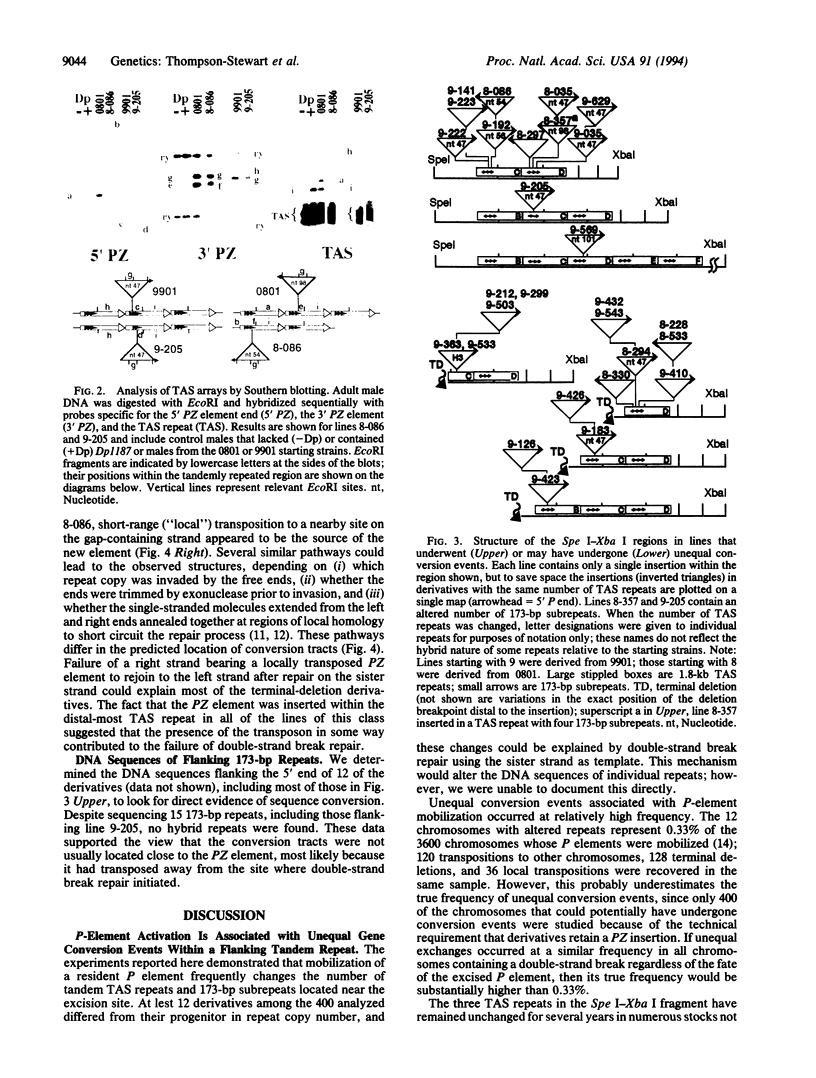
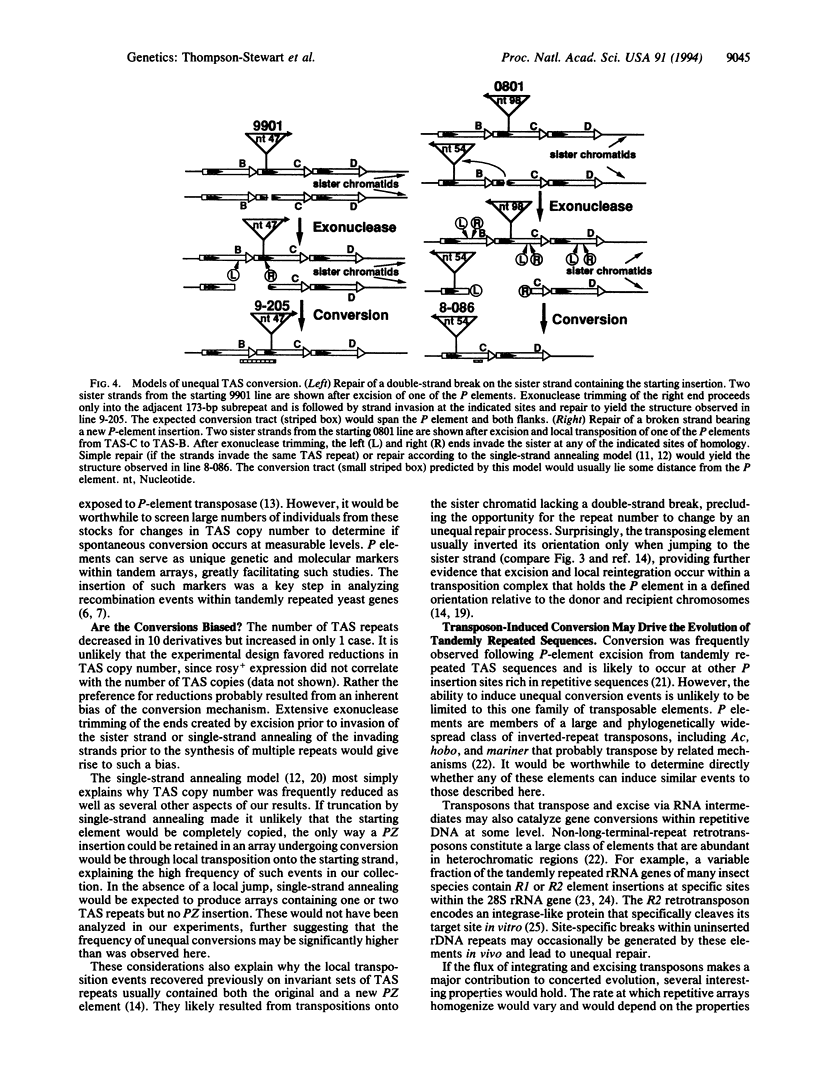
Abstract
Recombination and conversion have been proposed to drive the concerted evolution of tandemly repeated DNA sequences. However, specific correction events within the repeated genes of multicellular organisms have not been observed directly, so their nature has remained speculative. We investigated whether the excision of transposable P elements from tandemly repeated sequences would induce unequal gene conversion. Genetically marked elements located in a subtelomeric repeat were mobilized, and the structure of the region was analyzed in progeny. We observed that the number of repeats was frequently altered. Decreases were more common than increases, and this bias probably resulted from intrinsic mechanisms governing P element-induced double-strand break repair. Our results suggest that transposable elements play an important role in the evolution of repetitious DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ajioka J. W., Eanes W. F. The accumulation of P-elements on the tip of the X chromosome in populations of Drosophila melanogaster. Genet Res. 1989 Feb;53(1):1–6. doi: 10.1017/s0016672300027798. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Valgeirsdottir K., Lofsky A., Chin C., Ginther B., Levis R. W., Pardue M. L. HeT-A, a transposable element specifically involved in "healing" broken chromosome ends in Drosophila melanogaster. Mol Cell Biol. 1992 Sep;12(9):3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Sugimoto K. 5 S DNAs of Xenopus laevis and Xenopus mulleri: evolution of a gene family. J Mol Biol. 1973 Aug 15;78(3):397–415. doi: 10.1016/0022-2836(73)90464-6. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Cooley L., Kelley R., Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988 Mar 4;239(4844):1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Dover G. A. Evolution of genetic redundancy for advanced players. Curr Opin Genet Dev. 1993 Dec;3(6):902–910. doi: 10.1016/0959-437x(93)90012-e. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990 Aug 10;62(3):515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991 Sep 6;253(5024):1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Haber J. E. Exploring the pathways of homologous recombination. Curr Opin Cell Biol. 1992 Jun;4(3):401–412. doi: 10.1016/0955-0674(92)90005-w. [DOI] [PubMed] [Google Scholar]
- Jakubczak J. L., Xiong Y., Eickbush T. H. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990 Mar 5;212(1):37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Jakubczak J. L., Zenni M. K., Woodruff R. C., Eickbush T. H. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics. 1992 May;131(1):129–142. doi: 10.1093/genetics/131.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992 Nov;132(3):737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990 Oct 5;63(1):97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S., Saitou N., Ueda S. Concerted evolution of the primate immunoglobulin alpha-gene through gene conversion. J Biol Chem. 1992 Apr 15;267(11):7359–7367. [PubMed] [Google Scholar]
- Kuhner M. K., Peterson M. J. Genetic exchange in the evolution of the human MHC class II loci. Tissue Antigens. 1992 Apr;39(4):209–215. doi: 10.1111/j.1399-0039.1992.tb01937.x. [DOI] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Roy D., Mount S. M. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics. 1994 Mar;136(3):1001–1011. doi: 10.1093/genetics/136.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993 Dec 17;75(6):1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Pâques F., Wegnez M. Deletions and amplifications of tandemly arranged ribosomal 5S genes internal to a P element occur at a high rate in a dysgenic context. Genetics. 1993 Oct;135(2):469–476. doi: 10.1093/genetics/135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992 Feb;12(2):563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N., Spradling A. C. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993 Feb;133(2):347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988 Oct 21;55(2):235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Zhang P., Spradling A. C. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993 Feb;133(2):361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Spradling A. C. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3539–3543. doi: 10.1073/pnas.91.9.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]