ABSTRACT
Herpes simplex virus 2 (HSV-2) is a major global pathogen, infecting 16% of people 15 to 49 years old worldwide and causing recurrent genital ulcers. Little is known about viral factors contributing to virulence, and there are currently only two genomic sequences available. In this study, we determined nearly complete genomic sequences of six additional HSV-2 isolates, using Illumina MiSeq. We report that HSV-2 has a genomic overall mean distance of 0.2355%, which is less than that of HSV-1. There were approximately 100 amino-acid-encoding and indels per genome. Microsatellite mapping found a bias toward intergenic regions in the nonconserved microsatellites and a genic bias in all detected tandem repeats. Extensive recombination between the HSV-2 strains was also strongly implied. This was the first study to analyze multiple HSV-2 sequences, and the data will be valuable in future evolutionary, virulence, and structure-function studies.
IMPORTANCE HSV-2 is a significant worldwide pathogen, causing recurrent genital ulcers. Here we present six nearly complete HSV-2 genomic sequences, and, with the addition of two previously sequenced strains, for the first time genomic, phylogenetic, and recombination analysis was performed on multiple HSV-2 genomes. Our results show that microsatellite mapping found a bias toward intergenic regions in the nonconserved microsatellites and a genic bias in all detected tandem repeats and confirm that chimpanzee herpesvirus 1 (ChHV-1) is a separate species and that each of the HSV-2 strains is a genomic mosaic.
INTRODUCTION
Herpes simplex virus 2 (HSV-2) causes recurrent genital ulcers and is a significant global pathogen, with an estimated 536 million people between the ages of 15 and 49 years infected worldwide (1). Patients infected with HSV-2 experience a number of reactivation episodes following exposure, with a decreasing frequency of reactivations over time (2). Significantly, asymptomatic viral shedding occurs during up to 17% of days 10 years postinfection (3). Furthermore, HSV-2 seropositivity triples the risk of HIV coinfection (4). In developed countries, however, herpes simplex virus 1 (HSV-1) has been undergoing a changing epidemiology, with an increase in the number of genital infections; thus, HSV-2 is no longer the exclusive genital herpesvirus (5–7). As with HSV-1, there is no vaccine currently available. Sequencing of additional HSV-2 strains may aid vaccine development and therapeutic approaches by assessing protein variation and identifying possible targets.
HSV-2 infects the genital epithelial mucosa, and, unlike HSV-1, which establishes latency in the trigeminal ganglia, HSV-2 establishes latency in the lumbar-sacral ganglia (8). There is a paucity of information regarding factors influencing virulence, including the significance of the genetic makeup of individual isolates, which requires more investigation. Most studies involving HSV-2 have focused on seroprevalence, vaccine development, antiviral development, and characterization of drug resistance (4, 9–16).
HSV-2 is a large, double-stranded DNA (dsDNA) virus in the Alphaherpesvirinae subfamily. Phylogenetic analysis has shown that HSV-2 is more closely related to chimpanzee herpesvirus 1 (ChHV-1) than to HSV-1 (17, 18). Until recently, only one genomic HSV-2 sequence of a clinical isolate had been available, but another was made available recently (19, 20). Little is known about the sequence diversity in circulating strains; however, it has been estimated to be lower than that in strains of HSV-1 (21, 22). The HSV-2 genome is approximately 154,700 bp long and is divided into unique long (UL) and unique short (US) segments, which are flanked by inverted repeats. Variability in genome length is due to variable-nucleotide tandem repeats (VNTRs), which include microsatellites and tandem repeats.
We determined the sequence of six nearly complete HSV-2 genomes and included the two previously sequenced strains to begin to investigate HSV-2 genomic variability, phylogenetics, and recombination. Although the data set is small, genomic analysis of the strains suggested that HSV-2 has reduced nucleotide diversity compared to HSV-1. We also report that nonconserved microsatellites are biased toward intergenic areas, and tandem repeats were biased toward genic areas in the UL and US coding regions similarly to HSV-1. There were approximately 100 amino-acid-encoding single nucleotide polymorphisms (SNPs) and indels in each strain compared to the HG52 reference strain. Recombination analysis confirmed that each of the HSV-2 strains is a genomic mosaic. This is the first study to sequence and analyze multiple HSV-2 strains and is an important step in expanding the HSV-2 genomic database.
MATERIALS AND METHODS
Cells.
Vero cells were used to generate viral DNA stocks and were cultured in Dulbecco's modified Eagle's medium (DMEM) with 5% serum and antibiotics as described previously (23). For viral DNA isolation, the infections were carried out in DMEM with 2% serum and antibiotics.
Viruses.
HSV-2 strains 333 and HG52 are high-passage-number laboratory strains, while SD90e, 1192, CtSF, CtSF-R, COH 3818, and GSC-56 are low-passage-number clinical isolates. Strain 333 was isolated from a genital lesion in Texas, USA (24), and strain 1192 was isolated from a genital lesion in Wisconsin, USA, and was passaged three times prior to sequencing. The CtSF, CtSF-R, COH 3818, and GSC-56 strains were isolated from unknown locations in the United States. The genomic sequences for HSV-2 HG52 and SD90e (20) were obtained from GenBank, and these strains were from Scotland and South Africa, respectively. The genomes and associated accession numbers are shown in Table 1.
TABLE 1.
Genomes and accession numbers
| Virus | Strain | Host | Country of origin | GenBank accession no. |
|---|---|---|---|---|
| HSV-1 | 17 | Human | Scotland, United Kingdom | JN555585.1 |
| ChHV-1 | 105640 | Chimpanzee | United States | NC_023677.1 |
| HSV-2 | HG52 | Human | Scotland, United Kingdom | JN561323.2 |
| HSV-2 | SD90e | Human | South Africa | KF781518 |
| HSV-2 | 333 | Human | Texas, United States | KP192856 |
| HSV-2 | 1192 | Human | Wisconsin, United States | KP334095 |
| HSV-2 | COH 3818 | Human | United States | KP334096 |
| HSV-2 | CtSF | Human | United States | KP334097 |
| HSV-2 | CtSF-R | Human | United States | KP334093 |
| HSV-2 | GSC-56 | Human | United States | KP334094 |
Viral DNA preparation.
The viral DNA was prepared using a modification of our previous protocol (25). Briefly, a confluent TC100 plate of Vero cells was infected with virus and 4 ml of DMEM with 2% serum and then harvested when the cells reached a 100% cytopathic effect (CPE). The cells were scraped, and the whole preparation was subjected to three freeze-thaw cycles. The volume of the whole preparation was brought to 5 ml with DMEM with 2% serum. The viral preparation was then distributed equally onto five confluent TC100 plates of Vero cells. When the cells reached 100% CPE, the five plates were harvested, combined, and centrifuged at 600 × g for 10 min. The supernatant was removed, and then 5 ml of the supernatant was placed back into the tube containing the pellet and subjected to vortex mixing. The vortexed pellets were then subjected to three freeze-thaw cycles. The samples were then centrifuged at 600 × g for 10 min. The supernatants were combined and centrifuged at 600 × g for 20 min. The supernatant was layered onto a 36% sucrose cushion in phosphate-buffered saline (PBS) and centrifuged for 80 min at 24,000 × g. The resulting pellet was resuspended in 5 ml of PBS and layered onto another 36% sucrose cushion in PBS and centrifuged 80 min at 26,200 × g. The viral pellet was then resuspended in 3 ml of TE buffer (10 mM Tris [pH 7.4], 1 mM EDTA) with 0.15 M sodium acetate and 50 μg/ml RNase A and then incubated 30 min at 37°C. Proteinase K and SDS (50 μg/ml and 0.1%, respectively) were added, and the solution was incubated for 30 min at 37°C. The viral DNA was purified by phenol-chloroform extraction and ethanol precipitation, resuspended in deionized water, and stored at −20°C.
Construction and sequencing of Illumina libraries.
High-quality genomic DNA (500 ng) was submitted to the University of Wisconsin—Madison DNA Sequencing Facility for paired-end library preparation. Each library was generated using an Illumina TruSeq Nano LT sample preparation kit (Illumina Inc., San Diego, CA, USA) per the manufacturer's specifications, targeting 550-bp fragments. The quality and quantity of the DNA were assessed using an Agilent DNA High Sensitivity series chip assay (Agilent Technologies, Santa Clara, CA, USA) and a Qubit dsDNA kit (Life Technologies, Grand Island, NY, USA), respectively, and libraries were standardized to 2 nM. Paired-end sequencing (250 bp) was performed on an Illumina MiSeq system using version 2 kits and returning an average of 250,000 unique reads (125 Mb) per library. FASTQ reports were created using CASAVA 1.8.2.
Genomic assembly.
The paired-end sequencing reads from the HSV-2 333, 1192, CtSF, CtSF-R, COH 3818, and GSC-56 strains were aligned to reference HSV-2 strain HG52 (JN561323.2) using CLC-Bio Genomic Workbench (version 6.0). De novo assembly was also performed; however, we found it had no significant advantage over the reference assembly (data not shown). A consensus sequence generated from the aligned reads was extracted with a minimum threshold of 4× coverage. Regions with less than 4× coverage were represented with “N′s.” Gaps in the sequence were filled with “N′s” rather than being filled with a proxy sequence, as has been done with HSV-1 (26, 27). Following annotation, the sequences were submitted to GenBank.
Genomic sequence alignments.
Two genomic alignments of the eight HSV-2 strains were generated, one with the eight HSV-2 strains alone and one using HSV-1 and ChHV-1 as outgroups. All alignments were produced with ClustalW (28) from the Mega 6 software package (29). The two alignments are available as FASTA files at sites. ophth. wisc. edu/brandt/.
DNA polymorphism and DNA variability analysis.
To detect DNA polymorphisms across the eight HSV-2 genomes, DNAsp (30) was used to analyze the genomic HSV-2 alignment. Alignment gaps were excluded from the analysis with a sliding window of 100 bp and a step size of 25 nucleotides. To calculate nucleotide distance in the HSV-2 genomes, the HSV-2 alignment was first subjected to DNA substitution model testing using Mega 6, with the alignment gaps removed. The Tamura-Nei substitution model resulted in the lowest Bayesian inference criterion (BIC) score and was used to calculate overall mean and pairwise distances (Mega 6).
Microsatellite and tandem repeat detection.
To detect microsatellites and tandem repeats, HSV-2 strain HG52 was first analyzed as a baseline with Msatcommander (31, 32) and Tandem Repeat Finder (TRF) (v. 4.07b) (33). Msatcommander was configured to detect mononucleotide-to-hexanucleotide repeats, with a mononucleotide length of 10, a dinucleotide repeat length of 6, and the remaining parameters using a repeat length of 4. Tandem Repeat Finder (TRF) was used with the following parameters: match, 2; mismatch, 5; delta, 5; PM, 80; minscore, 40; maxperiod, 500. The remaining HSV-2 strains were also scanned with Msatcommander and TRF.
Phylogenetic analysis.
For genomic phylogenetic analysis, the HSV-1 strain 17 and ChHV-1 genome sequences were retrieved from GenBank and aligned to the eight HSV-2 genomic sequences. Nucleotide substitution modeling was performed on the data set using Mega 6, and this resulted in the GTR+G model having the lowest BIC score. A maximum likelihood tree was generated using GTR+G, with five gamma categories and 1,000 bootstrap replicates, and using Mega 6. Gaps were excluded from the alignment, resulting in a total of 135,376 positions in the data set.
Recombination analysis.
Two different methods were used to detect recombination within the eight HSV-2 strains. First, Splitstree (v. 4.13.1) (34) was used to generate a neighbor network based the whole-genome alignment of HSV-2, which included the HSV-1 and ChHV-1 genomes as outgroups. The Kimura 2-parameter substitution model (35) was used, with alignment gaps excluded from the data set. Next, individual Bootscan plots for each of the strains were produced, where one strain was used as the reference genome and scanned against the remaining genomes. The plots were created using whole-genome HSV-2 alignment (minus outgroups) and the RDP4 package (36), with a sliding window of 1,500 nucleotides, a step size of 750 bp, and the Kimura 2-parameter substitution model.
Nucleotide sequence accession numbers.
The genomes and associated accession numbers are shown in Table 1.
RESULTS AND DISCUSSION
Sequencing and genomic assembly.
The total number of sequence reads for each viral isolate ranged from 321,344 for strain 1192 to 494,046 for strain CtSF-R, with average read lengths of 216 and 219 bp for strains 1192 and COH 3818, respectively (Table 2). The average genomic sequence coverage ranged from 140× for strain 333 to 203× for strain CtSF. The coverage distribution was similar to that seen in HSV-1 (37), with lower coverage in the large repeat regions and higher coverage in the UL and US coding regions (Fig. 1). The areas of lower coverage generally corresponded to high GC content (Fig. 1). The sequence gaps were cataloged and are available for download at sites. ophth. wisc. edu/brandt/.
TABLE 2.
MiSeq sequencing statistics of HSV-2 isolates
| Viral isolate | No. of reads | No. of reads mapped to reference | Avg mapped read length (bp) | Avg coverage | Gapped genome length (bp) |
|---|---|---|---|---|---|
| 333 | 475,138 | 99,906 | 217 | 140× | 154,764 |
| 1192 | 321,344 | 109,545 | 216 | 152× | 154,668 |
| CtSF | 452,640 | 144,919 | 218 | 203× | 154,709 |
| CtSF-R | 494,046 | 140,481 | 216 | 190× | 154,705 |
| COH 3818 | 416,512 | 128,195 | 219 | 181× | 154,739 |
| GSC-56 | 407,234 | 110,159 | 219 | 156× | 154,703 |
FIG 1.

DNA polymorphism, microsatellite, and tandem repeat analysis of HSV-2. A schematic of the HSV-2 genome has been placed at the top of the figure, with a GC percentage plot (purple line) below. DNA polymorphisms in the eight-strain HSV-2 data set are plotted along the genome (green line), with genome coordinates underneath. Low-sequencing-coverage areas are denoted by red blocks. Microsatellite and tandem repeat sites are plotted underneath. These include conserved and nonconserved short repeats in the UL and US regions. Short sequence repeats based on the HG52 reference genome are also plotted in the inverted repeat regions.
Nucleotide polymorphism analysis.
To identify nucleotide differences between the HSV-2 genomes, we first mapped DNA polymorphisms across the alignment to distinguish whether the polymorphisms were evenly distributed or whether there were polymorphism hot spots. The subsequent analysis showed that the large terminal and internal inverted repeat regions contain a large amount of variability; however, this is mainly artifactual, due to lower coverage in these regions (Fig. 1). Additionally, the analysis showed a generally even distribution across the UL and US coding regions (Fig. 1). Four polymorphism spikes were detected in the UL coding region at approximately 72,000 bp, 89,000 bp, 99,000 bp, and 106,000 bp. The polymorphism spikes at approximately 72,000 bp and 106,000 bp are artifacts due to low sequencing coverage (Fig. 1). The polymorphism spike at 89,450 bp is a highly covered region which corresponds to an approximately 200-bp region in the reference HG52 strain that is variable compared to the remaining seven sequences. The cause of the nucleotide variability in this region of the HG52 strain is unclear. A spike occurring at an approximately 99,000-bp spike was also highly covered and appears to represent a small area of DNA variability among the HSV-2 strains. The genomic alignment shows an area of variability between bases 99452 and 99623 of the whole-genome alignment. This area corresponds to mainly a noncoding sequence between the UL45 and UL46 genes. The significance of this finding is not clear. Further studies may determine whether this 159-bp region is an area of low evolutionary constraint or an important variable regulatory region.
Short sequence repeat characterization.
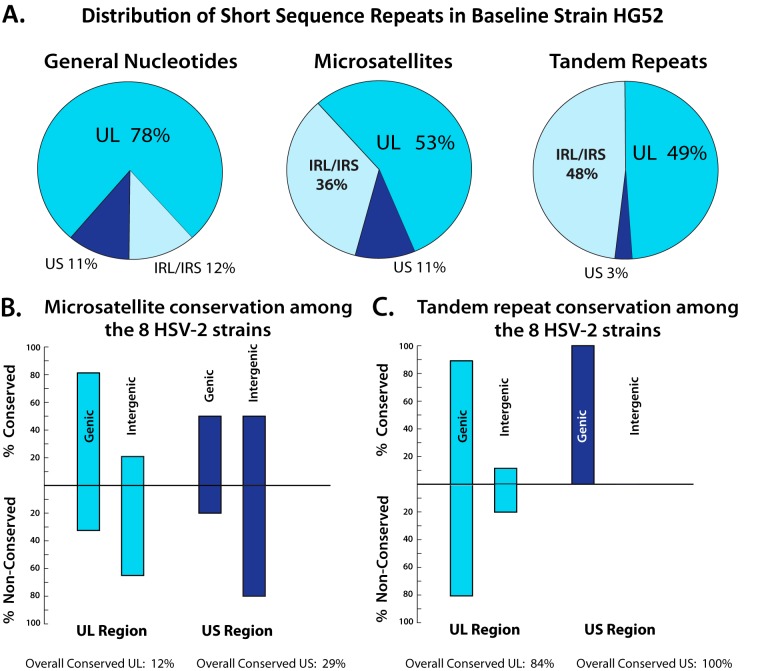
Two categories of short sequence repeats (SSRs), microsatellites and tandem repeats, were next examined. The HG52 reference sequence was scanned for microsatellites and tandem repeats and used as a baseline. The baseline distribution of the microsatellite and tandem repeats is plotted in Fig. 1 and categorized in Fig. 2A. The categorization of the repeats excluded the terminal repeat long/terminal repeat short (TRL/TRS) regions to prevent over counting. The baseline distribution analysis showed that 53% of the microsatellites were located in the UL coding region, 36% percent in the internal repeat long/internal repeat short (IRL/IRS) region, and 6% in the US coding region. The baseline tandem repeat distribution revealed that 49% were in the UL coding region, 48% were in the IRL/IRS repeats, and 3% were in the US coding region.
FIG 2.
Distribution of microsatellites and tandem repeats in HSV-2. (A) Distribution of microsatellite and tandem repeats in the reference, baseline HSV-2 HG52 strain. (B) Microsatellite conservation distribution in the full eight-strain HSV-2 data set. (C) Tandem repeat conservation distribution in the full eight-strain HSV-2 data set.
The remaining seven HSV-2 strains were then analyzed for microsatellites and tandem repeats; however, the terminal and internal inverted repeat regions were excluded from the analysis because of low coverage sequencing limitations in the terminal and the internal inverted repeat segments. The genome coordinates of the SSRs detected by the analysis were normalized to the HG52 baseline genome and plotted (Fig. 1). The microsatellites and tandem repeats were then classified into genic and intergenic as well as conserved and nonconserved categories (Fig. 2B and C). Microsatellite detection found that, overall, only 12% of the sites were conserved in the UL region and 29% were conserved in the US region (Fig. 2B). The analysis detected a genic bias of 80% in the conserved microsatellite sites in the UL region. However, the nonconserved microsatellite sites displayed an intergenic bias in both the UL and US regions, with intergenic bias values of 68% and 80%, respectively (Fig. 2B).
The tandem repeat detection determined that, overall, 84% of sites were conserved in the UL region and 100% were conserved in the US region (Fig. 2C). The tandem repeat sites showed a bias toward genic locations in both conserved and nonconserved sites in both the UL and US coding regions (Fig. 2C). The conserved tandem repeat sites were biased 89% toward genic sites in the UL region and 100% in the US region. The nonconserved tandem repeat sites were biased 80% toward genic regions in the UL region, with no nonconserved sites in the US region. The high percentage of conserved tandem repeat sites within genic areas is unsurprising, given that the majority of genic sequences encode proteins. The full lists of microsatellite and tandem repeats are available for download at the laboratory website sites. ophth. wisc. edu/brandt/.
The distribution of the short sequence repeats detected in this study was broadly similar to the distribution of those detected in HSV-1 (27). Microsatellite and tandem repeats have been used in the past to characterize HSV-1 and HSV-2 strains (38–40), and genomic analysis of the multiple strains in this study may allow precise short sequence repeat targeted analysis for the rapid characterization of viral strains.
Genomic distances.
To investigate HSV-2 genetic diversity, the genomic pairwise distances for the sequence data set were calculated (data not shown). The greatest calculated genetic distance was between strains HG52 and 333 (0.361%), while the smallest distance was between COH 3818 and CtSF-R (0.061%) (data not shown). The genetic distances were low, with the overall distance calculated at 0.2355%, compared to 0.8% with HSV-1 (41). The lower observed overall distance reported here corresponds to previous reports of lower genetic diversity in HSV-2 than in HSV-1 (21, 22). It is possible, however, that the low genetic diversity observed in this study was simply the result of the use of a low sample size. Only as more genomic sequences become available will it be feasible to perform a direct comparison of interstrain diversity between HSV-1 and HSV-2.
SNP and indel variant analysis.
SNP and indel variant analysis was performed on the six HSV-2 strains sequenced in this study to create a catalog of SNPs and indels. The HG52 genome was used as a reference genome, with a threshold of 4× coverage and 30% variable base frequencies. The results are shown in Table S1 in the supplemental material. The variant detection found a range of from 250 SNPs in strain 1192 to 279 SNPs in strain CtSF-R. Additionally, the analysis found a range of from 58 indels in strain GSC-56 to 69 indels in strain COH 3818. Of the SNP and indels detected, 94 protein-coding SNPs and indels were found in GSC-56 and up to 109 protein-coding SNPs and indels were found in strain CtSF-R. The variant analysis detected several complex SNPs and indels (Table S2 in the supplemental material) in each of the six strains, with a minimum of four in strain COH 3818 and a maximum of nine in strain 333. A total of three frameshift mutations were found; however, all three were the result of complex deletions. Two were found in strain 333 (UL32 [Ala271fs] and UL47 [Thr586fs]) and one in strain GSC-56 (UL32 [Ala271fs]).
Phylogenetic and recombinational analysis.
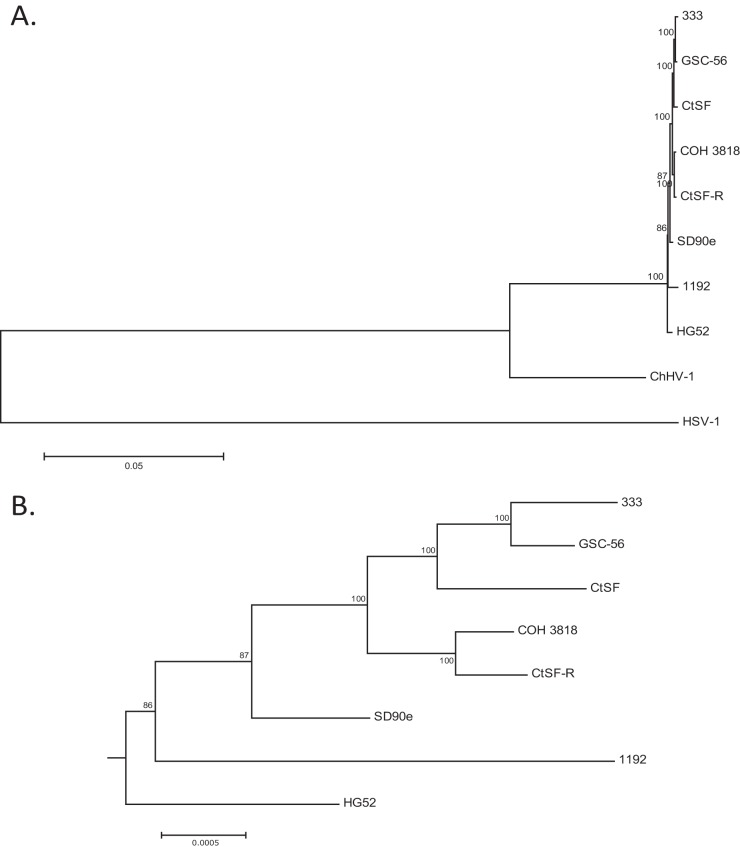
To investigate the phylogenetic structure of the eight HSV-2 strains, a maximum-likelihood-based tree which included HSV-1 and ChHV-1 as outgroups was generated (Fig. 3A), with an expansion of the HSV-2-specific node (Fig. 3B). The data suggest that, while they are closely related, HSV-2 and ChHV-1 are separate species, with a mean distance of approximately 8% (data not shown) between the ChHV-1 and the HSV-2 strains collectively. Both the phylogenetic tree data (Fig. 3B) and neighbor network data (Fig. 4A) show the presence of some clustering structures (e.g., clusters of 333, CtSF, and GSC-56 and of COH 3818 and CtSF-R), while the remaining strains are phylogenetically distinct.
FIG 3.
Maximum likelihood tree of eight genomic HSV-2 sequences. (A) Maximum likelihood tree using the GTR+G substitution model with five gamma categories, 1,000 bootstrap replicates, and complete deletion of alignment gaps. HSV-1 and ChHV-1 were used as outgroups. (B) Expansion of the HSV-2-specific node.
FIG 4.
Phylogenetic network, and Bootscans, based on genomic alignments suggesting recombination events in HSV-2. (A) Whole-genome phylogenetic network of the eight HSV-2 strains, with HSV-1 and ChHV-1 as outgroups. Only the HSV-2-specific node is shown. Alignment gaps were deleted, with distances corrected with the Kimura 2-parameter distance substitution model. (B) Three representative whole-genome-based Bootscans (minus outgroups) using HSV-2 strains HG52, 333, and 1192 as reference sequences, scanned against the remaining HSV-2 genomic sequences. A 1,500-bp sliding window with a 750-bp step size and the Kimura 2-parameter nucleotide substitution model were used. The dotted line within the plot indicates the 70% bootstrap cutoff.
The use of small numbers of genes has been effective for examining the deeper evolutionary roots of herpesviruses and has shown that herpesviruses generally codiverge with their hosts (42). However, using small gene sets to determine interstrain phylogenies is more difficult due to recombination. For example, in HSV-1, using the gG, gE, and gI genes, three genotypes have been described, with each gene tree showing various topologies, implying recombination (43). Subsequent research with HSV-1 whole genomes suggested the presence of several geography-based, phylogenetic interstrain groupings (27, 41); however, phylogenetic trees based on 5-kb segments of the genome resulted in various topologies and did not reflect the geographic origins of the samples (41). Thus, it is critical to use genomes that are as complete as possible in HSV-1 phylogenetic analysis. A previous study examining HSV-2 gG, gE, and gI genes from Norway, Sweden, and Tanzania reported extensive recombination (21). The study also concluded that there were two main genotypes, genotypes A and B. It is, then, highly likely that this is an underestimate of phylogenetic groupings and that sequencing and analysis of additional HSV-2 strains will also result in additional geography-based, phylogenetic interstrain groupings.
To investigate genomic recombination among the eight HSV-2 strains, two methods were used. First, a neighbor network was constructed using a whole-genome sequence alignment which included the eight HSV-2 strains (plus outgroups). The HSV-2-specific node is shown in Fig. 4A and highlights the phylogenetic dissonance in the data set, implying recombination. Finally, a whole-genome sequence alignment, which included the eight HSV-2 strains (minus outgroups) was used to perform Bootscan recombination analysis. Bootscan analysis is used to detect inconsistent phylogenetic signals across a sequence alignment, and the results imply recombination. Three representative scans are shown in Fig. 4B, with the remaining scans found in Fig. S1 in the supplemental material. The variable patterns observed in comparing the individual Bootscan plots and the phylogenetic dissonance results observed in the neighbor network suggest that each HSV-2 strain is a genetic mosaic, as expected. Additionally, the Bootscan recombination analysis suggests that recombination appears to occur randomly in the genome, with no obvious recombination hot spots or cold spots detected (Fig. 4B; see also Fig. S1).
Summary.
In conclusion, we determined the nearly complete genomic sequences of six HSV-2 strains using Illumina MiSeq SNP/indel analysis of the six HSV-2 strains and detected approximately 100 protein-coding SNPs and indels per genome. Genomic bioinformatic analysis of the 6 sequenced strains along with the genomes of two HSV-2 strains obtained from GenBank found a small amount (0.235%) of interstrain diversity. Microsatellite mapping of the full eight-strain data set found a bias toward intergenic regions in the nonconserved microsatellites and a genic bias in all detected tandem repeats. Recombination analysis further suggested that each HSV-2 strain is a genomic mosaic, as expected. Additional genomes will need to be sequenced to determine the clade structure and determine whether geographic clustering is present as has been shown for HSV-1. This will require an expansion of our strain collection. For the first time, the genomes of multiple HSV-2 isolates were analyzed, and these data will be highly useful in future evolutionary, virulence, and structure-function studies.
Supplementary Material
ACKNOWLEDGMENTS
We express appreciation to the University of Wisconsin—Madison Next-Gen Sequencing Core and to Kevin Hunt for their assistance and to David Knipe, Harvard University, for providing HSV-2 strains HG52 and SD90e.
This study was supported by a grant from the NIH (R01EY023292), a Core Grant for Vision Research (P30EY016665), and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness, Inc.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00416-15.
REFERENCES
- 1.Looker KJ, Garnett GP, Schmid GP. 2008. An estimate of the global prevalence and incidence of herpes simplex virus type 2 infection. Bull World Health Organ 86:805–812. doi: 10.2471/BLT.07.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedetti JK, Zeh J, Corey L. 1999. Clinical reactivation of genital herpes simplex virus infection decreases in frequency over time. Ann Intern Med 131:14–20. doi: 10.7326/0003-4819-131-1-199907060-00004. [DOI] [PubMed] [Google Scholar]
- 3.Phipps W, Saracino M, Magaret A, Selke S, Remington M, Huang ML, Warren T, Casper C, Corey L, Wald A. 2011. Persistent genital herpes simplex virus-2 shedding years following the first clinical episode. J Infect Dis 203:180–187. doi: 10.1093/infdis/jiq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. 2006. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS 20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 5.Kortekangas-Savolainen O, Orhanen E, Puodinketo T, Vuorinen T. 2014. Epidemiology of genital herpes simplex virus type 1 and 2 infections in southwestern Finland during a 10-year period (2003–2012). Sex Transm Dis 41:268–271. doi: 10.1097/OLQ.0000000000000101. [DOI] [PubMed] [Google Scholar]
- 6.Pereira VS, Moizeis RN, Fernandes TA, Araujo JM, Meissner RV, Fernandes JV. 2012. Herpes simplex virus type 1 is the main cause of genital herpes in women of Natal, Brazil. Eur J Obstet Gynecol Reprod Biol 161:190–193. doi: 10.1016/j.ejogrb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Peña KC, Adelson ME, Mordechai E, Blaho JA. 2010. Genital herpes simplex virus type 1 in women: detection in cervicovaginal specimens from gynecological practices in the United States. J Clin Microbiol 48:150–153. doi: 10.1128/JCM.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafferty WE, Coombs RW, Benedetti J, Critchlow C, Corey L. 1987. Recurrences after oral and genital herpes simplex virus infection. Influence of site of infection and viral type. N Engl J Med 316:1444–1449. [DOI] [PubMed] [Google Scholar]
- 9.Delaney S, Gardella C, Saracino M, Magaret A, Wald A. 2014. Seroprevalence of herpes simplex virus type 1 and 2 among pregnant women, 1989–2010. JAMA 312:746–748. doi: 10.1001/jama.2014.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley H, Markowitz LE, Gibson T, McQuillan GM. 2014. Seroprevalence of herpes simplex virus types 1 and 2—United States, 1999–2010. J Infect Dis 209:325–333. doi: 10.1093/infdis/jit458. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P, Xie L, Balliet JW, Casimiro DR, Yao F. 2014. A herpes simplex virus 2 (HSV-2) glycoprotein D-expressing nonreplicating dominant-negative HSV-2 virus vaccine is superior to a gD2 subunit vaccine against HSV-2 genital infection in guinea pigs. PLoS One 9:e101373. doi: 10.1371/journal.pone.0101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awasthi S, Shaw C, Friedman H. 2014. Improving immunogenicity and efficacy of vaccines for genital herpes containing herpes simplex virus glycoprotein D. Expert Rev Vaccines 13:1475–2488. doi: 10.1586/14760584.2014.951336. [DOI] [PubMed] [Google Scholar]
- 13.Awasthi S, Lubinski JM, Shaw CE, Barrett SM, Cai M, Wang F, Betts M, Kingsley S, Distefano DJ, Balliet JW, Flynn JA, Casimiro DR, Bryan JT, Friedman HM. 2011. Immunization with a vaccine combining herpes simplex virus 2 (HSV-2) glycoprotein C (gC) and gD subunits improves the protection of dorsal root ganglia in mice and reduces the frequency of recurrent vaginal shedding of HSV-2 DNA in guinea pigs compared to immunization with gD alone. J Virol 85:10472–10486. doi: 10.1128/JVI.00849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrel S, Aime C, Hermet L, Ait-Arkoub Z, Agut H, Boutolleau D. 2013. Surveillance of herpes simplex virus resistance to antivirals: a 4-year survey. Antiviral Res 100:365–372. doi: 10.1016/j.antiviral.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Frobert E, Burrel S, Ducastelle-Lepretre S, Billaud G, Ader F, Casalegno JS, Nave V, Boutolleau D, Michallet M, Lina B, Morfin F. 2014. Resistance of herpes simplex viruses to acyclovir: an update from a ten-year survey in France. Antiviral Res 111:36–41. doi: 10.1016/j.antiviral.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Wald A, Corey L, Timmler B, Magaret A, Warren T, Tyring S, Johnston C, Kriesel J, Fife K, Galitz L, Stoelben S, Huang ML, Selke S, Stobernack HP, Ruebsamen-Schaeff H, Birkmann A. 2014. Helicase-primase inhibitor pritelivir for HSV-2 infection. N Engl J Med 370:201–210. doi: 10.1056/NEJMoa1301150. [DOI] [PubMed] [Google Scholar]
- 17.Luebcke E, Dubovi E, Black D, Ohsawa K, Eberle R. 2006. Isolation and characterization of a chimpanzee alphaherpesvirus. J Gen Virol 87:11–19. doi: 10.1099/vir.0.81606-0. [DOI] [PubMed] [Google Scholar]
- 18.Severini A, Tyler SD, Peters GA, Black D, Eberle R. 2013. Genome sequence of a chimpanzee herpesvirus and its relation to other primate alphaherpesviruses. Arch Virol 158:1825–1828. doi: 10.1007/s00705-013-1666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. 1998. The genome sequence of herpes simplex virus type 2. J Virol 72:2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colgrove R, Diaz F, Newman R, Saif S, Shea T, Young S, Henn M, Knipe DM. 2014. Genomic sequences of a herpes simplex virus 2 clinical isolate and its plaque-purified derivative strain. Virology 450–451:140–145. doi: 10.1016/j.virol.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norberg P, Kasubi MJ, Haarr L, Bergstrom T, Liljeqvist JA. 2007. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J Virol 81:13158–13167. doi: 10.1128/JVI.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaoka H, Kawana T, Grillner L, Aomori T, Yamaguchi T, Saito H, Fujinaga K. 1987. Genome variations in herpes simplex virus type 2 strains isolated in Japan and Sweden. J Gen Virol 68(Pt 8):2105–2116. [DOI] [PubMed] [Google Scholar]
- 23.Grau DR, Visalli RJ, Brandt CR. 1989. Herpes simplex virus stromal keratitis is not titer-dependent and does not correlate with neurovirulence. Invest Ophthalmol Vis Sci 30:2474–2480. [PubMed] [Google Scholar]
- 24.Westmoreland D, Rapp F. 1976. Host range temperature-sensitive mutants of herpes simplex virus type 2. J Virol 18:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kintner RL, Brandt CR. 1994. Rapid small-scale isolation of herpes simplex virus DNA. J Virol Methods 48:189–196. doi: 10.1016/0166-0934(94)90118-X. [DOI] [PubMed] [Google Scholar]
- 26.Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol 84:5303–5313. doi: 10.1128/JVI.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J Virol 88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozas J, Rozas R. 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci 11:621–625. [DOI] [PubMed] [Google Scholar]
- 31.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 32.Faircloth BC. 2008. msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 8:92–94. doi: 10.1111/j.1471-8286.2007.01884.x. [DOI] [PubMed] [Google Scholar]
- 33.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 36.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolb AW, Adams M, Cabot EL, Craven M, Brandt CR. 2011. Multiplex sequencing of seven ocular herpes simplex virus type-1 genomes: phylogeny, sequence variability and SNP distribution. Invest Ophthalmol Vis Sci 52:9061–9073. doi: 10.1167/iovs.11-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deback C, Boutolleau D, Depienne C, Luyt CE, Bonnafous P, Gautheret-Dejean A, Garrigue I, Agut H. 2009. Utilization of microsatellite polymorphism for differentiating herpes simplex virus type 1 strains. J Clin Microbiol 47:533–540. doi: 10.1128/JCM.01565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deback C, Luyt CE, Lespinats S, Depienne C, Boutolleau D, Chastre J, Agut H. 2010. Microsatellite analysis of HSV-1 isolates: from oropharynx reactivation toward lung infection in patients undergoing mechanical ventilation. J Clin Virol 47:313–320. doi: 10.1016/j.jcv.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Burrel S, Ait-Arkoub Z, Voujon D, Deback C, Abrao EP, Agut H, Boutolleau D. 2013. Molecular characterization of herpes simplex virus 2 strains by analysis of microsatellite polymorphism. J Clin Microbiol 51:3616–3623. doi: 10.1128/JCM.01714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolb AW, Cécile A, Brandt CR. 2013. Using HSV-1 genome phylogenetics to track past human migrations. PLoS One 8:e76267. doi: 10.1371/journal.pone.0076267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeoch DJ, Dolan A, Ralph AC. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol 74:10401–10406. doi: 10.1128/JVI.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norberg P, Bergstrom T, Rekabdar E, Lindh M, Liljeqvist JA. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol 78:10755–10764. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.