ABSTRACT
All well-characterized positive-strand RNA viruses[(+)RNA viruses] induce the formation of host membrane-bound viral replication complexes (VRCs), yet the underlying mechanism and machinery for VRC formation remain elusive. We report here the biogenesis and topology of the Beet black scorch virus (BBSV) replication complex. Distinct cytopathological changes typical of endoplasmic reticulum (ER) aggregation and vesiculation were observed in BBSV-infected Nicotiana benthamiana cells. Immunogold labeling of the auxiliary replication protein p23 and double-stranded RNA (dsRNA) revealed that the ER-derived membranous spherules provide the site for BBSV replication. Further studies indicated that p23 plays a crucial role in mediating the ER rearrangement. Three-dimensional electron tomographic analysis revealed the formation of multiple ER-originated vesicle packets. Each vesicle packet enclosed a few to hundreds of independent spherules that were invaginations of the ER membranes into the lumen. Strikingly, these vesicle packets were connected to each other via tubules, a rearrangement event that is rare among other virus-induced membrane reorganizations. Fibrillar contents within the spherules were also reconstructed by electron tomography, which showed diverse structures. Our results provide the first, to our knowledge, three-dimensional ultrastructural analysis of membrane-bound VRCs of a plant (+)RNA virus and should help to achieve a better mechanistic understanding of the organization and microenvironment of plant (+)RNA virus replication complexes.
IMPORTANCE Assembly of virus replication complexes for all known positive-strand RNA viruses depends on the extensive remodeling of host intracellular membranes. Beet black scorch virus, a necrovirus in the family Tombusviridae, invaginates the endoplasmic reticulum (ER) membranes to form spherules in infected cells. Double-stranded RNAs, the viral replication intermediate, and the viral auxiliary replication protein p23 are all localized within such viral spherules, indicating that these are the sites for generating progeny viral RNAs. Furthermore, the BBSV p23 protein could to some extent reorganize the ER when transiently expressed in N. benthamiana. Electron tomographic analysis resolves the three-dimensional (3D) architecture of such spherules, which are connected to the cytoplasm via a neck-like structure. Strikingly, different numbers of spherules are enclosed in ER-originated vesicle packets that are connected to each other via tubule-like structures. Our results have significant implications for further understanding the mechanisms underlying the replication of positive-strand RNA viruses.
INTRODUCTION
Positive-strand RNA viruses [(+)RNA viruses] include numerous human, animal, and plant pathogens. A highly conserved feature of (+)RNA viruses is that they modify the host endomembrane system to assemble viral replication complexes (VRCs) (1). Typical rearrangements of intracellular membrane structures include membrane invaginations and proliferations (2, 3). Different viruses assemble their VRCs in different organelles, such as the endoplasmic reticulum (ER), mitochondria, peroxisomes, chloroplasts, and tonoplasts (4–6). These remodeled intracellular membranes offer a scaffold for VRC assembly and protect viral RNAs from degradation by cellular nucleases associated with the host innate immune responses, among others (7).
Specific cytopathological alterations arising from viral infection, in many cases, have been ascribed to the expression of virus-encoded proteins, as well as the participation of various host factors (8, 9). For example, p6 or nsp3-nsp4-nsp6 of the severe acute respiratory syndrome coronavirus (SARS-CoV) have the ability to induce double-membrane vesicles that are similar to those induced during SARS-CoV infection (10, 11), and open reading frame (ORF) 3a is both necessary and sufficient for SARS-CoV-induced Golgi apparatus fragmentation (12). Expression of Brome mosaic virus (BMV) 1a or CP, in the absence of other viral proteins, could induce ER membrane vesiculation similar to that present during BMV infection (13, 14). Expression of Tomato bushy stunt virus (TBSV) 33-kDa replication protein alone could lead to the aggregation of peroxisomes, resulting in the formation of peroxisomal ghosts, although they are distinct from peroxisomal spherules induced by viral infection (15). These results suggest that expression of one or several viral proteins, rather than all components, could rearrange intracellular membranes, thereby paving the way for the establishment of initial viral replication (5, 16).
Beet black scorch virus (BBSV) is one of the rhizomania-causing viruses that severely threaten the yield and quality of sugar beet production in the northern provinces of China (17). Since the first report of BBSV in China, it has been reported in North America, the Middle East, and Europe (18–21). BBSV, a member of the genus Necrovirus in the family Tombusviridae, possesses a positive-sense RNA genome of 3,644 nucleotides (nt) including six ORFs. p23 is an auxiliary replication protein, while p82 is the translational readthrough product of p23 and serves as the RNA-dependent RNA polymerase (RdRp). p7a, p7b, and p5′ are essential for cell-to-cell movement of BBSV in infected plants, and p24 is the capsid protein. Although BBSV is classified as a member of the Tombusviridae, it shows low sequence identity with other viruses in the family. This is exemplified by 61% identity at the nucleotide sequence level to the most closely related virus, Tobacco necrosis virus D (TNV-D) (21), implying that BBSV may produce cytopathological effects distinct from those of other viruses in the family.
In the present study, we investigated the cytopathological effects caused by BBSV infection. Our study revealed that BBSV infection induced the formation of ER-derived vesicle packets (VPs), which enclose various numbers of 50- to 70-nm vesicles. Both viral p23 and the viral replication intermediate double-stranded RNA (dsRNA) were associated with such vesicles, which we termed viral spherules. Further study indicated that rearrangement of the ER membrane by BBSV was mediated by the expression of p23. Electron tomographic analysis indicated that the spherules are invaginations of the ER membranes, with an ∼6-nm-wide neck connecting the interior of the viral spherule to the cytoplasm.
MATERIALS AND METHODS
Molecular cloning and plasmid construction.
All plasmids were constructed according to standard molecular cloning methods (22). Briefly, BBSV p23 was amplified from the pUBF52 plasmid (23) by PCR using different primer combinations listed in Table S1 in the supplemental material, and the resulting fragments flanked by appropriate restriction sites were digested and then ligated into similarly digested vectors, including pGD, pGDG, pGDGm, and pGDRm (24, 25), to generate the ultimate constructs: 35S-p23, green fluorescent protein (GFP)-p23, p23-GFP, and p23-red fluorescent protein (RFP), respectively. For bimolecular fluorescence complementation (BiFC) assays, p23 and p82 were amplified from the pUBF52 and pUBF52G647C plasmids, respectively; digested; and then ligated into similarly digested pSPYNE-35S and pSPYCE-35S vectors (26). NbrbcL was cloned from the cDNA of Nicotiana benthamiana and constructed into the BiFC vectors as described above. Detailed sequence information for all the primers used in this study is available in Table S1 in the supplemental material, and all constructs generated by PCR were verified by sequencing.
Virus inoculation and agroinfiltration of N. benthamiana leaves.
Virus inoculation was conducted as previously reported (23). Briefly, purified BBSV virions were diluted with an equal volume of 2× GKP buffer (50 mM glycine, 30 mM K2HPO4 [pH 9.2], 1% bentonite, 1% celite) to a final concentration of 100 ng/μl. Wild-type N. benthamiana or line 16C was mechanically inoculated with BBSV and then transferred into a growth chamber and maintained at 18°C under 16 h light.
All binary vectors were transformed into Agrobacterium tumefaciens EHA105 by the freeze-thaw method (27). Wild-type N. benthamiana or line 16C was agroinoculated with A. tumefaciens carrying the expression cassette according to a procedure described previously (28). The plants were then incubated at 18°C for 3 to 8 days prior to confocal laser scanning microscopy (CLSM) and immunoblot analysis.
Confocal laser scanning microscopy.
Fluorescence microscopy was performed using an Olympus FV1000 microscope (Olympus Corporation, Tokyo, Japan). GFP, mCherry, RFP, and chlorophyll autofluorescence was excited using 488-, 543-, 543-, and 633-nm laser lines, respectively. A sequencing scanning mode was used to minimize the cross talk between two partially overlapping emission spectra. Image acquisition was conducted at a resolution of 1,024 by 1,024 pixels and a scan rate of 12.5 μs/pixel.
Mesophyll protoplasts of N. benthamiana were prepared from agroinfiltrated or BBSV-inoculated 16C leaves as described previously (29) with some modifications. At 8 days postinfection (dpi), approximately 0.5 g leaf tissue was chopped, transferred to 15 ml enzyme solution (1.25% cellulase Onozuka R-10 [Yakult Honsha, Tokyo, Japan], 0.3% macerozyme R-10 [Yakult Honsha], 8.2% d-mannitol, 20 mM morpholineethanesulfonic acid [MES], 20 mM KCl, 10 mM CaCl2, and 0.1% bovine serum albumin [BSA], pH 5.5), and incubated for 3 h at room temperature (25°C) in the dark with gentle shaking (45 rpm). Released protoplasts were collected by filtration through 70-μm nylon mesh and immediately visualized with a confocal microscope as described above. All the raw image data were processed using Imaris software (version 7.2.3; Bitplane AG).
For BiFC assays, Agrobacterium cultures harboring BiFC constructs, mCherry fused with the ER retention signal (HDEL), and p19 plasmids were diluted to a final optical density at 600 nm (OD600) of 0.2, 0.2, and 0.1, respectively. The mixtures were then used to infiltrate 5- to 6-week-old N. benthamiana leaves. Images were taken with excitation at 514 nm (yellow fluorescent protein [YFP]) and 543 nm (mCherry) via CLSM.
Membrane flotation assays.
Membrane flotation assays were conducted as previously described (30). In brief, 1 g N. benthamiana leaf tissue showing characteristic symptoms of BBSV infection or agroinfiltrated with binary vectors was gently ground in 1.5 ml of homogenization buffer (50 mM Tris-HCl, pH 8.0, 10 mM KCl, 3 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1% BSA, 0.3% dextran, 13% [wt/vol] sucrose, 1% protease inhibitor cocktail [Sigma]), followed by filtering through Miracloth filter gauze (Merck/Calbiochem, Darmstadt, Germany) and centrifugation at 3,700 × g at 4°C for 10 min to remove unlysed cells, plastids, nuclei, and large debris (P3 fraction). The supernatant (S3) was centrifuged at 30,000 × g at 4°C for 20 min to obtain the soluble (S30) and pellet (P30) fractions, respectively. First, to examine whether p23 is an integral membrane protein, the P30 fraction was incubated with buffers as described previously (30). Second, the P30 fraction was resuspended with a volume of homogenization buffer equal to that of the corresponding supernatant. Next, 0.3 ml P30 fraction was placed at the bottom of a centrifuge tube by mixing with 1.6 ml of 85% (wt/vol) sucrose in NTE buffer (100 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1% protease inhibitor cocktail [Sigma]) and then overlaid with 7 ml of 65% and 3.1 ml of 10% sucrose in NTE buffer. Equilibrium centrifugation was carried out at 100,000 × g for 18 h at 4°C in a Hitachi (Japan) Himac CP-80β ultracentrifuge. Ten 1.2-ml fractions were sequentially collected from the top of the tube for immunoblotting analysis as described previously (31).
Transmission electron microscopy (TEM).
For conventional electron microscopy, leaf tissue fragments (about 1 to 2 mm2) from N. benthamiana were excised in a drop of 0.05 M phosphate buffer (pH 7.2) containing 2.5% glutaraldehyde and vacuum infiltrated in the same fixative for about 15 min to let all the pieces of leaves sink to the bottom of the tube. The samples were then fixed at 4°C for at least 1 day before postfixation in 2% osmium tetroxide (OsO4) for 2 h at 4°C. After dehydration through a graded series of ethanol and acetone, the leaf pieces were embedded in Spurr's resin (SPI Supplies). Ultrathin sections (70 nm) were cut with a diamond knife on an ultramicrotome (EM UC7; Leica, Germany) and mounted on Formvar-coated 100-mesh copper grids. The sections were double stained with saturated aqueous uranyl acetate for 30 min in the dark, followed by Reynolds' lead citrate for 5 min at room temperature, and then viewed with a Hitachi H-7650 (Hitachi, Japan) or a JEM-1230 (JEOL Co. Ltd., Japan) transmission electron microscope operating at 80 kV.
Immunoelectron microscopy was performed according to a previously described method (32) with minor modifications. Briefly, N. benthamiana leaves were fixed and embedded in Spurr's resin as described above. Ultrathin sections prepared from these blocks were placed on Formvar-coated 150-mesh nickel grids. The grids were etched on drops of 4% sodium metaperiodate for 10 min to unmask antigenic sites on the surface of the section (32). The grids were then rinsed three times on successive drops of double-distilled H2O (ddH2O) and placed on drops of 0.01 M phosphate-buffered saline (PBS) (pH 7.2) containing 0.02 M glycine for 10 min to quench residual aldehyde groups. Sections were then placed on a drop of blocking solution consisting of 0.01 M PBS (pH 7.2), 0.01% Triton X-100, 0.01% Tween 20, and 1% BSA for 10 min to reduce nonspecific binding of antibodies. Rabbit anti-p23 antibody and J2 mouse monoclonal anti-dsRNA (English & Scientific Consulting) were diluted 1:500 and 1:200, respectively, in blocking solution. The grids were incubated with the primary antibody solutions overnight at 4°C in a moist chamber. After extensive washing with blocking solution (six times for 5 min each), the sections were incubated with goat anti-rabbit or goat anti-mouse secondary antibodies conjugated with 10-nm gold particles (Sigma; both were diluted 1:200 in blocking solution) for 1 to 2 h at 28°C. After consecutive rinses in blocking solution and ddH2O, the sections were stained for 20 min with saturated aqueous uranyl acetate in the dark, followed by 3 min in Reynolds' lead citrate. Finally, the samples were examined under an electron microscope. Tissues from mock-inoculated plants of comparable age were processed similarly to serve as controls.
Electron tomography.
Epon 812 resin-embedded blocks of BBSV-infected leaf tissues were cut to a thickness of 200 nm and collected on Formvar-carbon-coated copper 100-mesh grids. The sections were poststained with saturated aqueous uranyl acetate, followed by Reynolds' lead citrate. The grids were placed in the single-tilt tomography holder on a Tecnai G2 Spirit BioTwin LaB6 transmission electron microscope (FEI) operated at 120 kV. Images were acquired as a single-axis tilt series across a range of −70° to +70° in 2° angular increments and were recorded with a cooled slow-scan charge-coupled-device (CCD) camera (4k Eagle; FEI) in 2-fold binning mode using FEI software. The electron microscope magnification was ×30,000, corresponding to a pixel size of 0.74 nm at the specimen level. The alignment, computation and reconstruction of electron tomograms were performed with the FEI software package Inspect 3D using the iterative routine SIRT. Rendering of the three-dimensional (3D) surface of the tomograms was performed using the Imaris software (version 7.2.3; Bitplane AG). Models were generated by manually tracing the areas of interest from unfiltered tomograms, followed by smoothing labels and gap filling. The size of the voxels in the tomograms corresponds to 1.489 nm.
RESULTS
BBSV infection reshapes the ER structure and generates membrane-associated spherules.
Previous studies indicated that viruses from the genus Necrovirus in the family Tombusviridae often cause cytopathic effects on the tonoplast (33) or ER (34, 35). Based upon this, we agroinfiltrated mCherry-fused γ-TIP, a plant tonoplast marker (36), into N. benthamiana leaves that had been infected by BBSV. No apparent morphological abnormalities were observed in tonoplasts by CLSM (data not shown). In contrast, in BBSV-infected plant epidermal cells of line 16C plants, transgenic N. benthamiana plants expressing an ER-localized GFP (37), the ER was rearranged, with formation of punctate structures in both inoculated and systemically infected leaves (Fig. 1A). Conversely, the ER presented a regular network in the mock-inoculated 16C plants (Fig. 1A). In order to examine the ER morphology in mesophyll cells, the inoculated leaves and upper leaves exhibiting chlorosis symptoms were processed by enzyme digestion, and the resulting protoplast was observed using CLSM. Punctate structures or aggregations along the ER network were clearly seen in the mesophyll cells derived from both locally and systemically infected leaves, in sharp contrast to the healthy cells, which maintained a regular, web-like ER structure (Fig. 1B). Taken together, these results indicated that BBSV infection was associated with remodeling of the ER network, implying that morphological changes of the ER are probably related to the establishment of viral infection.
FIG 1.
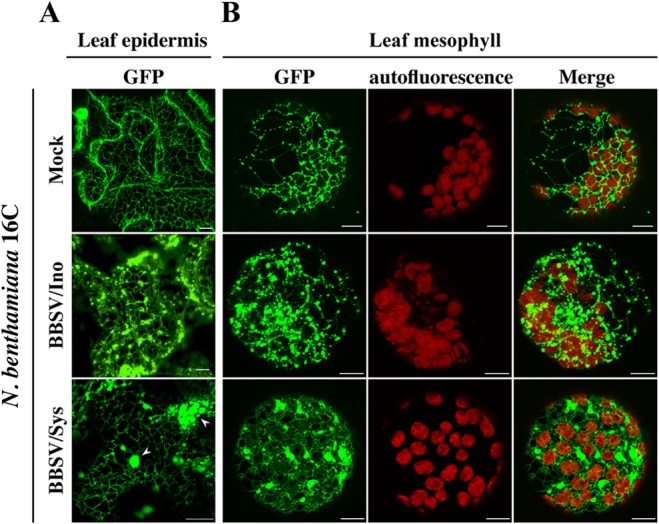
BBSV infection induced severe morphological changes in the endoplasmic reticulum. BBSV virions were inoculated onto N. benthamiana line 16C, in which the ER is decorated with GFP (37). The treatment is indicated on the left of each row. Both leaf epidermis (A) and mesophyll (B) cells were analyzed by CLSM. N. benthamiana 16C leaves were inoculated with BBSV virions (BBSV/Ino). At 3 dpi, the leaf epidermis was peeled from these leaves, and the remainder was treated with enzymes to liberate protoplasts from the mesophyll. Systemically infected leaves (BBSV/Sys) were harvested at about 8 dpi and similarly processed for CLSM analysis. Mock-inoculated leaves (Mock) from line 16C were processed following the same procedures. The arrowheads indicate the ER aggregates. Bars = 10 μm.
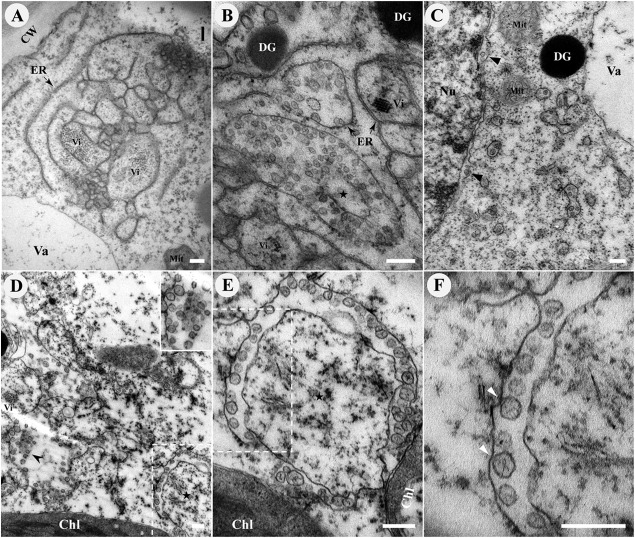
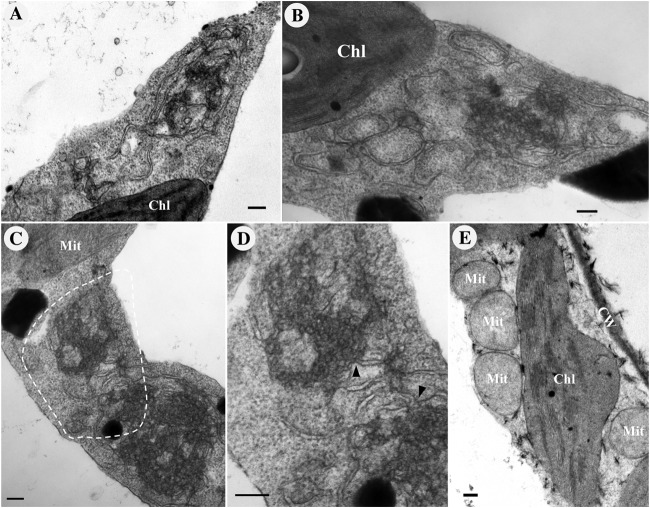
To investigate the changes in cytological structure caused by BBSV infection in detail, we used TEM to analyze the ultrastructure of BBSV-infected leaf tissues (Fig. 2). Large numbers of virus particles scattered in the cytoplasm were readily identified. In some regions of the cytoplasm, virus aggregated in irregular crystalline arrays (data not shown). In addition to the presence of virus particles, the most striking cytopathic effects were structural changes in the ER membranes, which proliferated and became convoluted. Large amounts of viral particles were intermingled within the extended ER cisternae (Fig. 2A). Meanwhile, many pockets of vesicles, along with the dilated ER, were frequently present, and crystalline arrays of virus particles occasionally appeared around them (Fig. 2B). These cytopathological features induced by BBSV infection were absent in the healthy cells (data not shown). Furthermore, pockets of vesicles were also found in the cytoplasm, as well as the perinuclear membrane (Fig. 2C). We termed these vesicle-containing large bubbles vesicle packets. In most cases, the vesicles were located near the ER membrane; however, some vesicles appeared to be free in the cytoplasm (Fig. 2D). Higher-magnification images revealed that the membrane-associated spherules were bound by a single lipid bilayer, with diameters that ranged between 50 and 70 nm (58.3 ± 6.9 nm [standard deviation {SD}]; n = 121) (Fig. 2E). The spherules were primarily adjacent to the outer ER membrane, and in many cases, the spherule membrane appeared to be continuous with the outer ER membrane, forming an invagination connected by a neck (Fig. 2F). Moreover, most of the spherules contained condensed or fibrillar materials, similar to those usually interpreted as nucleic acids (Fig. 2E and F) (38). Collectively, BBSV infection induced vesiculation of the ER membrane, resulting in the formation of ER-derived spherules, which possibly provide the sites for the assembly of VRCs.
FIG 2.
BBSV remodels the ER membranes. TEM was carried out to analyze the cytopathological changes during BBSV infection of N. benthamiana. (A) BBSV infection led to ER aggregation and large amounts of viral particles intermingled within the convoluted ER membrane. The arrow indicates the ER. (B to F) BBSV-induced vesiculation of the ER and VPs were often present in the dilated ER cisternae. (B) VPs were observed in the aggregates of branched ER cisternae (⭑), and a virus crystal (Vi) was in proximity to the VPs. Electron-dense globules (DG) were occasionally visible in the cytoplasm. The arrow indicates the ER. (C) Different numbers of spherules were enclosed in the VPs, and sometimes spherules appeared in the lumen of the perinuclear membrane (arrowheads). (D) Some spherules seem to be suspended in the cytoplasm (arrowheads). The enlarged image (top right corner) shows the region pointed to by the arrowhead. (E) Higher magnification of an area in panel D (dashed rectangle) shows the presence of fine fibrils in the VPs. (F) Membrane-associated spherules (further enlargement of the dashed rectangle in panel E) showing potential connections between the spherules and the outer ER membrane (arrowheads). Chl, chloroplast; Vi, virus particles or virus crystals; Mit, mitochondria; CW, cell wall; Nu, nucleus; Va, vacuole. Bars, 200 nm.
BBSV replication occurs in ER-derived spherules.
In order to demonstrate whether BBSV replicates on the ER-derived membrane spherules, we used immunogold electron microscopy to localize the site for BBSV replication. p23 and the dsRNA were selected as the targets for immunogold detection. Both of them are essential components of the VRC, and in particular, dsRNA is commonly considered a replication intermediate whose localization is usually thought to be the place where viral replication occurs (39). Initially, we prepared the embedded blocks following a commonly used method. Leaf tissues were first prefixed with 4% paraformaldehyde (PFA) and a low concentration of glutaraldehyde (GA), followed by postfixation without OsO4 to preserve the antigenicity of the target molecules. As shown in Fig. S1 in the supplemental material, in BBSV-infected leaf tissues, p23 was specifically localized on the ER, as evidenced by the gold particles present along the ER (see Fig. S1A in the supplemental material). In addition, large numbers of gold particles were present around the outside of vesicle-like structures (see Fig. S1B to D in the supplemental material). These vesicles, in different sizes, showed high similarity with the vesicle packets encompassing various numbers of spherules (Fig. 2; see Fig. S1E to F). Only a few gold particles were randomly distributed over the tissue sections derived from healthy control leaves (see Fig. S1G). Nevertheless, membranes were not well preserved due to the omission of OsO4 fixation, and thus, it was difficult to further explore the ER membrane-associated spherules.
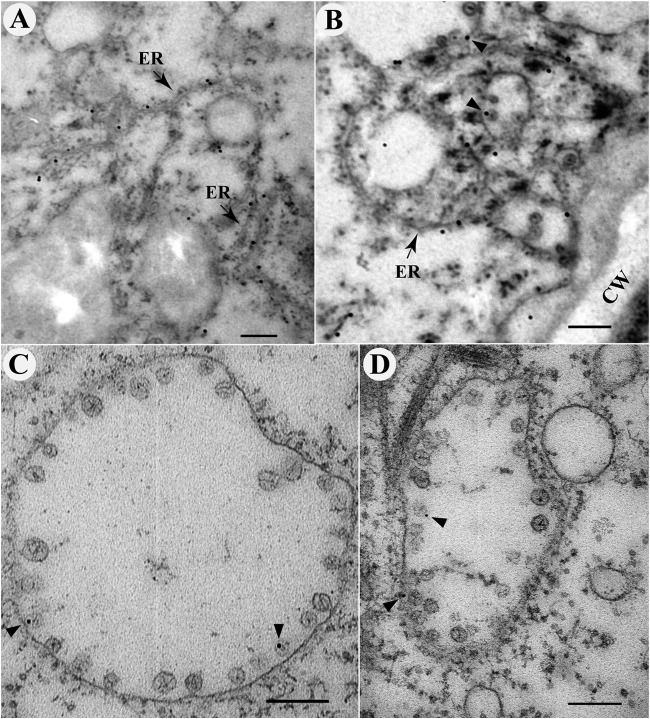
To overcome the above-mentioned limitations and to further validate that the ER-derived membrane spherules were the sites for BBSV replication, we employed an approach in which the antigenicity of target molecules was partially recovered while rendering the membrane ultrastructures recognizable (32). Before immunogold labeling, ultrathin sections from blocks prepared by a conventional chemical fixation procedure with 1% osmium tetroxide were etched in sodium metaperiodate to partially remove the osmium from the sections (40). As shown in Fig. 3A, gold particles representing the position of p23 could specifically colocalize with the ER (Fig. 3A). In addition, some gold particles were localized adjacent to the membrane-associated spherules (Fig. 3B). Statistical analysis revealed that more than 80% of p23-targeted gold particles were associated with the ER (82.0% ± 9.9%; [mean ± SD]; n = 111). For dsRNA detection, although the density of labeling is relatively low compared to that of p23, the association of gold particles with ER-derived membrane spherules could be clearly visualized (Fig. 3C and D). We also quantitated the percentage of VP-localized gold particles among the total dsRNA-targeted gold particles according to a previously described method (41). The results showed more than 75% of them were associated with the VPs (75.5% ± 10.7% [mean ± SD]; n = 102). For the mock-inoculated control, very few gold particles could be observed in the sections used for immunogold labeling with either p23 or dsRNA antibodies (see Fig. S2 in the supplemental material), confirming the specificity of these antibodies.
FIG 3.
BBSV replication occurs in ER membrane-associated spherules. (A and B) Immunogold labeling of the auxiliary replication protein p23 in BBSV-infected N. benthamiana cells. (A) p23 is localized to the ER. (B) p23 is localized to the aggregated ER or in close proximity to the spherules. The arrow indicates the ER. The arrowheads point to spherule-associated 10-nm gold particles. (C and D) Examples of immunogold labeling of dsRNA in membrane-associated spherules. The arrowheads indicate areas of spherules labeled with 10-nm gold particles. Note that the distance between the gold particles and the target epitopes may span up to 20 nm, as reported previously (44). CW, cell wall. Scale bar, 200 nm.
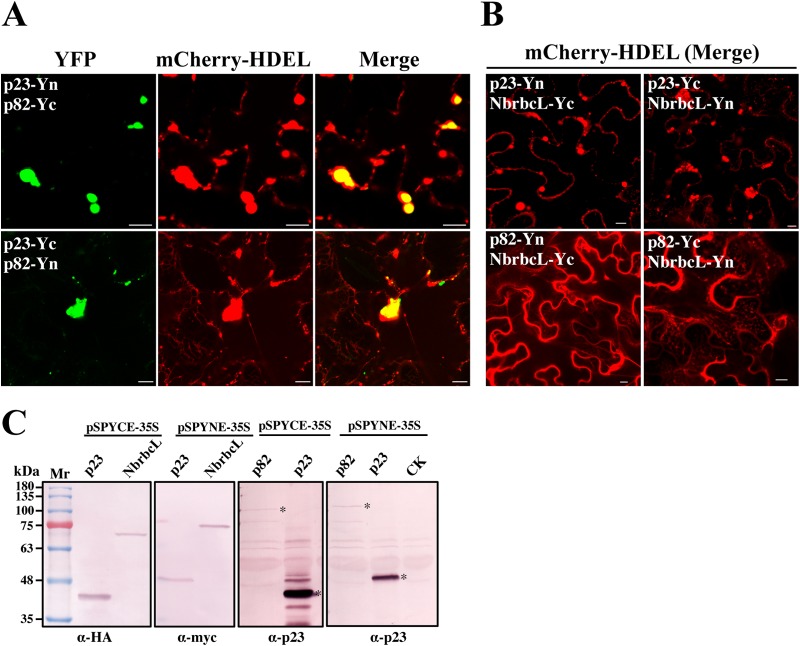
To further verify that establishment of BBSV VRCs was associated with the ER rather than membranes derived from other organelles, we performed the BiFC assays to localize the interaction between p23 and p82 (RdRp). The results showed that p23-p82 complexes colocalize well with the ER aggregates (Fig. 4A), reinforcing the notion that the BBSV replication compartment is ER associated. For the control, no fluorescence was detected, despite efficient expression of corresponding proteins in the agroinfiltrated leaves (Fig. 4B and C). Taking the data together, we concluded that the ER-associated membrane vesicles are the sites for VRC assembly and BBSV replication.
FIG 4.
Subcellular localization of p23-p82 complexes. (A) The BiFC assay indicates that p23 interacts with p82 and that the complexes colocalize with the aggregated ER. (B) No fluorescence is observed in leaves agroinfiltrated with control plasmid combinations. Bars = 10 μm. (C) Western blot analyses of protein expression in control agroinfiltrated leaves. The asterisks indicate the target bands.
ER rearrangement during BBSV infection was associated with expression of the auxiliary replication protein p23, an ER-localized membrane protein.
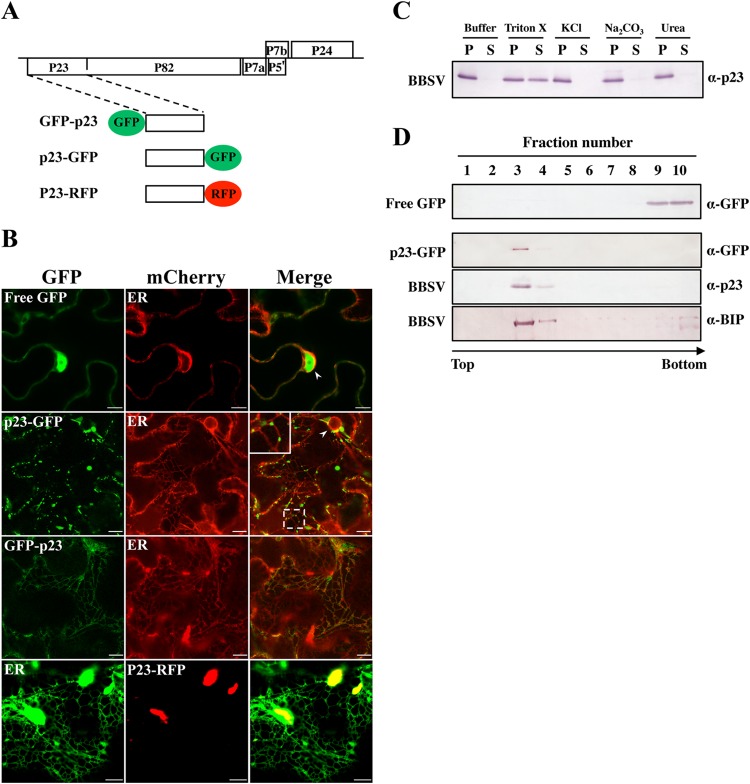
Previous reports showed that certain viral proteins were responsible for the formation of corresponding cytopathological structures (8, 9). Therefore, we attempted to evaluate which BBSV protein might play a key role in remodeling the ER structure. Our previous studies showed that there were no obvious changes in the ER membrane when viral movement proteins or coat protein was transiently expressed (42, 43). We noted that transient expression of p23 in N. benthamiana cells was always accompanied by ER aggregation (Fig. 4A and B), whereas the ER showed little change in comparison to the control upon expression of p82 (Fig. 4B), implying that p23 may play an important role in the establishment of BBSV VRCs. To further investigate the role of p23, p23 was fused with fluorescent proteins (Fig. 5A). Initially, the p23 ORF was fused to the C terminus of GFP to generate GFP-p23, which merged well with the ER, albeit with a relatively weak signal. However, GFP-p23 failed to induce ER aggregation and punctate structures (Fig. 5B). In contrast, when p23 was fused with the N terminus of GFP (p23-GFP), punctate structures colocalizing with the ER and the perinuclear membrane were readily detected (Fig. 5B). Western blotting revealed a higher expression level of p23-GFP than of GFP-p23 (see Fig. S3A in the supplemental material), agreeing well with a weaker signal of GFP-p23 under CLSM analysis. As expected, free GFP displayed typical nuclear-cytoplasmic distribution without entering the nucleolus, a pattern distinguished from that of GFP-p23 or p23-GFP (Fig. 5B).
FIG 5.
p23, an ER-localized membrane protein, induces the formation of ER aggregates in N. benthamiana cells. (A) Schematic diagram of BBSV genome organization and fluorescent fusion proteins of p23 used for subcellular localization. (B) CLSM analysis of the subcellular localization of p23 protein in agroinfiltrated leaves of N. benthamiana at 3 dpi. The arrowheads point to the fluorescent halo of perinuclear ER-mCherry. The region in the dashed rectangle in the merged image of p23-GFP is enlarged in the top left corner. Bars = 10 μm. (C) Extraction of p23 from the membrane fraction. The P30 precipitate of BBSV-infected leaf homogenate was treated with either an extraction solution (2% Triton X-100) or elution solutions (4 M urea, 0.1 M Na2CO3 [pH 10.5], or 1 M KCl), followed by Western blotting of the membrane (P) and the solubilized (S) fractions with anti-p23 (α-p23) antibody. (D) Membrane flotation assays revealed that p23 cofractionates with the ER marker BiP. The P30 precipitates derived from leaves agroinfiltrated with p23-GFP or infected by BBSV were subjected to differential centrifugation fractionation. As a control, free GFP in the S3 fraction was also subjected to a membrane flotation assay. The primary antibodies used for immunoblot analyses are indicated on the right.
To further verify that p23 was an ER-associated or integral membrane protein. Cell organelles extracted from BBSV-inoculated N. benthamiana leaves were separated into the following four fractions: S3, P3, S30, and P30 (30). The P30 fraction is enriched in intracellular membranes and membrane-associated proteins, as well as other large aggregates. The P30 membrane fraction was treated with either an extraction solution (detergent) or an elution solution (urea, high pH, or high salt), but only treatment with 2% Triton X-100 led to partial solubilization of p23, suggesting that p23 is an integral membrane protein (Fig. 5C). Furthermore, a membrane flotation assay was performed to separate membrane fractions (P30) from soluble fractions (Fig. 5D). The resuspended P30 fraction was loaded at the bottom of a gradient, and upon ultracentrifugation, the membrane floated to the top fractions while soluble fractions remained at the bottom. p23, either fused with GFP or from BBSV-infected leaves, could be detected only in the membrane fractions and was barely detectable in soluble fractions (Fig. 5D), a distribution pattern similar to that of the ER marker protein BiP (Fig. 5D). As a control, immunoblot analysis indicated that free GFP was distributed only in the bottom soluble fraction (Fig. 5D). These results, combined with the above-mentioned optical and electron microscopy analyses, indicated that BBSV p23 is an ER-localized membrane protein.
In addition, expression of RFP-fused p23 (p23-RFP) alone gives rise to ER aggregation in the epidermal cells, forming punctate structures similar to those caused by BBSV infection (Fig. 5B), and similar results were obtained when expressing p23 in the mesophyll cells (see Fig. S3B and C in the supplemental material). For the control leaves that were agroinoculated with empty vector (pGDG or pGD) (24), the ER structure was not affected and remained a regular network (Fig. 5B; see Fig. S3B in the supplemental material). These results indicate that expression of p23 alone in the N. benthamiana cells leads to the ER aggregation. To examine the effects of p23 expression on the ER membrane in detail, leaf tissues transiently expressing p23 were subjected to TEM analysis. The results showed that the ER was proliferated and convoluted (Fig. 6A and B), similar to what was displayed in BBSV-infected leaves (Fig. 2A). However, expression of p23 alone cannot form evident membrane-associated spherules similar to those in BBSV infection (Fig. 6C and D). In addition, expression of p23 often led to the formation of electron-dense materials in the cytoplasm, and the peripheries of these areas were linked with the ER (Fig. 6C and D), suggesting that these dense structures likely originated from ER membranes. With respect to the control leaf tissues, no changes in the ER structure were observed throughout the experiment (Fig. 6E). Our results indicated that expression of p23 alone alters the membrane structure of the ER in ways that resemble that in BBSV infection, although it is not sufficient to form the spherules seen during BBSV infection.
FIG 6.
p23 is at least partially responsible for ER rearrangement during BBSV infection. Agroinfiltrated leaf tissues were embedded, ultrasectioned, and observed under TEM. (A and B) Transient expression of p23 alone in N. benthamiana leaf tissues induces convolution and aggregation of the ER. (C) Electron-dense structures were also frequently observed in leaf tissues agroinfiltrated with p23. (D) Higher magnification of the area enclosed by the dashed line in panel C. The electron-dense areas are connected to the ER (arrowheads). (E) There was no obvious abnormality in the ER structure in leaves agroinfiltrated with empty vector. The expression of p23 in the agroinfiltrated leaves was confirmed by Western blotting (see Fig. S3C in the supplemental material, 35S-p23). Chl, chloroplast; Mit, mitochondria; CW, cell wall. Scale bars = 200 nm.
Three-dimensional architecture of the BBSV-induced replication factory.
Conventional two-dimensional (2D) transmission electron microscopy of random sections has inherent limitations in examining the stereo structures of the altered membrane morphology (44). To overcome such limitations, we further visualized the BBSV-induced membranous compartments using an electron tomography approach, a powerful tool to unravel the three-dimensional architecture of cellular structures and VRCs of various (+)RNA viruses (45, 46).
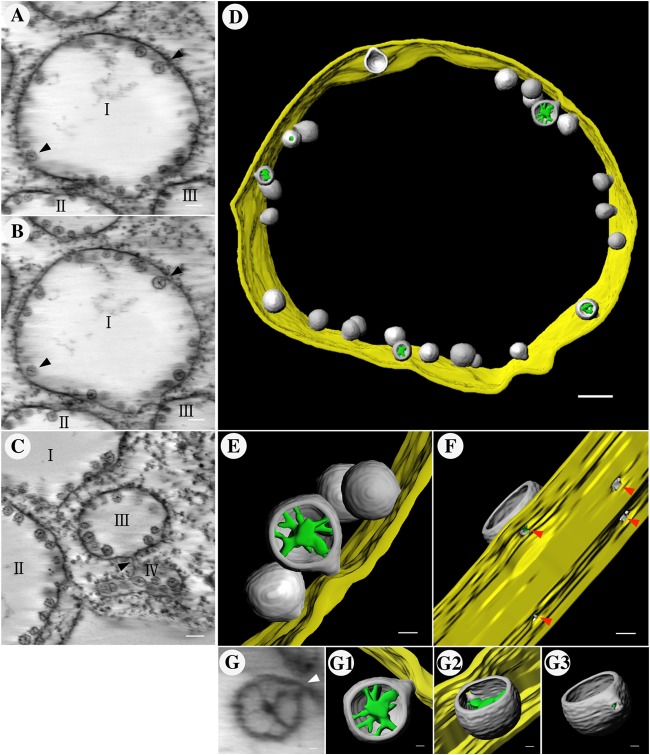
As shown in Fig. 7, some spherules seemed to be detached from the outer ER membranes and free in the cytoplasm in certain slices of the tomogram, while they became attached to the outer ER membrane in others (Fig. 7A and B; see Movie S1 in the supplemental material), similar to what was observed by Kopek et al. (44). To determine whether all or the majority of the spherules were connected to the outer ER membrane, 270 spherules from five independent electron tomographic experiments were examined, and 87% of them were found to be continuous with the outer ER membrane in successive slices of the tomograms (see Movies S1, S2, and S3 in the supplemental material). Another striking characteristic of the ultrastructural modifications induced by BBSV infection was that the vesicle packets were also connected with each other through a tubule-like structure with a diameter of 15 to 30 nm (Fig. 7C; see Movies S1 and S2). 3D surface rendering of the complete tomograms clearly showed that all the spherules lined a rounded ER outer membrane unit (Fig. 7D). Strikingly, in most cases, the spherules were connected to the ER outer membrane through neck-like structures (Fig. 7E and F; see Movie S4), indicating a connection between the interiors of spherules and the surrounding cytoplasm. These connections likely allow the import of cytoplasmic constituents (e.g., ribonucleotides) into the spherules and the export of newly synthesized viral RNAs to the cytoplasm (46). The mean diameter of the necked channel is 5.7 ± 0.6 nm (n = 36). In addition, the fibrillar materials in the spherules were also reconstructed into a 3D image. As shown in Fig. 7, these fibrillar materials were present in diverse states (Fig. 7G to G3; see Movie S5).
FIG 7.
Three-dimensional model of BBSV-induced, ER-derived spherules and vesicle packets by electron tomography. (A and B) A series of slices through a tomographic reconstruction show BBSV-induced ER membrane rearrangements (see Movie S1 in the supplemental material). The arrowheads indicate two spherules that appear to be free vesicles in one slice but connect to the outer membrane in the other slice (see Movie S1). I, II, and III represent different units of VPs. (C) Vesicle packets were also connected with each other through a tubule-like structure (arrowhead). (D) 3D models of the BBSV-modified ER and 3D surface reconstruction of the tomogram corresponding to the intact spherules in panels A and B. Gold, outer ER membrane; gray, BBSV-induced spherules; green, fibrillar materials inside the spherules. (E) Enlargement of the connections between the outer ER membrane and the spherules. (F) 90° counterclockwise rotation of panel E highlighting the pore-like openings (red arrowheads) (see Movie S4). (G to G3) 3D models of the fibrillar contents within the spherules. Panel G shows the electron density map from which the 3D surface-rendered models in panels G1 to G3 were derived. The arrowhead in panel G marks the channel connecting the interior of a representative spherule to the cytoplasm. Scale bars: 100 nm (A to D), 20 nm (E and F), and 10 nm (G to G3).
The 3D architecture of BBSV-induced membranous compartments was reconstructed using electron tomography, further confirming its function as the site for BBSV replication from the perspective of morphology.
DISCUSSION
In this study, we conducted a detailed analysis of the biogenesis and 3D architecture of the intracellular membrane structures induced by BBSV. Our results have implications for understanding the mechanisms underlying the replication of other plant and animal positive-strand RNA viruses.
BBSV remodels the ER membrane to form virus replication complexes.
The replication of all well-studied positive-strand RNA viruses is associated with intracellular membranes (8). Viruses always replicate their genomes in confined membranous compartments generated by extensive alteration of membrane structures (4, 5). Formation of large amorphous aggregates or small punctate spots is the typical change in the morphology of the ER during such viral infections. Some viruses, such as BMV, induce massive proliferation of the ER in cells, forming ER aggregates in the perinuclear region (47). In other cases, like those of Tobacco mosaic virus (TMV) and Red clover necrotic mosaic virus (RCNMV), the ER aggregates randomly disperse in the cytoplasm of the virus-infected cells (48, 49). In our study, BBSV could induce extensive rearrangement of the ER, forming punctate spots or patches in the cytoplasm (Fig. 1). Although the cytopathological effects caused by several viruses in the genus Necrovirus have been investigated, BBSV induced a cytopathological structure distinct from those of other viruses in the same genus. For example, TNV-A induces membranous vesicles with fibrillar material lining the tonoplast (33). BBSV elicits ER membrane dilations and invaginations, forming round outer membranes encompassing different numbers of vesicles in the cytoplasm, and small vesicles were occasionally observed in the perinuclear region (Fig. 2). TNV-W infection often produced rounded dilations of up to 400 nm in the ER cisternae (35), and Olive latent virus 1 (OLV-1) infections were characterized by the presence of tonoplast-associated vesicles and extensive vesiculation of the nuclear envelope (50). Moreover, fibrous structures, which were frequently observed in OLV-1 and TNV-W-infected cells (35, 50), were absent in BBSV-infected cells. In addition, convoluted membranes resulting from TNV-W, Leek white stripe virus, and BBSV infections differ from each other in terms of morphology (34, 35). Collectively, our results revealed a distinct pattern of cytopathology derived from BBSV infection, and the low sequence identity between BBSV and other viruses may be the reason for the diverse cytopathological structures.
To determine whether the membrane-associated spherules are the sites where BBSV generates progeny RNA, we examined the localization of dsRNA, a marker of viral replication complexes, in BBSV-infected cells using immunoelectron microscopy. The results clearly reveal that dsRNA is tightly associated with the spherules (Fig. 3), confirming their role in providing the sites for BBSV replication. Meanwhile, the auxiliary replication protein p23 was detected in both regular and convoluted ER membrane or showed an association with the spherules similar to that of dsRNA (Fig. 3). These results, together with the colocalization of p23-p82 complexes with the ER (Fig. 4), indicate that the membranous replication compartments of BBSV originate from the ER.
The expression of the auxiliary replication protein p23 contributes prominently to ER reorganization during BBSV infection.
One or several virus-encoded proteins usually play a major role in initiating membrane rearrangement and forming the scaffold for VRC assembly. Many animal virus-encoded proteins that contribute greatly to the formation of membranous replication sites have been identified (45). Sometimes these proteins alone are sufficient to induce cytopathological effects similar to that of cognate viral infection, such as poliovirus 2C or 2BC (51), hepatitis C virus 4B (52), and NSPs of SARS-CoV (10, 11), while in other cases, the expression of viral proteins in the absence of viral RNA synthesis could generate only intermediate membrane rearrangements, and the formation of parallel membrane structures requires full replication or viral RNA synthesis, as for flock house virus (FHV) and Semliki Forest virus (53, 54). For plant viruses, modification of the intracellular membrane structures was often associated with the expression of replication proteins. These morphological changes caused by the expression of replication proteins and viral infections often share great similarity. For example, BMV replication protein 1a is necessary and sufficient to induce perinuclear ER-bound spherules in yeast cells (13). In spite of intensive research on the small replicase proteins in the Tombusviridae (15, 55, 56), little is known about the characteristics of replication proteins in the genus Necrovirus; our results showed that expression of BBSV p23 alone induces ER proliferation and aggregation resembling that caused by BBSV infection (Fig. 1 and 5). Although expression of p23 alone is not sufficient to produce membranous spherules like those in BBSV infection (Fig. 2 and 6), these structures could be, at least, an important intermediate step for subsequent bona fide spherule formation. These results again emphasize the crucial roles of replication proteins in rearranging the intracellular membranes for VRC assembly and also hint at conserved strategies adopted by viruses during long-term evolution.
Topology and three-dimensional structure of BBSV-induced membrane spherules.
Since Kopek et al. generated the first 3D architecture of the FHV replication complex by using electron tomography (44), 3D structures of membranous replication factories have been characterized for multiple animal positive-strand RNA viruses within the Picornaviridae, Flaviviridae, Togaviridae, and Coronaviridae (45, 46, 57). However, 3D structural information on plant (+)RNA virus-infected cells is limited, although the replication mechanisms and virus-host interactions have been extensively studied for some plant viruses, such as BMV and TBSV (58, 59). To our knowledge, we present the first 3D model of ER membrane-associated replication complexes generated during replication of a plant (+)RNA virus in leaf tissues (Fig. 7). Despite marked differences in the hosts and genome organizations of viruses, the 3D structure of VRCs determined in our study shared some common features with those previously reported, such as convolution and invagination of the organelle membrane and the neck-like channels connecting the interiors of spherules to the cytoplasm (Fig. 7). The shared morphology of replication structures again suggested an evolutionarily conserved mechanism underlying the formation of VRCs across kingdoms (45, 46).
Besides these parallels, the BBSV-induced VRC also has its own characteristics. For example, unlike peroxisome- or mitochondrion-derived membrane vesicles, whose amounts were often dependent on the volume of those organelles (44, 60), the number of BBSV-induced membranous spherules in the vesicle packets varied greatly, with quantities ranging from one to several hundreds (Fig. 2C; see Movie S3 in the supplemental material). In addition, although the ER could be used as a platform for different viruses to synthesize their RNA, the morphologies of the replication sites differ very much between the virus groups. For instance, replication vesicles in West Nile virus (WNV)-infected cells are connected to each other directly via pores (61), whereas no such interconnections were observed in our study (Fig. 7D) or in FHV and other flaviviruses, including dengue virus and tick-borne encephalitis virus (62, 63). Moreover, the fibrillar materials in the spherules were also surface rendered (Fig. 7G; see Movie S5), which, to our knowledge, represents the first 3D visualization of thread-like structures in virus-infected plant cells. Additionally, the 3D surface-rendered model of the fibrillar contents within the BBSV-induced vesicles differs from that of WNV in its morphology and spatial distribution (61). The 3D model of BBSV-induced membranous vesicles broadens our knowledge about current VRCs and should provide valuable implications for further understanding the architecture and properties of the replication factories generated by other positive-strand RNA viruses.
In summary, we systemically investigated the biogenesis of the BBSV replication complex and present the first 3D model of an ER-derived membranous replication factory in virus-infected leaf cells. We propose that our findings will provide new insight into the replication of many other positive-strand RNA viruses and suggest new molecular targets for therapeutic intervention in RNA virus replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Arturo Diaz at Le Sierra University and Benjamin Kopek at Hope College for their critical reviews of the manuscript. We appreciate the helpful and stimulating discussions with Feng Qu (Ohio State University). We thank Jialin Yu and Chenggui Han at China Agricultural University for valuable discussions during the course of this work. We also thank Wei Ding and Jianxin Zhang (HPC-Service Station in the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences) for their help in 3D tomographic reconstruction and Yanli Zhang and Yingying Gao (Center for Biomedical Imaging Research of Tsinghua University) for their assistance with image processing.
This work was supported by the National Natural Science Foundation of China (31470253 and 31370176).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00401-15.
REFERENCES
- 1.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 3.Cottam E, Pierini R, Roberts R, Wileman T. 2009. Origins of membrane vesicles generated during replication of positive-strand RNA viruses. Future Virol 4:473–485. doi: 10.2217/fvl.09.26. [DOI] [Google Scholar]
- 4.Laliberté J-F, Sanfaçon H. 2010. Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48:69–91. doi: 10.1146/annurev-phyto-073009-114239. [DOI] [PubMed] [Google Scholar]
- 5.Verchot J. 2011. Wrapping membranes around plant virus infection. Curr Opin Virol 1:388–395. doi: 10.1016/j.coviro.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Netherton CL, Wileman T. 2011. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol 1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belov GA, van Kuppeveld FJM. 2012. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol 2:740–747. doi: 10.1016/j.coviro.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy PD, Barajas D, Pogany J. 2012. Host factors with regulatory roles in tombusvirus replication. Curr Opin Virol 2:691–698. doi: 10.1016/j.coviro.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Ferraro D, Zhao J, Hussain S, Shao J, Trujillo J, Netland J, Gallagher T, Perlman S. 2010. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J Virol 84:3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. 2013. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4:e00524-13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freundt EC, Yu L, Goldsmith CS, Welsh S, Cheng A, Yount B, Liu W, Frieman MB, Buchholz UJ, Screaton GR, Lippincott-Schwartz J, Zaki SR, Xu X-N, Baric RS, Subbarao K, Lenardo MJ. 2010. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J Virol 84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz M, Chen JB, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 14.Bamunusinghe D, Seo J-K, Rao ALN. 2011. Subcellular localization and rearrangement of endoplasmic reticulum by brome mosaic virus capsid protein. J Virol 85:2953–2963. doi: 10.1128/JVI.02020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilsner J, Oparka KJ. 2012. Missing links? The connection between replication and movement of plant RNA viruses. Curr Opin Virol 2:705–711. doi: 10.1016/j.coviro.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Cui X. 1988. An icosahedral virus found in sugar beet. J Xinjiang Shihezi Agric College 10:73–78. [Google Scholar]
- 18.Weiland JJ, Van Winkle D, Edwards MC, Larson RL, Shelver WL, Freeman TP, Liu HY. 2007. Characterization of a US isolate of beet black scorch virus. Phytopathology 97:1245–1254. doi: 10.1094/PHYTO-97-10-1245. [DOI] [PubMed] [Google Scholar]
- 19.Koenig R, Valizadeh J. 2008. Molecular and serological characterization of an Iranian isolate of Beet black scorch virus. Arch Virol 153:1397–1400. doi: 10.1007/s00705-008-0121-y. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Vazquez M, Ayala J, Garcia-Arenal F, Fraile A. 2009. Occurrence of Beet black scorch virus infecting sugar beet in Europe. Plant Dis 93:21–24. doi: 10.1094/PDIS-93-1-0021. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y, Cai Z, Ding Q, Li D, Han C, Yu J, Liu Y. 2002. The complete nucleotide sequence of Beet black scorch virus (BBSV), a new member of the genus Necrovirus. Arch Virol 147:2431–2435. doi: 10.1007/s00705-002-0896-1. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Yuan X, Cao Y, Xi D, Guo L, Han C, Li D, Zhai Y, Yu J. 2006. Analysis of the subgenomic RNAs and the small open reading frames of Beet black scorch virus. J Gen Virol 87:3077–3086. doi: 10.1099/vir.0.81928-0. [DOI] [PubMed] [Google Scholar]
- 24.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31:375–383. doi: 10.1046/j.1365-313X.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 25.Fan H, Sun H, Wang Y, Zhang Y, Wang X, Li D, Yu J, Han C. 2014. Deep sequencing-based transcriptome profiling reveals comprehensive insights into the responses of Nicotiana benthamiana to Beet necrotic yellow vein virus infections containing or lacking RNA4. PLoS One 9:e85284. doi: 10.1371/journal.pone.0085284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 27.Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J. 1978. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 28.Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto-Katou A, Katou S, Yoshioka H, Doke N, Kawakita K. 2006. Nitrate reductase is responsible for elicitin-induced nitric oxide production in Nicotiana benthamiana. Plant Cell Physiol 47:726–735. doi: 10.1093/pcp/pcj044. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Sanfaçon H. 2003. Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J Virol 77:523–534. doi: 10.1128/JVI.77.1.523-534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Li J, Pu H, Jin J, Zhang X, Chen M, Wang B, Han C, Yu J, Li D. 2010. Development of Tobacco necrosis virus A as a vector for efficient and stable expression of FMDV VP1 peptides. Plant Biotechnol J 8:506–523. doi: 10.1111/j.1467-7652.2010.00500.x. [DOI] [PubMed] [Google Scholar]
- 32.Bendayan M, Zollinger M. 1983. Ultrastructural localization of antigenic sites on osmium-fixed tissues applying the protein A-gold technique. J Histochem Cytochem 31:101–109. doi: 10.1177/31.1.6187796. [DOI] [PubMed] [Google Scholar]
- 33.Kassanis B, Vince DA, Woods RD. 1970. Light and electron microscopy of cells infected with tobacco necrosis and satellite viruses. J Gen Virol 7:143–151. doi: 10.1099/0022-1317-7-2-143. [DOI] [PubMed] [Google Scholar]
- 34.Lot H, Rubino L, Delecolle B, Jacquemond M, Turturo C, Russo M. 1996. Characterization, nucleotide sequence and genome organization of leek white stripe virus, a putative new species of the genus Necrovirus. Arch Virol 141:2375–2386. doi: 10.1007/BF01718638. [DOI] [PubMed] [Google Scholar]
- 35.Appiano A, Redolfi P. 1993. Ultrastructure and histochemistry of Phaseolus leaf tissues infected with an isolate of tobacco necrosis virus inducing localized wilting. Protoplasma 174:116–127. doi: 10.1007/BF01379043. [DOI] [Google Scholar]
- 36.Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz MT, Voinnet O, Baulcombe DC. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937–946. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC144041/pdf/100937.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatta T, Francki RIB. 1981. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J Gen Virol 53:343–346. doi: 10.1099/0022-1317-53-2-343. [DOI] [Google Scholar]
- 39.Kovalev N, Pogany J, Nagy PD. 2014. Template role of double-stranded RNA in tombusvirus replication. J Virol 88:5638–5651. doi: 10.1128/JVI.03842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skepper JN, Powell JM. 1 June 2008. Immunogold staining of epoxy resin sections for transmission electron microscopy (TEM). CSH Protoc 2008:pdb.prot5015. doi: 10.1101/pdb.prot5015. [DOI] [PubMed] [Google Scholar]
- 41.Rabouille C. 1999. Quantitative aspects of immunogold labeling in embedded and nonembedded sections. Methods Mol Biol 117:125–144. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Zhang X, Niu S, Han C, Yu J, Li D. 2011. Nuclear localization of Beet black scorch virus capsid protein and its interaction with importin alpha. Virus Res 155:307–315. doi: 10.1016/j.virusres.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Zhang Y, Xu J, Shi L, Fan H, Han C, Li D, Yu J. 2012. The R-rich motif of Beet black scorch virus P7a movement protein is important for the nuclear localization, nucleolar targeting and viral infectivity. Virus Res 167:207–218. doi: 10.1016/j.virusres.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero-Brey I, Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Restrepo-Hartwig MA, Ahlquist P. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol 70:8908–8916. http://jvi.asm.org/content/70/12/8908.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner KA, Sit TL, Callaway AS, Allen NS, Lommel SA. 2004. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320:276–290. doi: 10.1016/j.virol.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Reichel C, Beachy RN. 1998. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci U S A 95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castellano MA, Di Franco A, Martelli GP. 1987. Electron microscopy of two olive viruses in host tissues. J Submicrosc Cytol 19:495–508. [Google Scholar]
- 51.Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol 71:8962–8972. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC230196/pdf/718962.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J Virol 83:11378–11384. doi: 10.1128/JVI.01122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kallio K, Hellstrom K, Balistreri G, Spuul P, Jokitalo E, Ahola T. 2013. Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J Virol 87:9125–9134. doi: 10.1128/JVI.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopek BG, Settles EW, Friesen PD, Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J Virol 84:12492–12503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navarro B, Rubino L, Russo M. 2004. Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J Virol 78:4744–4752. doi: 10.1128/JVI.78.9.4744-4752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rochon DA, Singh B, Reade R, Theilmann J, Ghoshal K, Alam SB, Maghodia A. 2014. The p33 auxiliary replicase protein of Cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 452:133–142. doi: 10.1016/j.virol.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 57.Risco C, de Castro IF, Sanz-Sánchez L, Narayan K, Grandinetti G, Subramaniam S. 2014. Three-dimensional imaging of viral infections. Annu Rev Virol 1:453–473. doi: 10.1146/annurev-virology-031413-085351. [DOI] [PubMed] [Google Scholar]
- 58.Nagy PD, Richardson CD. 2012. Viral replication: in search of the perfect host. Curr Opin Virol 2:663–668. doi: 10.1016/j.coviro.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Diaz A, Wang X. 2014. Bromovirus-induced remodeling of host membranes during viral RNA replication. Curr Opin Virol 9:104–110. doi: 10.1016/j.coviro.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 60.Gómez-Aix C, García-García M, Aranda MA, Sánchez Pina MA. 2015. Melon necrotic spot virus replication occurs in association with altered mitochondria. Mol Plant Microbe Interact 28:387–397. doi: 10.1094/MPMI-09-14-0274-R. [DOI] [PubMed] [Google Scholar]
- 61.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miorin L, Romero-Brey I, Maiuri P, Hoppe S, Krijnse-Locker J, Bartenschlager R, Marcello A. 2013. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol 87:6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.