or 17 hpi for viral yield assessment. Treatment with si-H5 resulted in a >90% decrease in H5 protein levels compared to those achieved by treatment with TKO alone (Fig. 3A, inset) and a 1 log reduction in viral yield (Fig. 3A). To assess if H5 depletion affected DNA replication, Southern dot blot quantification of viral DNA was performed. To our surprise, siRNA-mediated depletion of H5 resulted in a very modest, albeit reproducible, reduction in DNA accumulation, with the accumulated levels of DNA being 70 to 84% of the control levels from 8 to 17 hpi (Fig. 3B; compare open circles to filled circles). Therefore, in this context, it appears that if H5 plays an important role in DNA replication, the residual H5 (present at levels ∼8% of WT levels) is sufficient to support nearly WT levels of DNA synthesis. Since this modest reduction in DNA levels would not be expected to reduce the viral yield 10-fold, H5 depletion must have a more significant impact on postreplicative events.
However, the possibility of incomplete transfection efficiency is an important caveat of these experiments. If H5 is an essential protein, we would have expected the viral yield to be reduced far more than 10-fold if it had been depleted in all cells and wondered whether, in fact, ∼10% of the cells had remained untransfected and therefore produced wild-type levels of virus. To visualize H5 depletion and DNA replication on a per cell basis, BSC40 cells were mock transfected (Fig. 3C, TKO) or treated with si-H5 for 18 h prior to infection with WT virus in the presence of BrdU. At 7 hpi, samples were processed for immunofluorescence microscopy. Anti-BrdU was used to label nascent viral DNA, and anti-H5 was used to visualize H5. In the majority of si-H5-treated cells, a marked reduction in the size and staining intensity of the BrdU-positive (BrdU+) H5-positive (H5+) viral factories was seen (Fig. 3C, open arrowheads). However, some cells presented with BrdU+ H5+ foci whose size and brightness were comparable to those visualized in the TKO-treated cells (Fig. 3C, filled arrowheads). Thus, the residual presence of H5 (present at levels ∼10% of WT levels) and the 1 log reduction in viral yield can most likely be attributed to the productive infection of the ∼10% of cells that did not receive H5-specific siRNA.
Generation and characterization of the CV-1–coH5 cell line.
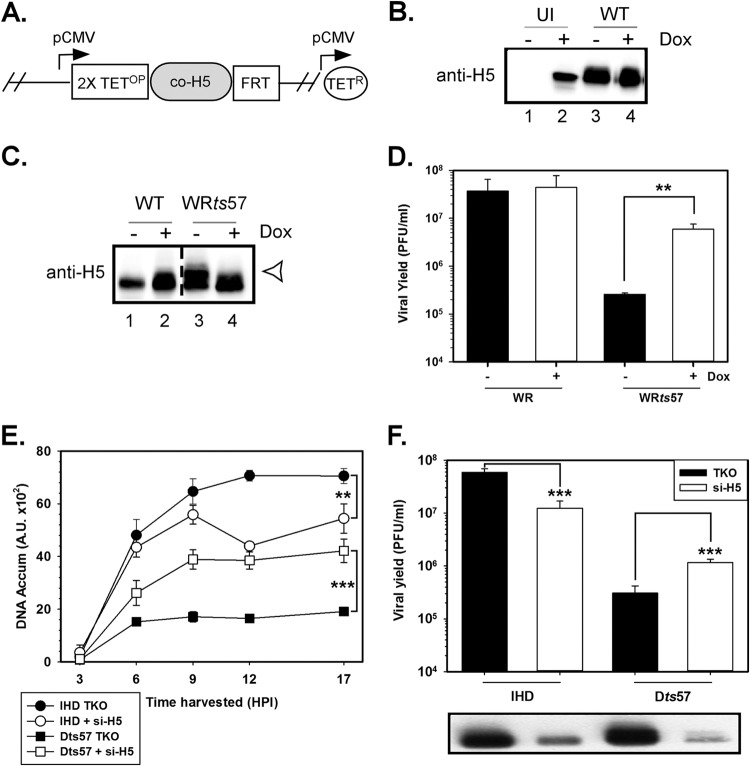
The most incisive approach to deciphering H5's essentiality during viral infection would be to generate an H5 deletion virus (vΔH5). For this purpose, we constructed a complementing H5-expressing cell line that supported and enabled the construction and replication of such a virus. We used the Flp–In–CV-1 system to establish a cell line containing a single, CMV-driven, doxycycline-inducible H5 transgene integrated into a constant genomic locus (Fig. 4A). To enable doxycycline-inducible H5 expression, we made a clonal derivative of the Flp–In–CV-1 cell line that expressed TetR (Flp–In–CV-1–TetR). After cotransfection with plasmids carrying the Flp recombinase and an H5 allele that had been codon optimized for expression in primate/human cells (coH5), cells in which the coH5 transgene had integrated into the FRT site (Fig. 4A) were expanded in selective medium. Immunoblot analysis (Fig. 4B) was used to assess the regulated expression of the H5 protein and to compare the levels of expression to those seen in a WT virus infection. The Dox-inducible codon-optimized H5 CV-1 (CV-1–coH5) cells were left untreated or incubated with 50 ng/ml Dox for 18 to 24 h and then left uninfected or infected with WT virus for 7 h. As shown in Fig. 4B, H5 accumulation was dependent upon the inclusion of doxycycline in the growth medium, with a >100-fold increase in H5 levels being seen after induction with doxycycline (compare lane 2 with lane 1 in Fig. 4B). The level of H5 in the induced cell line reproducibly plateaued at ∼50% of that which accumulated during a WT virus infection (Fig. 4B; compare lanes 2 and 3).
FIG 4.
Characterization of a CV-1– cell line expressing doxycycline-inducible, codon-optimized H5 (the CV-1–coH5 cell line). (A) Schematic representation of the genomic organization of CV-1–coH5 cells. A single copy of the coH5 ORF was inserted into a common genomic locus using Flp-mediated recombination into an FRT site; the parental cell line also expresses the Tet repressor (TetR). The transgene is under the regulation of a CMV promoter (pCMV) and two copies of the tetracycline operator (TetOP). (B) CoH5 is induced and accumulates in CV-1–coH5 cells upon doxycycline induction. CV-1–coH5 cells were induced with doxycycline (lanes +) or left untreated (lanes −). Twenty-four hours later, cells were left uninfected (UI) or infected with WT virus with or without Dox induction. After an additional 24 h, whole-cell lysates were prepared and subjected to immunoblot analysis with anti-H5. (C and D) H5 expressed from CV-1–coH5 cells can complement WRts57. coH5 cells were left untreated or treated with Dox for 18 h prior to infection with WR or WRts57 for 17 h. (C) For immunoblot analysis, parallel samples were maintained at the nonpermissive temperature (39°C), and cell lysates were then prepared and subjected to immunoblot analysis with anti-H5 serum. Open arrowhead, hyperphosphorylated H5. (D) For the analysis of viral yield, infections were performed at the nonpermissive temperature in the absence or presence of Dox. The average from 3 experiments is plotted, and standard errors are shown. Statistical analysis was performed using the Student t test. **, P ≤ 0.01. (E and F) siRNA-mediated depletion of the Dts57 H5 protein partially restores DNA replication and viral yield. Cells were incubated with transfection reagent (TKO) or si-H5 for 18 h prior to infection with IHD-W or Dts57 at 39°C. Cells were harvested at various times (in hours) postinfection for analysis of DNA accumulation (E) or viral yield (F). An immunoblot of the virally infected cell lysates probed for H5 is shown under the graph in panel F. Statistical analysis was performed using the Student t test. **, P = 0.002, panel E; ***, P < 0.001, panels E and F.
To verify that the H5 expressed from the cell line would be competent to support a viral infection, we assessed its ability to complement a WRts57 infection at the nonpermissive temperature. CV-1–coH5 cells (treated or not treated with Dox) were infected with WT or WRts57 and incubated at the nonpermissive temperature (39°C) for 18 h prior to harvesting for immunoblot analysis and viral yield assessments.
Immunoblot analysis of the cell lysates confirmed that WT virus-infected cells produced more H5 in the presence than in the absence of Dox (Fig. 4C, lanes 1 and 2) and that WRts57-infected cells presented with a hyperphosphorylated species of H5 protein (Fig. 4C, lane 3; the slowed migration is marked by the open arrowhead) that appeared to be reduced with Dox treatment (Fig. 4C, lane 4; the slower-migrating protein population is absent).
Comparable amounts of infectious virus were produced from WT virus infections treated or not treated with Dox (Fig. 4D; compare the filled and open bars for WR). These data validate that the excess H5 present in infected, induced cells (which contained ∼1.5-fold more H5 than WR-infected cells not treated with Dox) is not inhibitory. As expected, WRts57 infections at the nonpermissive temperature performed on CV-1–coH5 cells in the absence of Dox produced ∼100-fold less virus than WT virus infections (Fig. 4D, filled bar, no Dox treatment). An ∼22-fold increase in viral yield was seen when Dox was included in the culture medium (Fig. 4D, open bar, Dox treatment). These data validate the biological competency of the coH5 protein.
Does siRNA-mediated depletion of Dts57 H5 impact viral infection?
Nevertheless, the viral yield produced during complemented strain WRts57 infections at the nonpermissive temperature did not reach that seen during WT virus infections. Although this might reflect the reduced levels of wild-type H5 produced by the cell line, it is also possible that the ts57 H5 protein, which is stable at the nonpermissive temperature (data not shown), exerts a partial dominant negative effect. Some support for this interpretation emerged from siRNA studies in which we depleted H5 during infections of BSC40 cells with IHD-W WT virus or Dts57. As was seen with the WT WR strain (Fig. 3B), depletion of H5 during IHD-W infections led to a modest reduction in the accumulation of viral DNA from 9 to 17 hpi (Fig. 4E, filled versus open circles). In contrast, siRNA-mediated depletion of H5 during Dts57 infections at the nonpermissive temperature actually led to a reproducible increase in DNA accumulation (Fig. 4E; compare filled and open squares). This trend was also seen when viral yields were monitored (Fig. 4F).
Generation of vΔH5 recombinant virus.
To assess the importance of H5 to viral infection and to define the earliest essential role of H5, we generated vΔH5, in which the majority of the H5-coding sequence was deleted and replaced with the mCherry ORF (40) (Fig. 5A). The endogenous H5 promoter was left in place to regulate the expression of mCherry, and the promoter for the downstream H6R gene (normally embedded in the H5 gene but now downstream of the mCherry translational stop codon) was left in place. Following introduction of the targeting plasmid into WT virus-infected cells, mCherry-expressing virus was subjected to serial plaque purification on CV-1–coH5 cells until a homogeneous population of virus was obtained. Sequence analysis confirmed the genetic identity of the vΔH5 recombinant.
FIG 5.
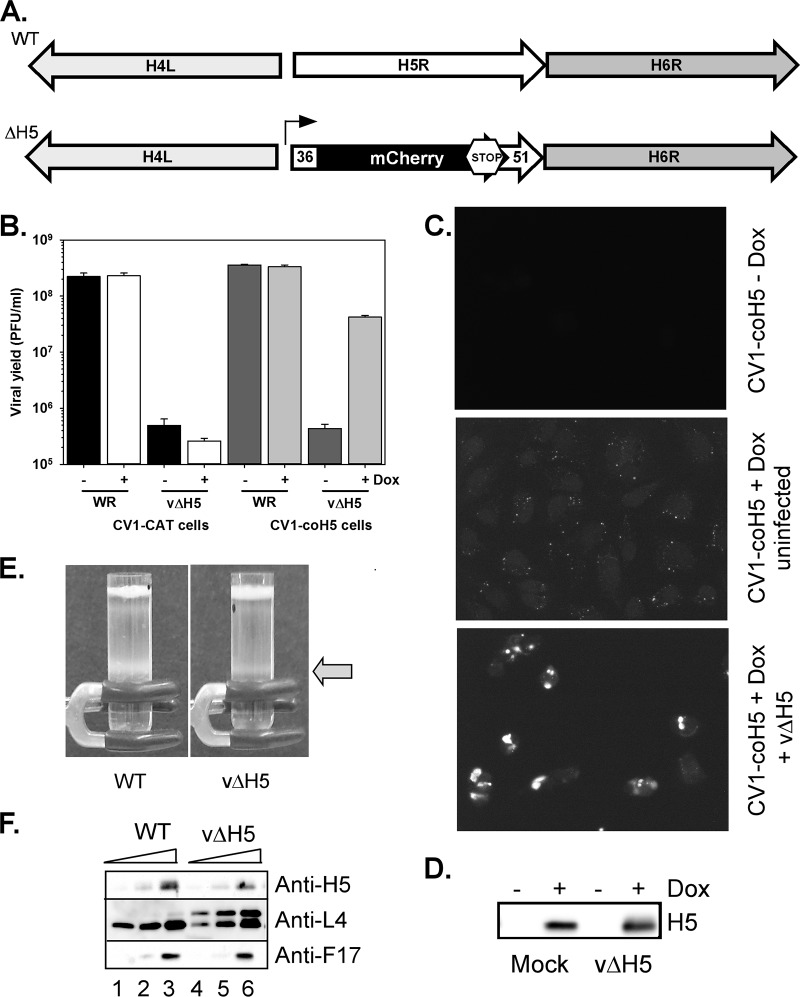
Generation and characterization of vΔH5. (A) A schematic representation of the genomic organization of the H5R ORF in WT virus and vΔH5. In vΔH5, sequences flanking the H5 ORF, including the endogenous H5 and H6 promoters, are retained, while the majority of the H5 coding sequence is replaced with an mCherry ORF containing a stop codon. (B) One-step growth analysis of vΔH5 under permissive and nonpermissive conditions. CV-1–CAT and CV-1–coH5 cells were left untreated or incubated with Dox for 18 h prior to infection with WR or vΔH5. At 18 hpi, infected cells were harvested and viral yields were assessed. Biological duplicates were titrated in duplicate, and the average values were plotted; standard errors are shown. (C and D) Visualization of the coH5 protein in CV-1–coH5 cells under various conditions. (C) coH5-expressing CV-1 cells were left untreated (top) or treated with Dox for 18 h and left uninfected (middle) or infected with vΔH5 for 7 h (bottom). H5 was visualized by immunofluorescence microscopy after staining with anti-H5 followed by incubation with an Alexa Fluor 488 GAR secondary antibody. (D) Immunoblot analysis was used to assess coH5 accumulation. (E) Sucrose gradient purification. WT virions were propagated in BSC40 cells, while the vΔH5 virions were grown in CV-1–coH5 cells treated with Dox for 24 h. Virions were purified from cytoplasmic extracts by sedimentation through a sucrose cushion, followed by banding on a 25 to 40% sucrose gradient. Arrow, the light-scattering bands of virus particles. (F) Immunoblot analysis of virion protein content. Virions at 0.5 (lanes 1 and 4), 1.0 (lanes 2 and 5), and 2.5 (lanes 3 and 6) μg were resolved and subjected to immunoblot analysis using antisera directed toward three abundant vaccinia virus core proteins: H5, L4, and F17.
Our success in generating vΔH5 by iterative plaque purification on CV-1–coH5 cells relied on the fact that this virus produces large plaques on the induced cell line (data not shown). One-step growth assays revealed that ∼300-fold more infectious virus was produced when vΔH5 infections were performed in the presence of Dox than in its absence (Fig. 5B, dark gray versus light gray bars), providing the first evidence that the absence of H5 greatly compromises infection. The viral yield from vΔH5 infections performed in the control CV-1–CAT cells was comparable to that in the uninduced CV-1–coH5 cells (Fig. 5B, black versus white bars [absence versus presence of Dox]), reinforcing the importance of H5 and confirming that the expression of H5 in CV-1–coH5 cells is fully dependent upon the inclusion of Dox in the medium. The yields of WT virus were not affected by the presence or absence of Dox (as we had seen previously; Fig. 4D) and were equivalent in CV-1–coH5 and CV-1–CAT cells (Fig. 5B).
Immunofluorescence microscopy of CV-1–coH5 cells provided independent confirmation that coH5 expression is tightly controlled by the omission of Dox from the tissue culture medium; in Fig. 5C, compare the top picture for cells not treated with Dox to the middle picture for cells treated with Dox. In uninfected, induced (Dox-treated) CV-1–coH5 cells, H5 was observed to be dispersed throughout the cell, including the nucleus, as well as in cytoplasmic puncta (Fig. 5C, middle). Upon infection with vΔH5, however, coH5 relocalized to cytoplasmic foci, which represent the sites of viral DNA replication (Fig. 5C, bottom). These foci colocalize with the viral DNA polymerase, the viral SSB protein, and nascent DNA synthesis (data not shown). The anti-H5 staining appeared to be brighter in the infected than in the uninfected (Dox-treated) CV-1–coH5 cells. To clarify whether H5 was stabilized upon infection or instead just appeared brighter due to localization in replication foci, we performed immunoblot analysis on these cells (Fig. 5D). Quantification of immunoblots from three biological duplicates revealed that, on average, there is a modest 20% more coH5 protein present in vΔH5-infected than uninfected CV-1–coH5 cells.
Because we (and others) have previously shown that H5 is encapsidated in infectious virions (31–35), we were interested in purifying and characterizing the mature virions (MVs) produced during permissive vΔH5 infections of CV-1–coH5 cells (Dox treated). As presented in Fig. 5E, both WT and permissive vΔH5 infections produced a light-scattering band (arrow) that sedimented to similar positions on a 25 to 40% sucrose gradient. Equal volumes of the two light-scattering bands were extracted via needle aspiration, concentrated by ultracentrifugation, and characterized further. Immunoblot analyses of increasing concentrations of virions indicated that the vΔH5 virions incorporated ∼50% of the amount of H5 seen in WT virions (Fig. 5F, lanes 6 versus 3). The levels of other abundant core proteins, such as L4 and F17, appeared to be unchanged. However, we did note an increase in the levels of full-length L4 that had not undergone proteolytic processing. The specific infectivity of the virions was also determined, and the particle/PFU ratio of the vΔH5 virions was found to be only 2-fold greater (54.8 ± 1.7) than that of the WT virions (27.3 ± 6.1). In sum, these data show that the virions produced upon infection of the induced CV-1–coH5 cells with vΔH5 sediment similarly to WT virions, contain levels of F17 and L4 comparable to those in WT virions, and contain encapsidated H5, although they contain more unprocessed L4 and their specific infectivity is ∼2-fold less than that of WT virions.
Analysis of vΔH5 in noncomplementing BSC40 cells: H5 is essential for viral DNA replication.
With vΔH5 in hand, the next and crucial step was to analyze its behavior in noncomplementing BSC40 cells. First, we performed a one-step growth curve and determined that the yield of infectious virus obtained from vΔH5-infected cells was reduced by nearly 4 logs (6,167-fold) compared to that obtained from WT virus-infected cells (Fig. 6A). These data confirm that expression of H5 during infection is vital for the production of infectious virus. As shown in Fig. 6B, immunoblot analysis demonstrated that several essential viral proteins involved in DNA replication (Pol, A20, D5, I3) accumulated to WT levels during vΔH5 infections, indicating that viral attachment, entry, early gene transcription, and early protein synthesis had proceeded normally. Because the incoming vΔH5 virions were produced in the presence of H5, we had anticipated that these early events of infection would not be impaired. The immunoblot analyses also confirmed that, in contrast to the findings for the other proteins examined, no H5 protein could be detected in these infections.
FIG 6.
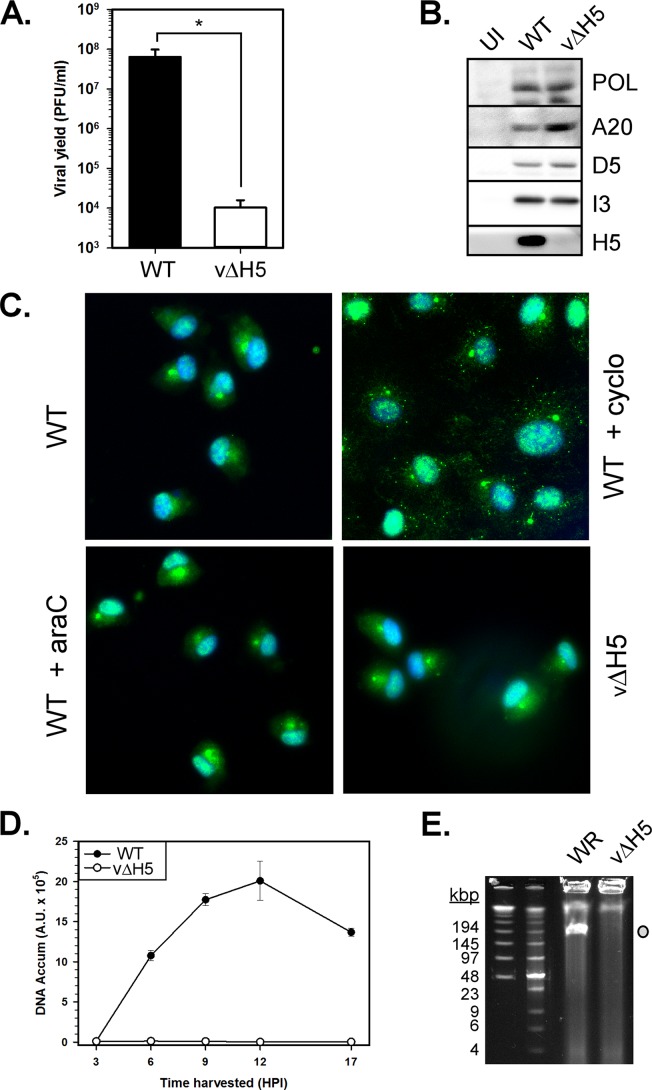
Characterization of vΔH5 infections in BSC40 cells at the nonpermissive temperature. (A) One-step viral growth assay comparing the infectivity of WT virus versus vΔH5 on BSC40 cells. Cells were infected with WT virus or vΔH5 and harvested at 17 hpi for viral yield assessment by plaque assay. The average values from 4 experiments are plotted, and standard errors are shown. Statistical analysis was performed using a Student t test. *, P ≤ 0.05. (B) Accumulation of key early viral proteins during WT virus and vΔH5 infections. Postnuclear supernatants from uninfected (UI) or WT virus- or vΔH5-infected cells were prepared at 6 hpi. Fifteen micrograms of the postnuclear supernatant was resolved and subjected to immunoblot analysis. (C) Analysis of core uncoating and disassembly. BSC40 cells were infected with WT virus, infected with WT virus and treated with cycloheximide (cyclo), infected with WT virus and treated with cytosine arabinoside (araC), or infected with vΔH5 for 3 h, at which time the cells were fixed. Cells were stained with an antibody specific for the A5 component of the core and visualized by incubation with an Alexa Fluor 488 GAR secondary antibody (green). The nucleus was visualized via DAPI (4′,6-diamidino-2-phenylindole) staining (blue). (D) Quantification of viral DNA accumulation by Southern dot blot hybridization. BSC40 cells were infected with WT virus or vΔH5 and harvested at various times (in hours) postinfection. Cytoplasmic viral DNA from 2 biological replicates was analyzed in quadruplicate; the average values are plotted, and standard errors are shown. (E) Visualization of accumulated viral genomes by pulsed-field gel electrophoresis. BSC40 cells were infected with either WT virus or vΔH5 and harvested at 10 hpi. Washed cell pellets were embedded in agarose, digested appropriately, and subjected to pulsed-field gel electrophoresis. The DNA was visualized by ethidium bromide staining; the electrophoretic positions of DNA molecular size markers are shown on the left, and their sizes (in kilobase pairs) are noted. Gray circle, the migration of full-length monomeric genomes.
We were next interested in assessing if viral uncoating proceeded normally; this step of core disassembly releases the genome into the cytosol, where it can undergo DNA replication. We therefore utilized immunofluorescence microscopy to assess core integrity versus disassembly (51). BSC40 cells were infected with WT virus in the absence or presence of cycloheximide or araC or were infected with vΔH5 (in the absence of drugs) (Fig. 6C). At 3 hpi, the cells were fixed and stained with an antibody to the A5 protein, a component of the core wall. If uncoating has not occurred, A5 is associated with the intact viral cores and discrete puncta are seen after staining with the Alexa Fluor 488-conjugated secondary antibody (a starry night pattern). If uncoating and core disassembly have occurred, A5 staining is dispersed throughout the cytoplasm. In WT-infected cells, the pattern of A5 staining was diffuse in the absence of drugs or the presence of araC, which has no impact on uncoating but impairs subsequent DNA replication. However, a very different result was seen when WT virus infections were performed in the presence of cycloheximide, an inhibitor of protein synthesis that has previously been shown to block core uncoating and genome release (52). Here, the characteristic starry night pattern of A5 staining, corresponding to intact viral cores, was seen. When BSC40 cells were infected with vΔH5, we observed a diffuse pattern of staining akin to that observed during infection with WT virus or WT virus plus araC treatment. Thus, uncoating and core disassembly occur normally during vΔH5 infections of BSC40 cells.
To assess if H5 is essential for DNA replication, BSC40 cells were infected with WT virus or vΔH5 for various times, and lysates were subjected to Southern dot blot analysis. In cells infected with WT virus, viral DNA replication occurred normally and increasing amounts of genomic DNA accumulated during the 17-h time course. In contrast, no accumulation of viral DNA could be detected at any time after infection with vΔH5 (Fig. 6D). Independent confirmation was obtained by harvesting cells after infection with WT virus or vΔH5 (10 hpi) and subjecting them to pulsed-field gel electrophoresis. Again, while WT virus infections produced high levels of the monomeric viral genome (Fig. 6E, gray circle), no genomic DNA could be observed in the vΔH5-infected cells. Taken together, these data provide conclusive evidence that H5 plays an essential role in the process of vaccinia virus DNA replication.
DISCUSSION
The work presented herein describes a variety of genetic approaches that we employed in an attempt to unravel the contributions of the H5 protein during the vaccinia virus life cycle. H5 is an abundant multifunctional protein that has been reported to interact with components of the DNA replication machinery as well as the postreplicative transcriptional machinery (24, 25, 28, 30). In addition, the H5 protein copurifies with and may be associated with an enzymatic activity capable of processing the 3′ termini of a subset of postreplicative viral mRNAs (53). In support of H5's contribution to transcription, it has also been shown to stimulate late transcription in a semipurified in vitro assay (54). Germane to each of these findings, the work of D'Costa, Condit, and colleagues has confirmed that the purified H5 protein exhibits nucleic acid binding activity, with the highest affinity being observed for dsDNA and double-stranded RNA (dsRNA) (Kds [dissociation constants] = 12 and 14 nM, respectively) (53).
Genetic analysis of H5 has also underscored the pleiotropic contributions of H5 to the viral life cycle. To identify viral proteins involved in transcriptional processivity, Condit and colleagues isolated viral mutants resistant to isatin-β-thiosemicarbazone (IBT) (55–60). IBT leads to the synthesis of run-on transcripts, which in turn generate excessive dsRNA that leads to the arrest of infection by the antiviral response. In addition to mapping IBT resistance to a variety of proteins known to be directly involved in transcription, they identified a mutation in the H5 gene (D36V) that conferred IBT resistance (61). Some years ago our laboratory isolated a virus (tsH5-4) with a mutated H5 allele (EEE73–75AAA) that conferred a dominant, temperature-sensitive phenotype (26). Somewhat unexpectedly, the biochemical progression of the infectious cycle was unimpaired, but a profound defect in early virion morphogenesis was observed.
As part of their thorough reassessment of the Dales collection of ts mutants, Condit and colleagues characterized Dts57, which was isolated after nitrosoguanidine and BrdU mutagenesis of the IHD-W strain of vaccinia virus (42). Marker rescue analysis indicated that recombination of the Dts57 genome with a WT H5 allele could rescue plaque formation at the nonpermissive temperature, and it was determined that the H5 gene of the mutant contained a single nucleotide change predicted to cause a G189R substitution (42). Briefly, their data indicated that Dts57 exhibited a strong defect in DNA replication at the nonpermissive temperature; furthermore, DNA accumulation stopped when infections were shifted from the permissive to the nonpermissive temperature at 9 hpi. These data suggest that the H5 protein encoded by the mutant is unable to provide a vital H5-dependent contribution to replication. Shift-up experiments indicated that Dts57 infections resulted in subsequent defects in gene expression and morphogenesis that could be revealed if genome replication was allowed to occur at the permissive temperature.
To study the impact of the G189R substitution in an otherwise WT background, we created WRts57 and confirmed that it retained a ts phenotype, albeit one much less severe than that for the Dts57 virus (Fig. 2). Moreover, there was significantly less of an impact on DNA replication, which continued at reduced but readily detectable levels. Because the H5 genes of the WR and IHD-W strains are identical, we posit that the original Dts57 virus might have had mutations elsewhere in the genome that contributed to the impaired DNA replication. Alternatively, since H5 is known to interact with several other viral proteins, polymorphisms in other components of the replication machinery might lessen or augment the impact of the G189R substitution. It should be noted, however, that the A20 genes of the IHD-W and WR strains are also identical.
The mutant H5 protein (G189R) is stable at the nonpermissive temperature, and therefore, the phenotype(s) seen represents a temperature-dependent diminution in some, but probably not all, of the protein's functions. We therefore turned to siRNA technology to assess the impact of H5 depletion on viral DNA replication and the subsequent phases of the viral life cycle (Fig. 3A and B). We were successful in achieving a >90% depletion of H5, as assessed by immunoblot analysis, and a 10-fold decrease in viral yield. Although these data superficially suggest that H5 might not be an essential protein, it is worth remembering that, if 10% of the cells were not successfully depleted of H5 and produced normal levels of infectious virus, this could account for the 10-fold decrease in viral yield. Indeed, our immunofluorescence data confirmed the presence of cells that retained H5 expression. Moreover, the cells in which H5 expression appeared to be normal also had undiminished levels of BrdU+ DNA. In contrast, the cells with little or no H5 expression had a comparable reduction in the level of BrdU positivity.
Realizing that siRNA depletion was also not going to reveal whether H5 makes a vital contribution to genome replication, we generated an H5-expressing CV-1 cell line as a prerequisite for generating a recombinant virus in which H5 was deleted (vΔH5). In this cell line, a single copy of a codon-optimized H5 allele is driven by the CMV promoter in a Dox-dependent manner (Fig. 4B). Upon induction, the levels of H5 approximate ∼1/2 of those seen in a WT virus infection. In the absence of infection, the coH5 protein appears to be dispersed throughout the cell. Cytoplasmic puncta of more intense staining are also seen; these puncta may represent higher-order H5 complexes or aggregates. Upon infection with the vΔH5 recombinant, H5 relocalizes to viral replication factories and is present in amounts sufficient to support the progression of the viral life cycle (Fig. 5C). The relocalization of H5 to replication foci is most likely facilitated by protein-protein interactions rather than solely by protein-nucleic acid interactions, because the 25-kDa H5 protein can diffuse freely into the nucleus (Fig. 5C, middle) and may be retained on cellular DNA. The virions that are produced in the induced cell line encapsidate nearly WT levels of H5 and have a particle/PFU ratio that is only 2-fold higher than that of WT virions produced in CV-1 cells (Fig. 5E and F). The increased particle/PFU ratio may represent a modest impairment of virion maturation, because the vΔH5 virions contain increased levels of unprocessed L4. Proteolytic processing of L4 by the I7 protease is one of the hallmarks of the immature virion-to-MV maturation step (62–65).
When vΔH5 stocks were used to infect noncomplementing BSC40 cells (Fig. 6), we observed that early gene expression proceeded normally; in fact, the key components of the genome replication machinery (E9 Pol, the I3 SSB protein, D5, A20 [and, by inference, D4 UDG]) accumulated to WT levels (Fig. 6B). We also verified that the subsequent uncoating of the core also proceeded normally (Fig. 6C).
Despite the presence of the replication machinery and the uncoating of the core, we observed a complete block to DNA replication in the absence of H5 expression (Fig. 6D and E). No viral DNA could be detected by Southern dot blot hybridization or pulsed-field gel electrophoresis. This striking result proves that the H5 protein is indeed essential for viral DNA replication. As would be expected in the absence of replication, we observed no expression of viral late proteins (not shown), and the yield of infectious virus was reduced by nearly 4 log units compared to that of WT virus (Fig. 6A).
The key question for future study will be to decipher how the abundant H5 protein supports viral replication. H5 has been shown to interact with the A20 processivity factor in both yeast two-hybrid and in vitro pulldown assays (28, 30). Herein, we show that H5 copurifies with the trimeric holoenzyme but not with the A20/UDG processivity factor, suggesting that it associates most stably with the holoenzyme (Fig. 1D). However, we know from several lines of investigation that H5 is not important for the stability of the replication proteins (Fig. 6B) or for the assembly or activity of the DNA Pol or the holoenzyme (data not shown). Purified preparations of DNA polymerase, as visualized by silver stain analysis, appear to be free of other proteins, and the processive holoenzyme can be assembled from separate baculovirus infections expressing Pol and A20/UDG (66). Moreover, active holoenzyme can be assembled using in vitro-transcribed/translated proteins (67).
How else might H5 contribute to genome replication? H5 is a DNA binding protein, and it is possible that newly synthesized H5 is essential to coat and protect the viral genome as it emerges from the core during uncoating. Highly sensitive assays would be needed to determine if the input genomes remain stable during infections with noncomplemented virus lacking H5. Alternatively, H5 might participate in the reorganization of endoplasmic reticulum membranes that enclose the replication paddocks; indeed, we have shown that H5 can associate co- or posttranslationally with microsomal membranes when synthesized using an in vitro transcription/translation reaction (data not shown). Alternatively, the high-affinity association of H5 with the viral genome might help to organize the genomes within the factory so that they are readily accessible to the replication machinery. H5 is highly abundant, and we know that it forms higher-order structures, with the tetrameric form being the most stable form in vitro (53). H5 might in fact form a lattice that serves as a scaffold on which the genomes and/or the holoenzyme might become arrayed for efficient synthesis. Modeling of the H5 protein and biophysical analyses suggest that much of the N-terminal 2/3 of the H5 protein is unstructured, whereas the C-terminal region is thought to form an amphipathic helix (53). It will be important to determine which regions of the protein mediate DNA binding, multimerization, and association with A20. With vΔH5 as a key genetic tool, we can perform extensive structure-function analyses with truncated and mutated versions of H5 and determine which regions or motifs of the protein are necessary or sufficient for genome replication.
Whereas H5 is present and highly conserved in chordopoxviruses, it is absent from the entomopoxvirus family. It is interesting to speculate about whether another viral or cellular protein can substitute for H5 in this context or whether the cytoplasmic environment of insect cells is structured in a manner that obviates the need for the functions that H5 provides.
Our studies provide an informative comparison of the opportunities and challenges presented by temperature-sensitive mutants, siRNA, and deletion mutants. Until recently, complementing cell lines have been rare in the poxvirus field, but the availability of codon-optimized alleles and tunable expression systems has enhanced their utility. Such cell lines enable the isolation of deletion mutants in the absence of selective pressure. In the case of the H5 protein, the residual H5 expressed in a subset of cells after siRNA treatment and the complex phenotype of ts mutants in which only some of the functions of H5 were perturbed were unable to provide a clear view of the function of H5. In contrast, the generation and phenotypic analysis of the vΔH5 recombinant have provided clear and incontrovertible evidence that H5 is an essential protein that plays a vital role in the replication of the viral genome.
ACKNOWLEDGMENTS
We thank members of the P. Traktman lab for lively discussions and their interest in this project. We thank Clive Wells for his expert assistance in preparing the samples for electron microscopy and Bethany Unger and Matthew Wiebe for their invaluable assistance with immunofluorescence microscopy.
This work was supported, in part, by NIH grant 5 R01 AI021758, awarded to P.T.
REFERENCES
- 1.Tolonen N, Doglio L, Schleich S, Krijnse-Locker J. 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol Biol Cell 12:2031–2046. doi: 10.1091/mbc.12.7.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle KA, Arps L, Traktman P. 2007. Biochemical and genetic analysis of the vaccinia virus D5 protein: multimerization-dependent ATPase activity is required to support viral DNA replication. J Virol 81:844–859. doi: 10.1128/JVI.02217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans E, Klemperer N, Ghosh R, Traktman P. 1995. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J Virol 69:5353–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans E, Traktman P. 1987. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J Virol 61:3152–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans E, Traktman P. 1992. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma 102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- 6.Gammon DB, Evans DH. 2009. The 3′-to-5′ exonuclease activity of vaccinia virus DNA polymerase is essential and plays a role in promoting virus genetic recombination. J Virol 83:4236–4250. doi: 10.1128/JVI.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greseth MD, Boyle KA, Bluma MS, Unger B, Wiebe MS, Soares-Martins JA, Wickramasekera NT, Wahlberg J, Traktman P. 2012. Molecular genetic and biochemical characterization of the vaccinia virus I3 protein, the replicative single-stranded DNA binding protein. J Virol 86:6197–6209. doi: 10.1128/JVI.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii K, Moss B. 2001. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J Virol 75:1656–1663. doi: 10.1128/JVI.75.4.1656-1663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemperer N, McDonald W, Boyle K, Unger B, Traktman P. 2001. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J Virol 75:12298–12307. doi: 10.1128/JVI.75.24.12298-12307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald WF, Traktman P. 1994. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr Purif 5:409–421. doi: 10.1006/prep.1994.1059. [DOI] [PubMed] [Google Scholar]
- 11.Punjabi A, Boyle K, DeMasi J, Grubisha O, Unger B, Khanna M, Traktman P. 2001. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J Virol 75:12308–12318. doi: 10.1128/JVI.75.24.12308-12318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochester SC, Traktman P. 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol 72:2917–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanitsa ES, Arps L, Traktman P. 2006. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J Biol Chem 281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- 14.Tseng M, Palaniyar N, Zhang W, Evans DH. 1999. DNA binding and aggregation properties of the vaccinia virus I3L gene product. J Biol Chem 274:21637–21644. doi: 10.1074/jbc.274.31.21637. [DOI] [PubMed] [Google Scholar]
- 15.Wiebe MS, Traktman P. 2007. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1:187–197. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buller RM, Smith GL, Cremer K, Notkins AL, Moss B. 1985. Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature 317:813–815. doi: 10.1038/317813a0. [DOI] [PubMed] [Google Scholar]
- 17.Child SJ, Palumbo GJ, Buller RM, Hruby DE. 1990. Insertional inactivation of the large subunit of ribonucleotide reductase encoded by vaccinia virus is associated with reduced virulence in vivo. Virology 174:625–629. doi: 10.1016/0042-6822(90)90119-C. [DOI] [PubMed] [Google Scholar]
- 18.Garcia AD, Moss B. 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol 75:6460–6471. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes SJ, Johnston LH, de Carlos A, Smith GL. 1991. Vaccinia virus encodes an active thymidylate kinase that complements a cdc8 mutant of Saccharomyces cerevisiae. J Biol Chem 266:20103–20109. [PubMed] [Google Scholar]
- 20.Kerr SM, Smith GL. 1991. Vaccinia virus DNA ligase is nonessential for virus replication: recovery of plasmids from virus-infected cells. Virology 180:625–632. doi: 10.1016/0042-6822(91)90076-N. [DOI] [PubMed] [Google Scholar]
- 21.Paran N, De Silva FS, Senkevich TG, Moss B. 2009. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe 6:563–569. doi: 10.1016/j.chom.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senkevich TG, Koonin EV, Moss B. 2009. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc Natl Acad Sci U S A 106:17921–17926. doi: 10.1073/pnas.0909529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaud G, Beaud R. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J Gen Virol 78(Pt 12):3297–3302. [DOI] [PubMed] [Google Scholar]
- 24.Black EP, Moussatche N, Condit RC. 1998. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology 245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- 25.Dellis S, Strickland KC, McCrary WJ, Patel A, Stocum E, Wright CF. 2004. Protein interactions among the vaccinia virus late transcription factors. Virology 329:328–336. doi: 10.1016/j.virol.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 26.DeMasi J, Traktman P. 2000. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J Virol 74:2393–2405. doi: 10.1128/JVI.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domi A, Beaud G. 2000. The punctate sites of accumulation of vaccinia virus early proteins are precursors of sites of viral DNA synthesis. J Gen Virol 81(Pt 5):1231–1235. [DOI] [PubMed] [Google Scholar]
- 28.Ishii K, Moss B. 2002. Mapping interaction sites of the A20R protein component of the vaccinia virus DNA replication complex. Virology 303:232–239. doi: 10.1006/viro.2002.1721. [DOI] [PubMed] [Google Scholar]
- 29.Kovacs GR, Moss B. 1996. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol 70:6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCraith S, Holtzman T, Moss B, Fields S. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci U S A 97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J Virol 80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grubisha O, Traktman P. 2003. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J Virol 77:10929–10942. doi: 10.1128/JVI.77.20.10929-10942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matson J, Chou W, Ngo T, Gershon PD. 2014. Static and dynamic protein phosphorylation in the vaccinia virion. Virology 452–453:310–323. doi: 10.1016/j.virol.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. 2007. Protein composition of the vaccinia virus mature virion. Virology 358:233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 35.Yoder JD, Chen TS, Gagnier CR, Vemulapalli S, Maier CS, Hruby DE. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol J 3:10. doi: 10.1186/1743-422X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon J, Mohandas A, Wilton S, Dales S. 1991. A prominent antigenic surface polypeptide involved in the biogenesis and function of the vaccinia virus envelope. Virology 181:671–686. doi: 10.1016/0042-6822(91)90901-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Costa SM, Bainbridge TW, Kato SE, Prins C, Kelley K, Condit RC. 2010. Vaccinia H5 is a multifunctional protein involved in viral DNA replication, postreplicative gene transcription, and virion morphogenesis. Virology 401:49–60. doi: 10.1016/j.virol.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elroy-Stein O, Fuerst TR, Moss B. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A 86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu K, Lemon B, Traktman P. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol 69:7823–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 41.Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J Virol 74:3682–3695. doi: 10.1128/JVI.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato SE, Moussatche N, D'Costa SM, Bainbridge TW, Prins C, Strahl AL, Shatzer AN, Brinker AJ, Kay NE, Condit RC. 2008. Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants. Virology 375:213–222. doi: 10.1016/j.virol.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traktman P, Boyle K. 2004. Methods for analysis of poxvirus DNA replication. Methods Mol Biol 269:169–186. doi: 10.1385/1-59259-789-0:169. [DOI] [PubMed] [Google Scholar]
- 44.Whitbeck JC, Foo CH, Ponce de Leon M, Eisenberg RJ, Cohen GH. 2009. Vaccinia virus exhibits cell-type-dependent entry characteristics. Virology 385:383–391. doi: 10.1016/j.virol.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald WF, Crozel-Goudot V, Traktman P. 1992. Transient expression of the vaccinia virus DNA polymerase is an intrinsic feature of the early phase of infection and is unlinked to DNA replication and late gene expression. J Virol 66:534–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaud G, Beaud R, Leader DP. 1995. Vaccinia virus gene H5R encodes a protein that is phosphorylated by the multisubstrate vaccinia virus B1R protein kinase. J Virol 69:1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livingston CM, DeLuca NA, Wilkinson DE, Weller SK. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J Virol 82:6324–6336. doi: 10.1128/JVI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle KA, Stanitsa ES, Greseth MD, Lindgren JK, Traktman P. 2011. Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J Biol Chem 286:24702–24713. doi: 10.1074/jbc.M111.222216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lackner CA, D'Costa SM, Buck C, Condit RC. 2003. Complementation analysis of the Dales collection of vaccinia virus temperature-sensitive mutants. Virology 305:240–259. doi: 10.1006/viro.2002.1745. [DOI] [PubMed] [Google Scholar]
- 50.Dales S, Milovanovitch V, Pogo BG, Weintraub SB, Huima T, Wilton S, McFadden G. 1978. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology 84:403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- 51.Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. 2012. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep 2:1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Joklik WK. 1964. The intracellular uncoating of poxvirus DNA. II. The molecular basis of the uncoating process. J Mol Biol 8:277–288. [DOI] [PubMed] [Google Scholar]
- 53.Kay NE, Bainbridge TW, Condit RC, Bubb MR, Judd RE, Venkatakrishnan B, McKenna R, D'Costa SM. 2013. Biochemical and biophysical properties of a putative hub protein expressed by vaccinia virus. J Biol Chem 288:11470–11481. doi: 10.1074/jbc.M112.442012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Costa SM, Bainbridge TW, Condit RC. 2008. Purification and properties of the vaccinia virus mRNA processing factor. J Biol Chem 283:5267–5275. doi: 10.1074/jbc.M709258200. [DOI] [PubMed] [Google Scholar]
- 55.Condit RC, Easterly R, Pacha RF, Fathi Z, Meis RJ. 1991. A vaccinia virus isatin-beta-thiosemicarbazone resistance mutation maps in the viral gene encoding the 132-kDa subunit of RNA polymerase. Virology 185:857–861. doi: 10.1016/0042-6822(91)90559-T. [DOI] [PubMed] [Google Scholar]
- 56.Condit RC, Xiang Y, Lewis JI. 1996. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implications for control of transcription. Virology 220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- 57.Cresawn SG, Prins C, Latner DR, Condit RC. 2007. Mapping and phenotypic analysis of spontaneous isatin-beta-thiosemicarbazone resistant mutants of vaccinia virus. Virology 363:319–332. doi: 10.1016/j.virol.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hassett DE, Lewis JI, Xing X, DeLange L, Condit RC. 1997. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology 238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- 59.Latner DR, Xiang Y, Lewis JI, Condit J, Condit RC. 2000. The vaccinia virus bifunctional gene J3 (nucleoside-2′-O-)-methyltransferase and poly(A) polymerase stimulatory factor is implicated as a positive transcription elongation factor by two genetic approaches. Virology 269:345–355. doi: 10.1006/viro.2000.0243. [DOI] [PubMed] [Google Scholar]
- 60.Meis RJ, Condit RC. 1991. Genetic and molecular biological characterization of a vaccinia virus gene which renders the virus dependent on isatin-beta-thiosemicarbazone (IBT). Virology 182:442–454. doi: 10.1016/0042-6822(91)90585-Y. [DOI] [PubMed] [Google Scholar]
- 61.Cresawn SG, Condit RC. 2007. A targeted approach to identification of vaccinia virus postreplicative transcription elongation factors: genetic evidence for a role of the H5R gene in vaccinia transcription. Virology 363:333–341. doi: 10.1016/j.virol.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ansarah-Sobrinho C, Moss B. 2004. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J Virol 78:6335–6343. doi: 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byrd CM, Bolken TC, Hruby DE. 2002. The vaccinia virus I7L gene product is the core protein proteinase. J Virol 76:8973–8976. doi: 10.1128/JVI.76.17.8973-8976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byrd CM, Bolken TC, Hruby DE. 2003. Molecular dissection of the vaccinia virus I7L core protein proteinase. J Virol 77:11279–11283. doi: 10.1128/JVI.77.20.11279-11283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ericsson M, Cudmore S, Shuman S, Condit RC, Griffiths G, Locker JK. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J Virol 69:7072–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sele C, Gabel F, Gutsche I, Ivanov I, Burmeister WP, Iseni F, Tarbouriech N. 2013. Low-resolution structure of vaccinia virus DNA replication machinery. J Virol 87:1679–1689. doi: 10.1128/JVI.01533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan H, Nuth M, Zhukovskaya N, Saw YL, Bell E, Isaacs SN, Ricciardi RP. 2014. A novel target and approach for identifying antivirals against molluscum contagiosum virus. Antimicrob Agents Chemother 58:7383–7389. doi: 10.1128/AAC.03660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]