ABSTRACT
Flavivirus NS4A protein induces host membrane rearrangement and functions as a replication complex component. The molecular details of how flavivirus NS4A exerts these functions remain elusive. Here, we used dengue virus (DENV) as a model to characterize and demonstrate the biological relevance of flavivirus NS4A oligomerization. DENV type 2 (DENV-2) NS4A protein forms oligomers in infected cells or when expressed alone. Deletion mutagenesis mapped amino acids 50 to 76 (spanning the first transmembrane domain [TMD1]) of NS4A as the major determinant for oligomerization, while the N-terminal 50 residues contribute only slightly to the oligomerization. Nuclear magnetic resonance (NMR) analysis of NS4A amino acids 17 to 80 suggests that residues L31, L52, E53, G66, and G67 could participate in oligomerization. Ala substitution for 15 flavivirus conserved NS4A residues revealed that these amino acids are important for viral replication. Among the 15 mutated NS4A residues, 2 amino acids (E50A and G67A) are located within TMD1. Both E50A and G67A attenuated viral replication, decreased NS4A oligomerization, and reduced NS4A protein stability. In contrast, NS4A oligomerization was not affected by the replication-defective mutations (R12A, P49A, and K80A) located outside TMD1. trans complementation experiments showed that expression of wild-type NS4A alone was not sufficient to rescue the replication-lethal NS4A mutants. However, the presence of DENV-2 replicons could partially restore the replication defect of some lethal NS4A mutants (L26A and K80A), but not others (L60A and E122A), suggesting an unidentified mechanism governing the outcome of complementation in a mutant-dependent manner. Collectively, the results have demonstrated the importance of TMD1-mediated NS4A oligomerization in flavivirus replication.
IMPORTANCE We report that DENV NS4A forms oligomers. Such NS4A oligomerization is mediated mainly through amino acids 50 to 76 (spanning the first transmembrane domain [TMD1]). The biological importance of NS4A oligomerization is demonstrated by results showing that mutations of flavivirus conserved residues (E50A and G67A located within TMD1) reduced the oligomerization and stability of the NS4A protein, leading to attenuated viral replication. A systematic mutagenesis analysis demonstrated that flavivirus conserved NS4A residues are important for DENV replication. A successful trans complementation of replication-lethal NS4A mutant virus requires wild-type NS4A in the context of the viral replication complex. The wild-type NS4A protein alone is not sufficient to rescue the replication defect of NS4A mutants. Intriguingly, distinct NS4A mutants yielded different complementation outcomes in the replicon-containing cells. Overall, the study has enhanced our understanding of flavivirus NS4A at the molecular level. The results also suggest that inhibitor blocking of NS4A oligomerization could be explored for antiviral drug discovery.
INTRODUCTION
Many flaviriruses are significant human pathogens, including the four serotypes of dengue virus (DENV). DENV infection causes an acute and systemic disease that may often be asymptomatic but can lead to clinical symptoms ranging from fever to life-threatening dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (1, 2). DENV is a global public health threat, with an estimated 390 million human infections per year, of which 96 million show clinical manifestations (3). Prior infection with a particular DENV serotype does not always protect against the other three serotypes (1) and is often associated with increased disease severity in the second infection (4). There are currently no licensed vaccines or antiviral drugs for either prophylaxis or treatment of DENV infection. In light of this lack of weaponry in our fight against DENV, there is an ongoing urgent need for the development of effective antivirals against the virus. Understanding the molecular details of viral replication will greatly facilitate vaccine and therapeutic development.
DENV is a positive-strand RNA virus with a genome approximately 11 kb in length. The viral genome has one open reading frame flanked by 5′ and 3′ untranslated regions (UTRs). The genome is expressed as a single polyprotein that is processed co- and posttranslationally by viral and host proteases to yield three structural proteins (capsid [C], membrane [M], and envelope [E] glycoproteins) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (5). The structural proteins form virus particles and are associated with important biological functions of the virus life cycle, such as receptor binding, virus fusion, and virion assembly. The nonstructural proteins function in viral replication and assembly and in evasion of the immune response (5–8). NS1 plays important roles in viral RNA replication (9) and the formation of immune complexes (10). NS3 functions as a protease (along with NS2B), an RNA helicase, and a nucleotide triphosphatase (11, 12). NS5 has methyltransferase and RNA-dependent RNA polymerase activities (13–16). The remaining three highly hydrophobic membrane proteins (NS2A, NS4A, and NS4B) are less well characterized. NS2A has been shown to be critical for virus replication and assembly (8, 17, 18). NS4B has been shown to be important in viral replication (19) and suppression of interferon response (7, 20).
DENV NS4A is a multifunctional transmembrane protein. It colocalizes with double-stranded RNA and promotes membrane rearrangements that harbor viral replication complexes (21). NS4A has also been shown to antagonize the host immune response (7) and to induce autophagy (22). Known interactors of DENV type 2 (DENV-2) NS4A include the host protein vimentin (23) and the viral protein NS4B (24). Miller and colleagues have proposed a membrane topology model of DENV NS4A (Fig. 1A) that contains an N-terminal region, three transmembrane domains (TMD1 to -3), and a C-terminal tail (21). Stern and coworkers recently reported that the cytosolic N-terminal 48 amino acids of DENV NS4A form an amphipathic helix that mediates oligomerization (25); however, the role of the rest of the molecule (i.e., amino acids 49 to 127) in NS4A oligomerization has not been studied. Furthermore, how NS4A oligomerization contributes to viral replication remains to be determined.
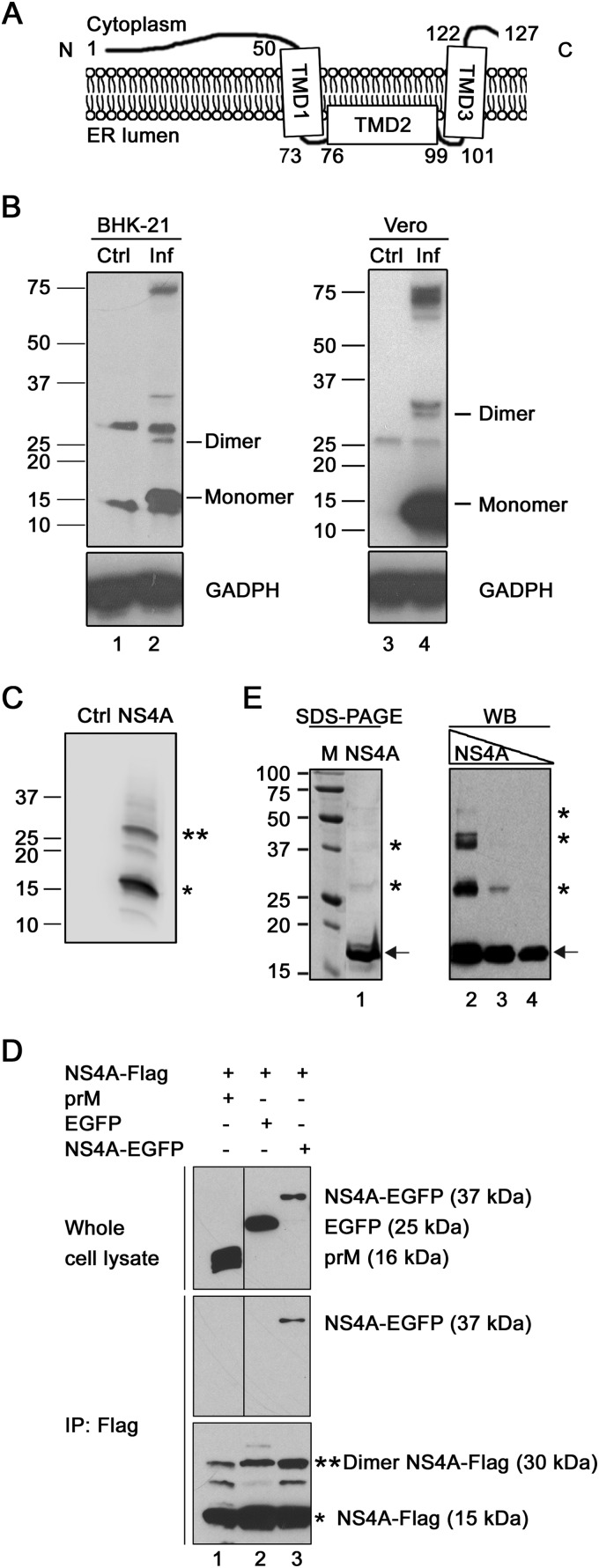
FIG 1.
Oligomerization analysis of DENV-2 NS4A. (A) Model for membrane topology of DENV-2 NS4A (21). (B) Oligomerization of NS4A in DENV-2-infected cells. BHK-21 cells were infected with DENV-2 at a multiplicity of infection (MOI) of 0.5 (lane 2) or left uninfected as a negative control (lane 1). Vero cells were also infected with DENV-2 at an MOI of 0.5 (lane 4) or left uninfected as a negative control (lane 3). At 48 h postinfection, the clarified cell lysates were incubated in LDS sample buffer without DTT at room temperature. Protein samples were analyzed by Western blotting using anti-NS4A mouse monoclonal antibody. The bands of 14 and 28 kDa are indicated as monomeric and dimeric forms, respectively. (C) Oligomerization of recombinant NS4A transiently expressed in 293T cells. The cells were transfected with a pXJ-NS4A-Flag plasmid (lane 2). The cell lysates were analyzed by Western blotting using anti-Flag antibody. The single and double asterisks indicate bands at 15 and 30 kDa, respectively. (D) Co-IP analysis of NS4A oligomerization. 293T cells were cotransfected with two plasmids, one encoding NS4A-Flag and the other encoding either prM, EGFP, or NS4A-EGFP. After 48 h, the cell lysates were subjected to co-IP as described in Materials and Methods. The whole-cell lysates and IP eluates were then analyzed by Western blotting using rabbit anti-EGFP, rabbit anti-Flag, and rabbit anti-prM antibodies. (E) Oligomerization of recombinant DENV-2 NS4A protein. (Left) SDS-PAGE analysis of purified recombinant NS4A protein. Lane M, protein standards (Bio-Rad) (the protein molecular masses [in kilodaltons] are shown on the left); lane 1, 3 μg NS4A protein. (Right) Western blot (WB) analysis of recombinant NS4A using an anti-NS4A mouse monoclonal antibody as the primary antibody. Threefold dilutions of NS4A proteins were loaded: 0.33 μg (lane 2), 0.11 μg (lane 3), and 0.037 μg (lane 4). The arrows and asterisks indicate bands corresponding to monomers and dimers/oligomers of the NS4A protein, respectively. Ctrl, control; Inf, infected.
Here, we report that in the context of full-length NS4A, TMD1 is the major determinant for NS4A oligomerization. Mechanistically, mutations within TMD1 reduced NS4A oligomerization and protein stability, leading to attenuated viral replication. In contrast, lethal replication-defective mutations outside TMD1 had no effect on NS4A oligomerization. trans complementation experiments showed that the replication defect of NS4A mutants could be partially rescued in cells expressing wild-type NS4A in the context of a viral replication complex but not in cells expressing wild-type NS4A alone. Interestingly, distinct NS4A mutants exhibited different outcomes of trans complementation in the replicon cells.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
BHK-21 cells and Vero cells were cultured in high-glucose Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), 1% penicillin-streptomycin (Invitrogen), and 4 mM l-glutamine. 293T cells were maintained in low-glucose DMEM (Invitrogen) supplemented with 10% FBS and 1% penicillin-streptomycin. BHK-21 cells expressing NS4A-enhanced green fluorescent protein (EGFP) in a Tet-off-inducible manner was maintained in the same medium as BHK-21 cells, supplemented with 500 μg/ml Geneticin (Life Technologies) and 200 μg/ml hygromycin. BHK-21 cells harboring the DENV-2 replicon were also cultured in the same medium as BHK-21 cells, with 1 mg/ml Geneticin as an additional supplement. Recombinant DENV-2 (strain NGC) was produced from its corresponding cDNA clone, pACYC-NGC-FL (26). The following antibodies were used in this study: in-house-generated mouse monoclonal antibody (MAb) against DENV-2 NS4A protein, anti-EGFP rabbit polyclonal antibody (PAb) (Abcam, Cambridge, United Kingdom), anti-Flag rabbit PAb (Sigma, St. Louis, MO), anti-premembrane (prM) protein rabbit PAb (GeneTex, Irvine, CA), anti-E mouse MAb 4G2 (ATCC, Manassas, VA), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) rabbit PAb (Sigma), anti-His mouse MAb (Qiagen, Netherlands), goat anti-mouse IgG conjugated with Alexa Fluor 488 or Alexa Fluor 568 (Life Technologies), and goat anti-mouse or goat anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (Sigma).
Plasmid construction.
Standard molecular biology procedures were performed for all plasmid constructions. A mammalian expression vector, pXJ, driven by a cytomegalovirus (CMV) promoter was used to express a recombinant full-length (FL) or truncated NS4A protein that was C-terminally tagged with either Flag or EGFP. Fragments encoding C-terminally Flag-tagged NS4A were amplified from pACYC-NGC-FL by PCRs with corresponding primer pairs. The PCR products were digested with restriction enzymes EcoRI and XhoI and inserted into the pXJ expression vector (27). For the C-terminally EGFP-tagged constructs, fragments of FL and truncated NS4A were also amplified from pACYC-NGC FL, digested with EcoRI and BamHI, and cloned into plasmid pXJ-EGFP.
For alanine scanning of NS4A, individual mutations were introduced into a subclone, TA-E (containing DENV-2 NGC cDNA from nucleotide 5426 to the 3′ end) using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) with corresponding primer pairs. The fragment containing each mutation was then inserted into pACYC-NGC-FL at either XhoI and BspEI or BspEI and NruI restriction sites. For construct pXJ-NS4A-EGFP or pXJ-NS4A-Flag, encoding mutant NS4A proteins, the corresponding NS4A fragments were amplified from the pACYC-NGC-FL plasmids containing the respective NS4A mutations. These fragments were then digested and inserted into either pXJ-EGFP or pXJ vectors as described above. All constructs were validated by DNA sequencing, and primer sequences are available upon request.
SDS-PAGE and Western blotting.
Samples were incubated in 4× lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) supplemented with 100 mM dithiothreitol (DTT) at 70°C for 15 min (unless otherwise indicated) and analyzed on a 15% polyacrylamide-SDS gel. The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane using the Trans-Blot Turbo Transfer apparatus (Bio-Rad, Hercules, CA) and incubated for 2 h at room temperature in blocking buffer, which contained 5% skim milk (Sigma) in TBST (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, and 0.1%Tween 20). The blots were then incubated in primary antibody diluted in blocking buffer overnight at 4°C. The following primary antibodies were used for Western blotting: anti-NS4A (1:100), anti-GAPDH (1:4,000), anti-Flag (1:2,000), anti-EGFP (1:2,000), and anti-prM (1:4,000). After three 5-min washes in TBST, the blots were incubated in either goat anti-rabbit (1:30,000) or goat anti-mouse (1:10,000) antibody conjugated to HRP in blocking buffer for 1 h. The blots were washed three times for 5 min each time in TBST buffer, and detection was done using the ECL Western blotting system (GE Healthcare).
Coimmunoprecipitation.
293T cells in 6-cm dishes were transfected with 2 μg of various pXJ constructs with X-tremeGene 9 DNA transfection reagent (Roche, Switzerland). The cells were harvested at 48 h posttransfection (p.t.) and incubated in 550 μl immunoprecipitation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5% n-dodecyl-β-d-maltopyranoside [DDM], 0.5 mM EDTA, 0.5 mM EGTA, EDTA-free protease inhibitor cocktail [Roche], and phosphatase inhibitor cocktail tablets [Roche]) with rotation for 1 h at 4°C. The cell lysates were clarified by centrifugation at 20,000 × g for 15 min at 4°C and used for coimmunoprecipitation (co-IP) with anti-Flag M2 magnetic beads (Sigma). The beads and lysates were incubated with rotation for 1 h at 4°C in a total volume of 450 μl containing 250 mM sodium chloride. The beads were then washed vigorously three times with phosphate-buffered saline (PBS) containing 0.1% Tween 20. The proteins were eluted from the beads using 4× LDS sample buffer supplemented with 100 mM DTT by heating at 70°C with shaking for 15 min and analyzed by Western blotting. To quantify the relative pulldown efficiency, each protein band was measured using densitometry (ImageJ): NS4A-EGFP truncates in the whole-cell lysate (a) and IP eluate (b) and FL NS4A-EGFP in the whole-cell lysate (c) and IP eluate (d). The pulldown efficiency for each truncate relative to the FL control was quantified using the formula (b/a)/(d/c).
Cross-linking experiment.
An NS4A construct containing residues 17 to 80 was expressed and purified into dodecylphosphocholine (DPC) micelles as described previously (24). Cross-linking of the purified NS4A using glutaraldehyde (GA) was carried out by a method described previously (28, 29). Briefly, 50 μM NS4A was reconstituted into a cross-linking buffer that contained 20 mM sodium phosphate, pH 6.5, 0.1 mM DTT, and 20 mM DPC. GA cross-linker from a 10% stock solution was added to the protein solution to 16 mM. The protein mixture was incubated at room temperature for 10 min, separated by SDS-PAGE, and analyzed by Western blotting using an anti-His antibody.
Nuclear magnetic resonance (NMR) measurements.
The 15N longitudinal or spin-lattice relaxation (T1) and transverse or spin-spin relaxation (T2) rates (30) and backbone 1H-15N steady-state nuclear Overhauser effect (NOE) (heteronuclear NOE [hetNOE]) (31) of the NS4A in DPC micelles were measured at 313 K on a Bruker Avance magnet with a proton frequency of 600 MHz. The relaxation delays for T1 measurements were 10, 50, 100, 200, 400, 800, 1,400, and 1,800 ms. The delays for T2 measurements were 16.9, 34, 51, 68, 85, 102, 119, 136, and 153 ms (32). The relaxation rate constants were obtained by fitting the peak intensities into a monoexponential function with NMRView (33). The hetNOEs were obtained using two data sets that were collected with and without initial proton saturation for a period of 3 s. To test the effect of the DPC-to-protein ratio on protein dimerization, NS4A was first purified into DPC micelles using 2 mM DPC micelles to get a sample with a DPC-to-protein ratio of 70. To make a sample with a DPC-to-protein ratio of 300, DPC (20%) was added to the sample with a low DPC-to-protein ratio. The 1H-15N heteronuclear single quantum coherence (HSQC) spectra of NS4A samples with different DPC-to-protein ratios were acquired and compared. To test residues that could be affected by the dimerization, the 1H-15N HSQC spectra of a uniformly 15N-labeled NS4A in DPC micelles in the absence and presence of an unlabeled NS4A peptide containing residues 40 to 76 were acquired and compared.
Alanine-scanning analysis.
DENV-2 NS4A mutations were engineered using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). FL DENV-2 RNAs were transcribed in vitro using the T7 mMessage mMachine kit (Ambion, Austin, TX) from cDNA plasmids prelinearized with restriction enzyme XbaI. BHK-21 cells were electroporated with 10 μg of RNA transcripts. The transfected cells were first cultured in 2% high-glucose DMEM at 37°C for 24 h and then transferred to 30°C for the next 4 days. The transfected cells were monitored for DENV envelope (E) protein expression by immunofluorescence assay (IFA) at 1 and 4 days p.t. using the anti-E mouse monoclonal antibody 4G2 and a goat anti-mouse IgG conjugated with Alexa Fluor 488 as the primary and secondary antibodies, respectively. Culture media from the transfected cells were collected 5 days p.t. (designated passage 0 [P0]), and the viral titers were quantified by plaque assay using BHK-21 cells. These P0 viruses were subsequently passaged for 5 rounds in Vero cells for adaptive mutations. The presence of revertants in each passage was checked by reverse transcription (RT)-PCR, plaque assay, and whole-genome sequencing.
Cycloheximide chase experiment.
293T cells in 6-cm dishes were transfected with 2 μg of either wild-type (WT), E50A, or G67A pXJ-NS4A-Flag plasmid using X-tremeGene 9 DNA transfection reagent. The pXJ-NS4A-Flag plasmid encodes a full-length DENV-2 NS4A protein fused with a C-terminal Flag epitope. At 24 h p.t., the cells were treated with 75 μg/ml cycloheximide (CHX) (Sigma) in dimethyl sulfoxide (DMSO) or DMSO alone as a negative control. Cells were harvested at 0, 0.5, 1, 2, 3, and 4 h post-CHX treatment and incubated in lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 0.5% DDM, and EDTA-free protease inhibitor cocktail) with rotation for 1 h at 4°C. Cell lysates were clarified by centrifugation at 20,000 × g for 15 min and analyzed by Western blotting using rabbit anti-Flag antibody to detect NS4A-Flag. The blots were also probed with a polyclonal rabbit anti-GAPDH antibody as a loading control.
trans complementation analysis.
BHK-21 cells expressing NS4A-EGFP with a Tet-off-inducible system were engineered using a Tet-Off Advanced Inducible Gene Expression kit (Clontech, Mountain View, CA). Expression of NS4A-EGFP was determined by IFA using both EGFP and mouse monoclonal anti-NS4A antibody. trans complementation experiments were performed as described previously (34). WT and mutant DENV-2 genome length RNAs were prepared as described under “Alanine-scanning analysis” above. BHK-21 cells expressing NS4A-EGFP (BHK-21-NS4A-EGFP) or BHK-21 cells harboring a DENV-2 replicon (BHK-21-Replicon) (35) were electroporated with RNA transcripts (10 μg). At 72 h p.t, the cells were fixed with methanol, and expression of viral E protein was detected by IFA using mouse MAb 4G2 as the primary antibody. The secondary antibodies used were goat anti-mouse IgG conjugated to Alexa Fluor 568 and Alexa Fluor 488 for BHK-21-NS4A-EGFP and BHK-21-Replicon cells, respectively.
RESULTS
Oligomerization of DENV-2 NS4A protein.
We performed four sets of experiments to demonstrate the oligomerization of DENV-2 NS4A.
(i) Cells infected with DENV-2.
As shown in Fig. 1B, Western blotting of lysates of BHK-21 cells infected with DENV-2 showed four distinct bands. The 14- and 28-kDa bands correspond to the molecular masses of monomeric and dimeric forms of NS4A, respectively; the 35- and 75-kDa bands may represent uncleaved viral polyprotein or higher-order oligomers of NS4A; and two nonspecific cellular proteins were also detected in the immunoblots (Fig. 1B, lane 2). The two bands representing the 14-kDa monomeric and 28-kDa dimeric NS4A forms were also detected in infected Vero cells (lane 4).
(ii) Cells transiently expressing NS4A with a C-terminal Flag epitope tag (NS4A-Flag).
Western blotting of 293T cells transfected with the NS4A-Flag-expressing plasmid revealed two bands of 15 and 30 kDa (Fig. 1C) that correspond to the monomeric and dimeric NS4A-Flag forms, respectively.
(iii) Co-IP of NS4A-Flag and NS4A with a C-terminal EGFP tag (NS4A-EGFP).
293T cells were cotransfected with two plasmids: one plasmid encoding NS4A-Flag and another plasmid encoding NS4A-EGFP. Anti-Flag antibody was able to pull down NS4A-Flag, together with NS4A-EGFP (Fig. 1D, lane 3). As negative controls, when NS4A-Flag was coexpressed with EGFP or DENV-2 prM protein, the anti-Flag antibody could pull down only NS4A-Flag, but not EGFP or prM (Fig. 1D, lanes 1 and 2).
(iv) Recombinant NS4A protein.
We purified DENV-2 NS4A protein using an Escherichia coli expression system, as previously reported (36). The recombinant protein consisted of an N-terminal His6 tag, a tobacco etch virus (TEV) protease cleavage sequence, a thrombin cleavage site, and a full-length NS4A protein. As shown in Fig. 1E (left), SDS-PAGE analysis revealed a major 17-kDa band corresponding to the calculated protein mass (indicated by an arrow); in addition, two weak bands migrating at 28 and 37 kDa were observed (Fig. 1E, lane 1, asterisks). Western blotting showed that both the 17-kDa band and the higher-molecular-mass bands were recognized by an anti-NS4A monoclonal antibody (Fig. 1E, lane 2), suggesting that the higher-molecular-mass bands are not derived from contaminating proteins; in addition, increasing protein concentrations shifted the 17-kDa band to higher-molecular-mass bands (Fig. 1E, lanes 2 to 4). The aberrant migration behavior of different oligomeric forms (17, 28, and 37 kDa) of recombinant NS4A could be due to varying amounts of detergent associated with the protein. Collectively, the above-described results demonstrate that DENV-2 NS4A oligomerizes in infected cells, when expressed alone in cells, and in purified form.
Mapping the determinant for NS4A oligomerization.
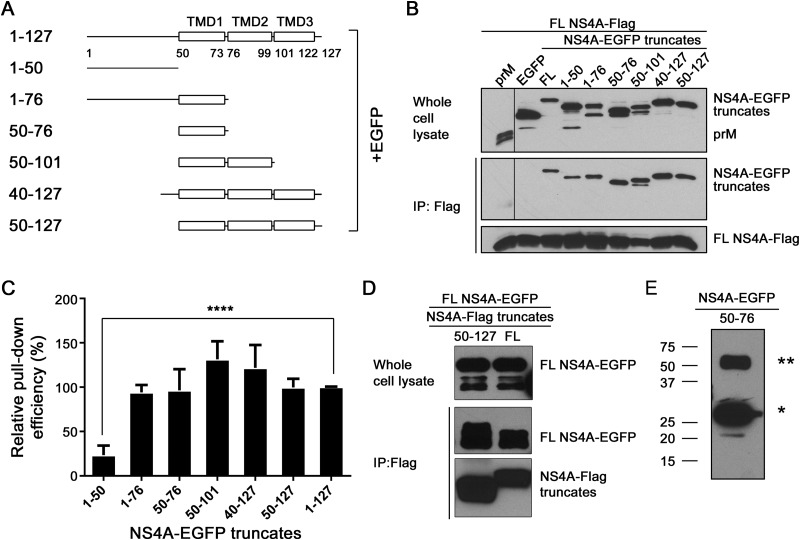
We used co-IP to map the regions responsible for NS4A oligomerization. 293T cells were cotransfected with two plasmids, one expressing a full-length NS4A-Flag and another expressing a truncated NS4A-EGFP (Fig. 2A). As shown in Fig. 2B (top), the protein expression levels were similar for various NS4A-EGFP truncates, except for constructs 1–50 and 50–76, whose expression levels were higher. Bands representing both intact and degraded proteins were observed for constructs 1–50, 1–76, 50–76, and 50–101 (Fig. 2B, top). Anti-Flag antibody was able to pull down full-length NS4A-Flag (Fig. 2B, bottom), together with various NS4A-EGFP truncates with differing efficiencies (Fig. 2B, middle). Compared to the full-length construct 1–127 NS4A-EGFP (set as 100%), the pulldown efficiencies for constructs 1–50, 1–76, 50–76, 50–101, 40–127, and 50–127 were estimated to be 20%, 94%, 110%, 130%, 120%, and 100%, respectively (Fig. 2C). Notably, when we performed the co-IP with a higher salt concentration (350 mM), the binding of construct 1–50 was negligible in comparison to the other constructs (data not shown). We also performed a reverse co-IP in which truncated NS4A-Flag was transiently coexpressed, along with full-length NS4A-EGFP. As shown in Fig. 2D, a 50–127 NS4A-Flag construct lacking the N-terminal 49 residues was able to pull down full-length NS4A-EGFP as efficiently as full-length NS4A-Flag. The results suggest that (i) construct 50–76, which comprise TMD1, constitutes the most critical segment for NS4A oligomerization and (ii) construct 1–50, representing the cytosolic N-terminal region, may also contribute to the oligomerization, but to a much lesser extent.
FIG 2.
Mapping the determinant for NS4A oligomerization. (A) NS4A truncate-EGFP constructs. FL NS4A consists of residues 1 to 127. The various truncates were designed according to the membrane topology model of NS4A. (B) Co-IP analysis of NS4A-EGFP truncates pulled down by FL NS4A-Flag. 293T cells were cotransfected with two plasmids encoding FL NS4A-Flag and NS4A truncate-EGFP, respectively. At 48 h posttransfection, the cell lysates were harvested and co-IP experiments were performed as described in Materials and Methods. (C) Relative pulldown efficiencies of NS4A truncate-EGFP by FL NS4A-Flag. The pulldown efficiency for each truncate relative to the FL control was quantified using the formula described in Materials and Methods. Means and standard errors were obtained from 3 to 5 independent experiments. ****, P < 0.0001 (unpaired two-tailed t test). (D) Reverse co-IP of FL NS4A-EGFP by NS4A-Flag truncate. 293T cells were cotransfected with two plasmids encoding FL NS4A-EGFP with either a 50–127 truncate or an FL construct of NS4A-Flag, respectively. The co-IP experiment was performed as described above. (E) Oligomerization analysis of an NS4A-EGFP truncate consisting only of TMD1 (residues 50 to 76). 293T cells were transfected with a plasmid encoding NS4A-EGFP truncate 50–76. The cell lysate was analyzed 48 h later by Western blotting using a rabbit anti-EGFP antibody. The single and double asterisks indicate bands of 28 and 56 kDa, respectively.
To confirm that TMD1 is the major determinant for NS4A oligomerization, we transiently expressed 50–76 NS4A-EGFP alone in 293T cells and directly probed its oligomerization status by Western blotting rather than co-IP (Fig. 2E). Indeed, Western blotting detected both monomeric and dimeric forms of 50–76 NS4A-EGFP. This result further demonstrates that TMD1 alone is competent to form dimers. Besides TMD1, we also attempted to probe the roles of TMD2 and TMD3 in NS4A oligomerization. Using the same co-IP approach, we expressed luminal TMD2 and TMD3 fused with an N-terminal signal peptide (sequence derived from Gaussia luciferase protein). The N-terminal signal peptide was engineered to ensure the correct orientation of the expressed TMD2 and TMD3 proteins. Unfortunately, these experiments were unsuccessful because the signal peptide itself resulted in nonspecific binding (data not shown).
Dynamic analysis of the NS4A (17–80) construct in detergent DPC micelles.
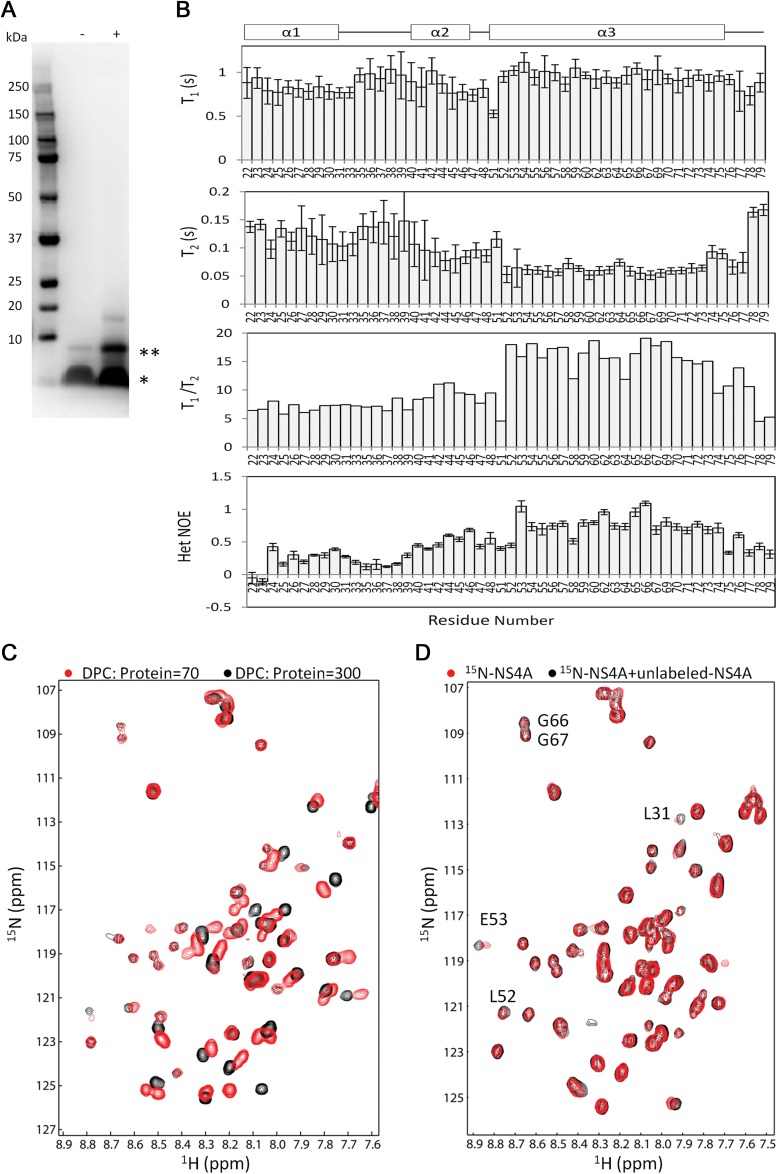
We recently showed that a construct containing residues 17 to 80 of NS4A is feasible for structural studies by NMR spectroscopy. The NMR analysis revealed the secondary structure of NS4A (17–80) with three α helices (24). SDS-PAGE analysis of this protein alone showed a dominant monomer and a minor dimer (Fig. 3A). In the presence of the cross-linker agent glutaraldehyde, the protein displayed more dimer bands and even higher-order oligomers. The results suggested that the NS4A (17–80) construct could form a dimer in vitro.
FIG 3.
Oligomerization analysis of NS4A (17–80). (A) Cross-linking of NS4A (17–80) in DPC micelles. The presence (+) or absence (−) of the cross-linker glutaraldedyde is indicated. The monomer (*) and dimer (**) species of NS4A (17–80) are also indicated. (B) Relaxation analysis of NS4A (17–80). The NS4A (17–80) construct was purified in DPC micelles. The 15N T1, T2, and hetNOE were measured at 313 K. The error bars indicate standard errors during curve fitting. (C) 1H-15N HSQC spectra of the NS4A (17–80) samples with different DPC-to-protein ratios. NS4A (17–80) was purified with a DPC-to-protein ratio of 70. To make a sample with a DPC-to-protein ratio of 300, DPC was added to the sample. The DPC concentration (millimolar) was calculated using the following formula: CDPC + n × CNS4A, where CDPC and CNS4A are the DPC concentration in the buffer and the NS4A concentration, respectively, and n is the aggregation number of DPC (54, obtained from the manufacturer). (D) 1H-15N HSQC spectra of NS4A (17–80) in the absence and presence of unlabeled NS4A. 15N-labeled NS4A (17–80) was concentrated to 0.5 mM in a buffer containing 20 mM sodium phosphate, pH 6.5, and 150 mM DPC. A synthetic peptide corresponding to residues 40 to 76 of NS4A was added to the sample at a 1:1 molar ratio. The spectra were superimposed.
Since construct 17–80 contains the major determinant, TMD1, for NS4A oligomerization, we used NMR to characterize its oligomerization. We conducted a relaxation experiment to understand its flexibility on the picosecond to nanosecond time scale. Figure 3B shows the T1, T2, and hetNOE results. The first two helices in the construct are flexible, as characterized by the low hetNOE and high T2 values. The transmembrane helix α3 is rigid, as evidenced by the high hetNOE and T1/T2 values. Interestingly, we did not identify the cross peak of E50, which may be due to the exchanges arising from its oligomerization. α2 was more stable than α1, as indicated by the higher T1/T2 and hetNOE values from α2. The high flexibility of α1 also suggests that its role in oligomerization might be less important than those of the other, more rigid regions, such as α2 and α3 (Fig. 3B).
Oligomerization of NS4A (17–80) in DPC micelles.
As the purified NS4A (17–80) was prepared in detergent micelles, changing the detergent-to-protein ratio can affect protein dimerization. Decreasing the detergent-to-protein ratio may favor protein dimerization in the transmembrane region. We compared the 1H-15N HSQC spectra of the NS4A (17–80) samples with different DPC-to-protein ratios (Fig. 3C). Line broadening and chemical shift perturbations were observed for several residues, which strongly suggests that a changing DPC-to-protein ratio affected the NS4A (17–80) environment. These changes may arise from protein dimerization. To further probe residues that are important for dimerization, the 1H-15N HSQC spectra of 15N-labeled NS4A (17–80) in the absence and presence of unlabeled NS4A peptide containing residues 40 to 76 were acquired and superimposed (Fig. 3D). Residues L31, L52, E53, G66, and G67 showed line broadening, suggesting that these residues are important for protein dimerization. It should be noted that the unlabeled NS4A (40–76) peptide was used for the above-described experiment because the NS4A (50–76) peptide was too hydrophobic for successful synthesis (data not shown).
Functional analysis of flavivirus conserved NS4A amino acids.
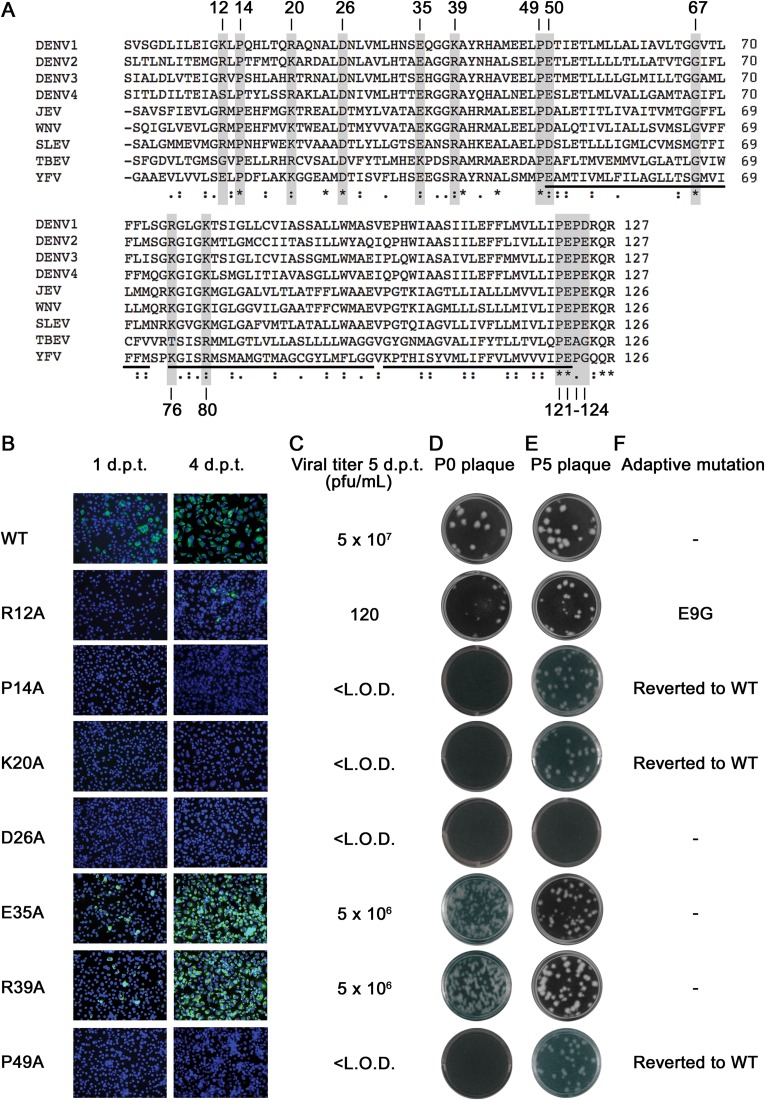
Using an infectious cDNA clone of DENV-2, we performed alanine scanning of 15 flavivirus conserved residues in NS4A (Fig. 4A). The mutated residues were R12, P14, K20, D26, E35, R39, P49, E50, G67, R76, K80, P121, E122, P123, and E124. These residues were selected because most of them contained charged side chains that may facilitate revertant analysis. To examine the effects of individual mutations on viral replication, we electroporated BHK-21 cells with equal amounts of WT or mutant genome length RNA. The electroporated cells were monitored for viral envelope protein expression using an immunofluorescence assay (Fig. 4B). Virus production in culture media at day 5 posttransfection was quantified using a plaque assay (Fig. 4C and D). Both the immunofluorescence assay and virus production results consistently showed that, except for mutants E35A and R39A, which retained the WT replication level, all the mutants reduced or abolished viral replication.
FIG 4.
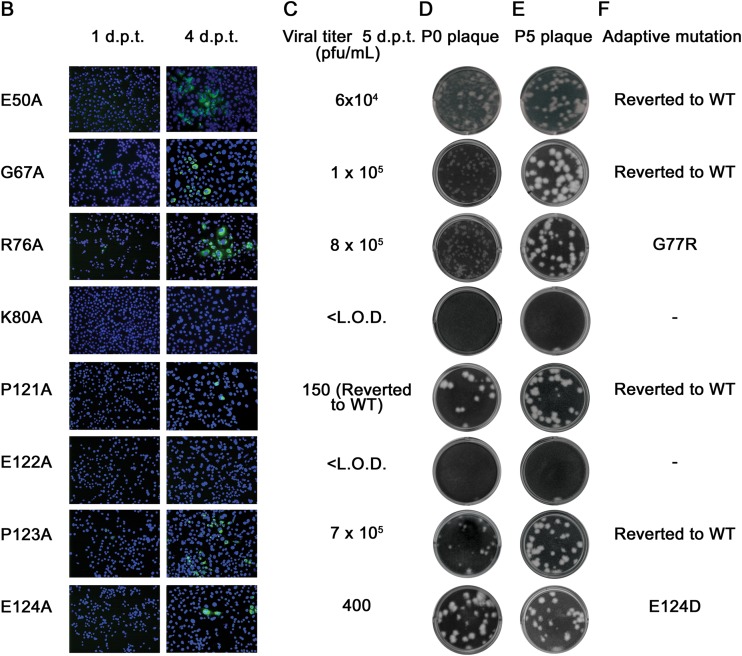
Functional analysis of flavivirus conserved NS4A amino acids. (A) Multiple-sequence alignment of various NS4A sequences from the genus Flavivirus using ClustalW software. The asterisks indicate identical, fully conserved residues, whereas the colons and periods mark strongly and weakly similar residues, respectively. The shading marks the positions of conserved residues selected for alanine-scanning mutagenesis. The black bars underlining the alignment mark the transmembrane domains. JEV, Japanese encephalitis virus; WNV, West Nile virus; SLEV, St. Louis encephalitis virus; TBEV, tick-borne encephalitis virus; YFV, yellow fever virus. (B to F) Analysis of NS4A alanine-scanning mutants in DENV-2 genome length RNA. Equal amounts of WT or mutant genome length RNAs were electroporated into BHK-21 cells. (B) IFA analysis. The transfected cells were monitored for DENV E protein expression at 1 and 4 days posttransfection (p.t.). Anti-E 4G2 MAb and goat anti-mouse IgG conjugated with Alexa Fluor 488 were used as the primary and secondary antibodies, respectively. Green and blue indicate E protein and nucleus staining, respectively. (C) Viral titer from plaque assay. Culture media from the transfected cells were collected at 5 days p.t., and the viral titers were quantified by plaque assay; the limit of detection (L.O.D.) was 10 PFU/ml. (D) P0 plaque morphology for WT and mutant viruses. For P0, the viruses were derived from the media collected on day 5 p.t. (E) P5 plaque morphology for WT and mutant viruses. The P0 viruses were subsequently passaged for 5 rounds in Vero cells for adaptive mutations, and the viral supernatant from P5 was tested by plaque assay. (F) Adaptive mutations from revertant analysis. The presence of revertants in each passage was checked by RT-PCR and whole-genome sequencing. The amino acid substitutions identified here were all within the NS4A gene. d.p.t, days posttransfection.
We sequenced each viable virus recovered on day 5 from the transfected cells. The sequencing results revealed that, except for mutant P121A, which had reverted to the WT Pro (Fig. 4C), all the viable mutants (R12A, E35A, R39A, E50A, G67A, R76A, P123A, and E124A) retained the engineered Ala substitutions without any extra mutation (data not shown). The results indicate that the plaques observed for mutant P121A (Fig. 4D) were derived from the revertant virus (equivalent to the WT virus).
Revertant analysis.
We performed revertant analysis for all mutant viruses. The culture media from transfected cells were continuously passaged on Vero cells for 5 rounds (5 to 6 days per round). During revertant selection, plaque assay and virus-specific RT-PCR were performed to monitor the improvement in viral fitness. Figure 4E shows the plaque morphology of P5 viruses. The P5 viruses were subjected to whole-genome sequencing. The results showed that no revertant viruses were recovered from three lethal mutants (D26A, K80A, and E122A) and two WT level replicative mutants (E35A and R39A). The other P5 mutants yielded adaptive mutations within the NS4A gene; no mutations outside the NS4A region were found (Fig. 4F). Specifically, mutant R12A acquired an E9G change, mutant R76A obtained a neighboring G77R substitution, mutant E124A changed to E124D, and all the other mutants (P14A, K20A, P49A, E50A, G67A, and P123A) reverted to their corresponding WT sequences.
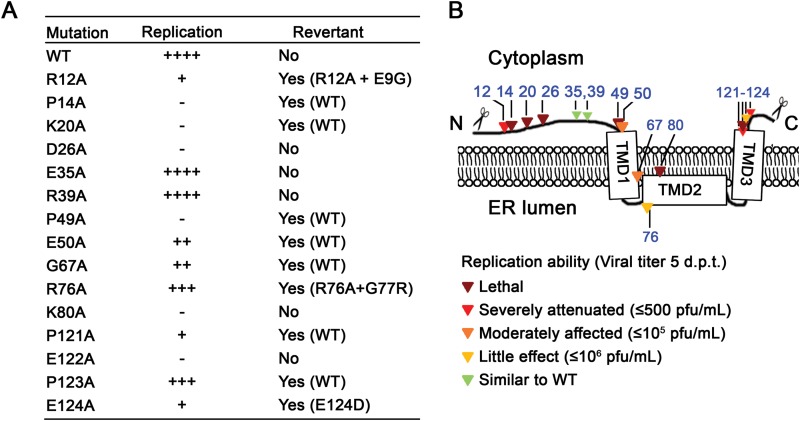
Figure 5A summarizes the mutational effects on viral replication and the revertant analysis results. Figure 5B depicts the mutated residues on an NS4A topology model (21) and their impacts on viral replication. Overall, the results demonstrate that most of the flavivirus conserved NS4A residues are important for DENV-2 replication.
FIG 5.
Summary of NS4A alanine-scanning analysis. (A) Replication abilities of alanine-scanning mutants. The replication abilities of the mutants are indicated as follows: −, lethal; +, severely attenuated (≤500 PFU/ml); ++, moderately affected (≤105 PFU/ml); +++, little effect (≤106 PFU/ml); ++++, similar to WT. (B) Mutated residues and their impacts on virus replication are depicted on a membrane topology model of NS4A. Mutations are indicated by triangles, and the colors indicate the effects of the mutations on virus replication as for panel A: dark red, −; red, +; orange, ++; yellow, +++; green, ++++. The scissors mark the site of viral protease cleavage.
Effects of TMD1 mutations on NS4A oligomerization.
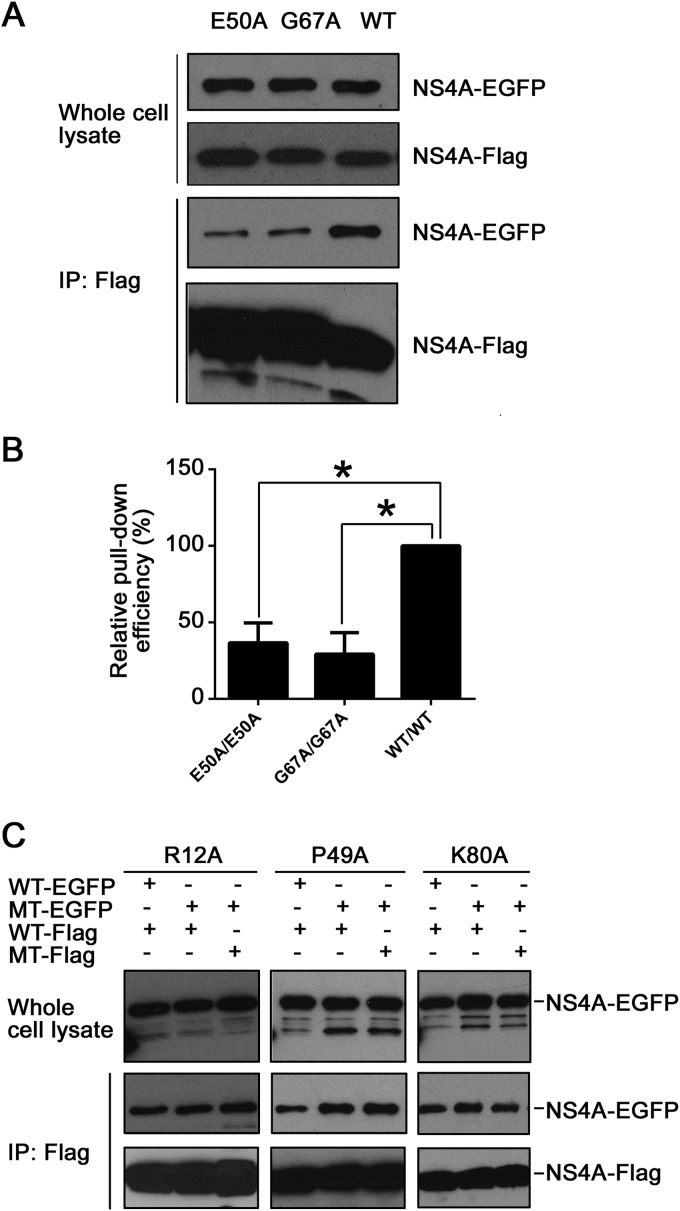
Among the 15 residues analyzed above, 2 amino acids (E50 and G67) are located in TMD1, a region critical for NS4A oligomerization. Ala substitution for both residues attenuated viral replication (Fig. 4 and 5). These results prompted us to test whether the attenuated viral replication was caused by a defect in NS4A oligomerization. A co-IP experiment was performed to address this question. 293T cells were transfected with two plasmids, one encoding NS4A-EGFP and another encoding NS4A-Flag. As shown in Fig. 6A, similar levels of protein expression were achieved for WT, E50A, and G67A NS4A-EGFP (Fig. 6A, top). Anti-Flag antibody pulled down comparable amounts of the mutants and slightly less WT NS4A-Flag (Fig. 6A, bottom), but the co-IP efficiencies of E50A and G67A NS4A-EGFP were much lower than that of WT NS4A-EGFP (Fig. 6A, middle). Quantitatively, the pulldown efficiencies of the E50A and G67A mutants were 37% and 29% of that of the WT (set as 100%), respectively (Fig. 6B). In contrast, the pulldown efficiencies of three replication-defective mutants containing changes outside TMD1 (R12A, P49A, and K80A) were similar to that of the WT (Fig. 6C). These results demonstrate that mutations E50A and G67A within TMD1 specifically attenuate NS4A oligomerization.
FIG 6.
Effects of TMD1 mutations on NS4A oligomerization. (A) Co-IP analysis of recombinant NS4A with mutations in TMD1 (E50A and G67A). Cells were cotransfected with two plasmids encoding NS4A-Flag and NS4A-EGFP. Both plasmids had either the E50A (lane 1) or G67A (lane 2) mutation, with the WT (lane 3) as a control. The co-IP experiments were performed as described in Materials and Methods. (B) Relative pulldown efficiencies of TMD1 mutants. The relative pulldown efficiency of mutant NS4A-EGFP by mutant NS4A-Flag was determined using the method described in Materials and Methods, normalized to the WT pair. Means and standard errors were obtained from 2 or 3 independent experiments. *, P < 0.05 (unpaired two-tailed t test). (C) Co-IP analysis of replication-defective mutants outside TMD1 (R12A, P49A, and K80A). Cells were cotransfected with two plasmids encoding WT NS4A-EGFP and WT NS4A-Flag, mutant NS4A-EGFP and WT NS4A-Flag, or mutant NS4A-EGFP and mutant NS4A-Flag. The co-IP experiments were performed as described in Materials and Methods. MT, mutant.
Destabilization of NS4A protein by weakened oligomerization.
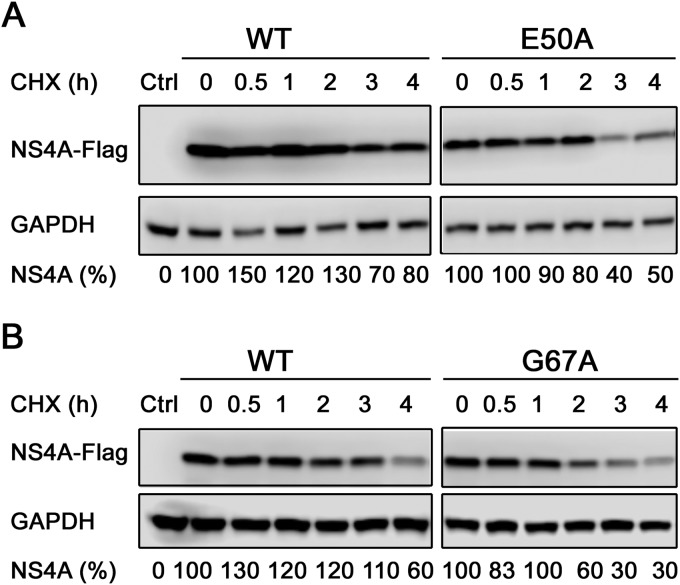
One possible consequence of weakened NS4A oligomerization is decreased protein stability, leading to reduced viral replication. To test this possibility, we compared the stabilities of E50A and G67A mutant and WT NS4A in cells. Equal amounts of plasmids encoding the WT and mutant NS4A-Flag proteins were transfected into 293T cells. At 24 h p.t., the cells were treated with CHX to inhibit protein synthesis. Western blotting was performed to quantify the amounts of WT and mutant NS4A proteins at various time points after CHX treatment. As shown in Fig. 7, the degradation rates of E50A and G67A mutant proteins were higher than that of the WT NS4A-Flag. In contrast, the stabilities of the cellular protein GAPDH were similar in the wild-type and mutant NS4A-Flag-expressing cells. The result suggests that mutations E50A and G67A decrease the stability of the NS4A protein.
FIG 7.
Differential responses of NS4A proteins following CHX treatment. 293T cells were transfected with NS4A-Flag with E50A (A) or G67A (B), with the WT construct as a control. At 24 h posttransfection, the cells were treated with 75 μg/ml CHX. Cell lysates were collected at the indicated time points post-CHX treatment and analyzed by Western blotting using a polyclonal rabbit anti-Flag antibody to detect NS4A-Flag. The blot was also probed with a polyclonal rabbit anti-GAPDH antibody as a loading control. The percentages below the blots indicate the amounts of NS4A measured by densitometry (ImageJ) normalized to the GAPDH control, with time zero set to 100%. The data shown are representative of the results of 2 or 3 independent experiments.
trans complementation analysis of NS4A.
To test if the replication defect of NS4A can be trans complemented, we engineered a BHK-21 cell line expressing WT NS4A fused with C-terminal EGFP (NS4A-EGFP) using a Tet-off system (BHK-21-NS4A-EGFP). The cellular expression of NS4A-EGFP after the removal of doxycycline was confirmed by IFA using both EGFP and anti-NS4A antibody (Fig. 8A). Three lethal NS4A mutants, D26A, K80A, and E122A, were selected for trans complementation analysis because these mutants were shown to be stable with no revertants after 5 rounds of passaging in Vero cells. We also included an additional mutant, L60A, in the trans complementation experiment because the mutant was recently shown to be replication lethal with no reversion (24).
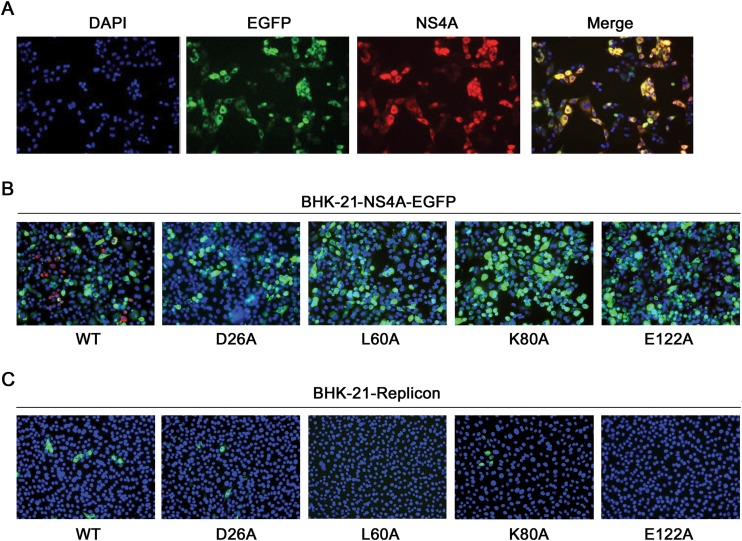
FIG 8.
trans complementation of NS4A. (A) BHK-21-NS4A-EGFP Tet-off cell line. Expression of NS4A-EGFP was detected with mouse anti-NS4A and Alexa Fluor 568-conjugated goat anti-mouse IgG as primary and secondary antibodies, respectively. (B and C) BHK-21-NS4A-EGFP cells (B) and BHK-21-Replicon cells (C) were electroporated with genome length RNAs encoding WT NS4A and NS4A D26A, L60A, K80A, and E122A. At 72 h posttransfection, the expression of the viral E protein was detected by IFA using a mouse 4G2 MAB as the primary antibody, with goat anti-mouse IgG conjugated with either Alexa Fluor 568 (B) or Alexa Fluor 488 (C) as the secondary antibody. Blue indicates nuclei stained with DAPI (4′,6-diamidino-2-phenylindole) in all three panels.
Upon transfection of WT or mutant genome length RNA into BHK-21-NS4A-EGFP cells, we examined viral E protein expression to monitor trans complementation. This is because the replicon RNA did not express viral structural proteins and only replicating genome length RNA could express viral E protein. As shown in Fig. 8B, the viral E protein was expressed in the WT-RNA-transfected cells but not in the mutant-RNA-transfected cells. This result indicates that the expression of NS4A alone is not able to trans complement the replication defect of NS4A mutants.
Next, we examined whether WT NS4A presented in the context of a replication complex is able to trans complement the replication defect of NS4A mutants. The WT and NS4A mutant genome length RNAs were individually transfected into BHK-21 cells harboring a DENV-2 replicon (BHK-21-Replicon). The replicon cells transfected with WT genome length RNA showed E-positive cells (Fig. 8C). However, the E-positive cells among the replicon cells were significantly fewer than those after transfecting the WT RNA into naive BHK-21 cells without replicon (compare Fig. 8C with 4B); such a difference is most likely caused by exclusion of superinfection (37).
The genome length RNA containing a D26A or K80A mutation also produced E-positive cells, albeit fewer than the WT RNA. In contrast, the L60A and E122A RNAs did not produce any E-positive cells. These results indicate that NS4A within a replication complex supplied in trans is able to complement only mutants D26A and K80A, but not L60A (located in TMD1) and E122A (within the PEPEKQR signal sequence required for the NS4A-2K junction cleavage).
DISCUSSION
The goal of the current study was to examine whether DENV NS4A oligomerizes and, if so, to define the determinants for the oligomerization and to characterize its functional relevance. Flavivirus NS4A is colocalized with double-stranded RNA (dsRNA) and other components of the viral replication complex. The expression of DENV NS4A alone has been shown to induce membrane remodeling (21). Therefore, flavivirus NS4A is the key viral protein to modulate the formation of vesicle packets, where the replication complex forms and synthesizes viral RNA. Two driving forces could enable the NS4A protein to promote membrane deformation. One driving force may derive from an active insertion of the amphipathic helix located within the N-terminal 48 amino acids (25) or the TMD2 helix into one leaflet of the lipid bilayer (Fig. 1A). Another driving force may result from the oligomerization of the NS4A protein. Both mechanisms are commonly observed to cause membrane curvature (38, 39). During hepatitis C virus replication, oligomerization of the viral NS4B protein was proposed to modulate membrane curvature (40).
Four lines of in vitro and in vivo evidence support the oligomerization of DENV NS4A (Fig. 1). (i) Both monomeric and dimeric forms of NS4A could be detected in DENV-2-infected BHK-21 and Vero cells. (ii) Transient expression of DENV-2 NS4A in the absence of other viral proteins generated monomeric and dimeric forms of NS4A. (iii) When coexpressed in cells, the NS4A protein tagged with Flag could pull down another NS4A protein tagged with EGFP. (iv) Recombinant NS4A protein forms monomers, dimers, and higher-order oligomers in a concentration-dependent manner. When interpreting these results, caution should be taken, because the oligomeric status of NS4A was judged by its mobility on SDS-PAGE, and anomalous migration of membrane protein has been observed on SDS-PAGE due to the difference in detergent binding (41), as well as the unknown effect of SDS on the stability of NS4A oligomerization. To exclude the possibility of such an artifact, we attempted to use gel filtration to estimate the oligomeric status of purified NS4A protein; unfortunately, the gel filtration result was not conclusive due to the presence of detergent (data not shown). Therefore, we took the NMR approach to confirm the oligomerization of the NS4A (17–80) construct (Fig. 3) (see below).
Co-IP experiments using various NS4A truncates revealed TMD1 (residues 50 to 76) as the major determinant for NS4A oligomerization. Specifically, TMD1 alone retained the full-length NS4A oligomerization activity, whereas the N-terminal 50-residue peptide exhibited only 20% of the WT NS4A oligomerization activity (Fig. 2C). The latter result agrees with a previous report that the N-terminal 48 amino acids of DENV-2 NS4A could mediate protein oligomerization (25); the discrepancy between the two studies is due to the fact that the previous study did not analyze regions beyond the first 48 amino acids of NS4A. Besides co-IP, we used two complementary approaches to confirm and characterize TMD1 as the major determinant for NS4A oligomerization. First, addition of the cross-linker glutaraldehyde to recombinant NS4A (17–80) increased protein oligomerization in solution (Fig. 3A). Second, the 1H-15N HSQC spectra of NS4A (17–80) were affected by changing the detergent-to-protein ratio (Fig. 3C) or by addition of unlabeled 40–76 peptide (Fig. 3D). Furthermore, the NMR experiment revealed line broadening of residues L31, L52, E53, G66, and G67, suggesting that these amino acids are important for NS4A oligomerization. Our conclusion that TMD1 is the major determinant for NS4A oligomerization is in agreement with a previous molecular dynamics simulation study predicting an intermolecular NS4A TMD1-TMD1 interaction (42). As mentioned above, since both the N-terminal region (residues 1 to 50) and TMD1 (residues 50 to 76) contribute to NS4A oligomerization, it is not unreasonable to speculate that one region (most likely TMD1) mediates NS4A dimerization while the other region (likely the N-terminal region) mediates higher-order oligomerization. However, the current results could not exclude the possibility that TMD2 and/or TMD3 may also facilitate NS4A oligomerization.
We performed systemic mutagenesis to analyze the function of DENV-2 NS4A in viral replication. Ala substitution for most of the flavivirus conserved NS4A residues attenuated or abolished viral replication (Fig. 4 and 5). Revertant analysis of these mutants yielded adaptive mutations within the NS4A gene. Among the 15 mutated residues, E50A and G67A are located in TMD1; both mutations reduced viral replication. Interestingly, NMR analysis indicated that G67 participates in TMD1-mediated NS4A oligomerization (Fig. 3D). Remarkably, we found that both the E50A and G67A mutations reduced NS4A oligomerization (Fig. 6A and B), leading to decreased stability of the mutant proteins in cells (Fig. 7). These results are in stark contrast to the replication-defective mutations outside TMD1 (R12A, P49A, and K80A), which did not show any effect on NS4A oligomerization. Collectively, our results clearly indicate the biological relevance of NS4A oligomerization in DENV replication.
Two cell lines (one expressing WT NS4A and one harboring a DENV-2 replicon) were used to study NS4A trans complementation (Fig. 8). None of the four lethal NS4A mutants (D26A, L60A, K80A, and E122A) could be trans complemented in cells expressing WT NS4A in the absence of other viral proteins. In contrast, two mutants (D26A and K80A) could be partially trans complemented in the replicon cells, whereas the other two mutants (L60A and E122A) could not be trans complemented. The results demonstrate that (i) the WT NS4A in the context of a replication complex is required in order to rescue the replication of NS4A mutants and (ii) the success of trans complementation depends on the location of the NS4A mutation. As shown in Fig. 5B, the D26A mutation is located in the flexible N-terminal cytoplasmic domain; the L60A mutation is located in TMD1 and was recently shown to be important for DENV NS4A-NS4B interaction (24); the K80A mutation resides in the luminal TMD2; and the E122A mutation is located within the C-terminal PEPEKQR signal sequence, which is critical for the NS4A-2K junction cleavage (43). The mechanism that governs the success of NS4A trans complementation remains to be determined. Since NS4A has distinct functions at different stages of the viral replication cycle, it is expected to interact with different viral and host factors in a spatially and temporally regulated manner. Different NS4A mutants with distinct functional defects could have different abilities to be trans complemented. As mentioned above, NS4A needs to oligomerize to induce membrane curvature before the replication complex forms; NS4A then needs to interact with NS4B to facilitate viral RNA synthesis. Since TMD1 of NS4A is required for both NS4A oligomerization and NS4A-NS4B interaction (24), the switch of TMD1 binding from NS4A to NS4B may modulate the transition from vesicle packet formation to viral replication complex formation. More experiments are needed to further define the above working hypothesis.
ACKNOWLEDGMENTS
We thank Julien Lescar and our Novartis colleagues for helpful discussions during the course of this study.
This project was partially supported by a grant from the TCR flagship “STOP Dengue” program of the National Medical Research Council in Singapore to the NITD. C.K. appreciates partial support from an A*STAR IAF grant (111105) and a JCO grant (1331A028).
REFERENCES
- 1.Simmons CP, Farrar JJ, Nguyen VV, Wills B. 2012. Dengue. N Engl J Med 366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 2.Endy TP, Anderson KB, Nisalak A, Yoon IK, Green S, Rothman AL, Thomas SJ, Jarman RG, Libraty DH, Gibbons RV. 2011. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl Trop Dis 5:e975. doi: 10.1371/journal.pntd.0000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2009. Dengue: guidelines for diagnosis, treatment, prevention and control. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM, Thiel HJ. 2007. Flaviviridae: the viruses and their replication, p 1101–1152. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Morrison J, Aguirre S, Fernandez-Sesma A. 2012. Innate immunity evasion by dengue virus. Viruses 4:397–413. doi: 10.3390/v4030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73:4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avirutnan P, Hauhart RE, Somnuke P, Blom AM, Diamond MS, Atkinson JP. 2011. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol 187:424–433. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warrener P, Tamura JK, Collett MS. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol 67:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wengler G, Czaya G, Farber PM, Hegemann JH. 1991. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol 72:851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- 13.Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 17.Xie XP, Zou J, Puttikhunt C, Yuan ZM, Shi PY. 2015. Two distinct sets of NS2A molecules are responsible for dengue virus RNA synthesis and virion assembly. J Virol 89:1298–1313. doi: 10.1128/JVI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol 76:4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J Biol Chem 281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 20.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 22.McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. 2011. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem 286:22147–22159. doi: 10.1074/jbc.M110.192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teo CS, Chu JJ. 2014. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J Virol 88:1897–1913. doi: 10.1128/JVI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, Yuan Z, Shi PY. 2015. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol 89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. 2013. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol 87:4080–4085. doi: 10.1128/JVI.01900-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou G, Chen YL, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi PY. 2011. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem 286:14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinogradova O, Badola P, Czerski L, Sonnichsen FD, Sanders CR II. 1997. Escherichia coli diacylglycerol kinase: a case study in the application of solution NMR methods to an integral membrane protein. Biophys J 72:2688–2701. doi: 10.1016/S0006-3495(97)78912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Wong YL, Huang Q, Kang C. 2014. Structural insight into the transmembrane domain and the juxtamembrane region of the erythropoietin receptor in micelles. Biophys J 107:2325–2336. doi: 10.1016/j.bpj.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM. 1990. Analysis of the backbone dynamics of interleukin-1 beta using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry 29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- 31.Kay LE, Torchia DA, Bax A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 32.Li Q, Wong YL, Ng HQ, Gayen S, Kang C. 2014. Structural insight into the transmembrane segments 3 and 4 of the hERG potassium channel. J Pept Sci 20:935–944. doi: 10.1002/psc.2704. [DOI] [PubMed] [Google Scholar]
- 33.Johnson BA. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, Shi PY. 2011. Inhibition of dengue virus by targeting viral NS4B protein. J Virol 85:11183–11195. doi: 10.1128/JVI.05468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng CY, Gu F, Phong WY, Chen YL, Lim SP, Davidson A, Vasudevan SG. 2007. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res 76:222–231. doi: 10.1016/j.antiviral.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Zou J, Xie X, Lee LT, Chandrasekaran R, Reynaud A, Yap L, Wang QY, Dong H, Kang C, Yuan Z, Lescar J, Shi PY. 2014. Dimerization of flavivirus NS4B protein. J Virol 88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of West Nile virus superinfection through RNA replication. J Virol 83:11765–11776. doi: 10.1128/JVI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 39.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu GY, Lee KJ, Gao L, Lai MM. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol 80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. 2009. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A 106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin MH, Hsu HJ, Bartenschlager R, Fischer WB. 2014. Membrane undulation induced by NS4A of dengue virus: a molecular dynamics simulation study. J Biomol Struct Dyn 32:1552–1562. doi: 10.1080/07391102.2013.826599. [DOI] [PubMed] [Google Scholar]
- 43.Ambrose RL, Mackenzie JM. 2011. A conserved peptide in West Nile virus NS4A protein contributes to proteolytic processing and is essential for replication. J Virol 85:11274–11282. doi: 10.1128/JVI.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]